Published online Aug 26, 2022. doi: 10.4252/wjsc.v14.i8.587
Peer-review started: April 7, 2022
First decision: May 11, 2022
Revised: May 26, 2022
Accepted: July 31, 2022
Article in press: July 31, 2022
Published online: August 26, 2022
Processing time: 141 Days and 7.6 Hours
Medulloblastomas (MBs) are the most prevalent brain tumours in children. They are classified as grade IV, the highest in malignancy, with about 30% metastatic tumours at the time of diagnosis. Cancer stem cells (CSCs) are a small subset of tumour cells that can initiate and support tumour growth. In MB, CSCs contribute to tumour initiation, metastasis, and therapy resistance. Metabolic differences among the different MB groups have started to emerge. Sonic hedgehog tumours show enriched lipid and nucleic acid metabolism pathways, whereas Group 3 MBs upregulate glycolysis, gluconeogenesis, glutamine anabolism, and glut
Core Tip: Considering the profound cellular metabolism rewiring associated with the tumorigenesis process, metabolic targeting for cancer treatment has gained a lot of attention in the last years. Interestingly, increasing evidence indicates that cancer stem cells (CSCs) show unique metabolic features within tumours. A deeper understanding of the metabolic vulnerabilities of medulloblastoma (MB) and MB-CSCs would accelerate the design of novel therapeutic strategies with the aim of improving the survival of paediatric patients suffering this disease.
- Citation: Martín-Rubio P, Espiau-Romera P, Royo-García A, Caja L, Sancho P. Metabolic determinants of stemness in medulloblastoma. World J Stem Cells 2022; 14(8): 587-598
- URL: https://www.wjgnet.com/1948-0210/full/v14/i8/587.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v14.i8.587
Medulloblastoma (MB) is the most common brain malignancy in young children. MB originates in the cerebellum and spreads via the cerebrospinal fluid to the brain and spine[1-3]. MB patients are classified in four different groups defined by the World Health Organization, based on the developmental pathway involved in their oncogenesis[2]: Wingless/integrated (WNT), sonic hedgehog (SHH), Group 3 and Group 4 (Table 1).
| MB group | Pathway | Prognosis | Cell of origin | CSC markers | Metabolism |
| WNT[2] | ↑ WNT signalling (Activated β-catenin)[4,5] | Good[4,5] | Progenitors in the dorsal brainstem[23] | CD133[20], Nestin[20], CD15[30], Sox2[30,31], ki67/nestin, Ki67/DCX, and Ki67/TUC-4[32] | ↑ Glycolysis[47] |
| SHH[2] | ↑ SHH signalling pathway[5] | Intermediate[4] | Immature cerebellar granule neuron precursors[19,20] or neural stem cells[21-23] | CD133[20], Nestin[20], CD15[30], Sox2[30,31], CD271 [28], ki67/nestin, ki67/DCX, and ki67/TUC-4[32] | ↑ Glycolysis[46]; ↑ Fatty acid synthesis[40]; ↓ Fatty acid oxidation[40] |
| Group 3[2] | ↑ MYCN gene[4,5] | Poor prognosis and high metastatic rate[4,5] | Cerebellar stem cells[25,26] | CD133[20], Nestin[20], CD15[30], Sox2[30,31], ki67/nestin, ki67/DCX, and ki67/TUC-4[32] | ↑ Glycolysis[47]; ↑ Fatty acid synthesis[51]; ↑ Gluconeogenesis[51]; ↑Glutamine anabolism[51] |
| Group 4[2] | Chromosome 17 abnormalities[4,5] | Intermediate[2] | Further investigation needed | CD133[20], Nestin[20], CD15[30], Sox2[30,31], ki67/nestin, ki67/DCX, and ki67/TUC-4[32] | ↑ Fatty acid synthesis[41]; ↑ One carbon pool by folate[41] |
The WNT group is the rarest, accounting for 10% of all MBs. These tumours occur in children over 3 years or teenagers, and generally have very good prognosis. WNT tumours show activating somatic mutations in β-catenin or germline mutations in adenomatous polyposis coli (APC), both leading to constitutive WNT signalling[4,5].
SHH group represents 30% of MB patients; most of them are either infants (3-years-old) or adults (> 15-years-old), with intermediate prognosis[4]. The molecular mechanism regulating this subtype implicates loss-of-function mutations in negative regulators or over-expression of different members of the SHH signalling pathway such as protein patched homolog 1 (PTCH1), smoothened (SMO), GLI family zinc finger 1 (GLI), and suppressor of fused homolog (SUFU)[5].
Group 3 accounts for tumours with a poor prognosis and high metastatic rate, occurring predominantly in males (2:1) and young adults (> 16-years-old). These tumours display frequent amplification or overexpression of the MYCN gene, and other genetic events such as orthodenticle homeobox 2
Group 4 includes predominantly males (3:1), of all ages. These tumours show intermediate prognosis due to their metastatic potential: 30%-40% of Group 4 MB cases are already metastatic at diagnosis[2]. No underlying common cause has been described for Group 4 tumours, but they show chromosome 17 abnormalities and neuronal differentiation transcriptomic profile[4,5].
Cancers are hierarchically organised structures with different levels of intratumoural heterogeneity. Indeed, distinct clone and cancer cell populations co-exist in a tumour. Among them, there is a subset of cancer cells, the so-called cancer stem cells (CSCs), which are at the origin of intratumoural heterogeneity[6]. On the one hand, CSCs display the ability to self-renew, allowing them to preserve their identity as stem cells[6,7]. On the other hand, these cells maintain intraclonal heterogeneity and give rise to all differentiated progenies within each cancer subclone in the primary tumour[8]. These differentiated progenies have limited or no tumour-initiating and metastatic capacities, despite their high proliferation rate[9]. Additionally, CSCs have the enhanced capacity to drive tumorigenesis and progression, resistance to conventional radiotherapy and chemotherapy, and invasiveness[6,10]. In fact, they are primarily responsible for tumour metastasis in secondary organs[6]. Indeed, CSCs and tumour-differentiated cells can acquire mobility thanks to processes such as epithelial-to-mesenchymal transition. Both subpopulations can migrate to other tissues through the bloodstream, but only those with stem-like features will be able eventually to initiate secondary lesions causing metastasis[9].
Evidence of CSCs was first identified in acute myelogenous leukaemia[11] and was verified sub
The origin of CSCs in the tumorigenesis process remains controversial, and it seems to be cancer type-specific. In general terms, two hypotheses have been formulated: The “stem cell hypothesis” and the “de-differentiation hypothesis”[7,15]. The “stem cell hypothesis” accounts for the similarities observed between normal tissue SCs and CSCs, postulating that malignant transformation driven by frequent genetic mutations in SCs[7,15] favours the aggressive behaviour of CSCs. The latter consists of a de-differentiation of tumour cells caused by the acquisition of stem-like abilities and tumour-initiating potential[7,16].
Although the “de-differentiation hypothesis” cannot be completely ruled out, it is clear that the stem or progenitor populations represent the cell-of-origin of MB tumours and their comprised CSCs (Table 1). Indeed, the main genes and pathways altered in MB [WNT, SHH, Notch, MYC, signal transducer and activator of transcription 3 (STAT3)] are key regulators of cell cycle and stemness[17]. Early studies demonstrated that MB tumours originate from immature cerebellar granule neuron precursors[18,19] or neural SCs[8,20,21] escaping self-renewal restriction upon activation of the SHH pathway. While this still holds true for the SHH subgroup, further studies have determined that the different subgroups arise from distinct developmental origins. Indeed, the WNT group originates from progenitors in the dorsal brainstem[22] and Group 3 tumours derive from cerebellar SCs[23,24]. However, the origin of Group 4 MB is still under debate.
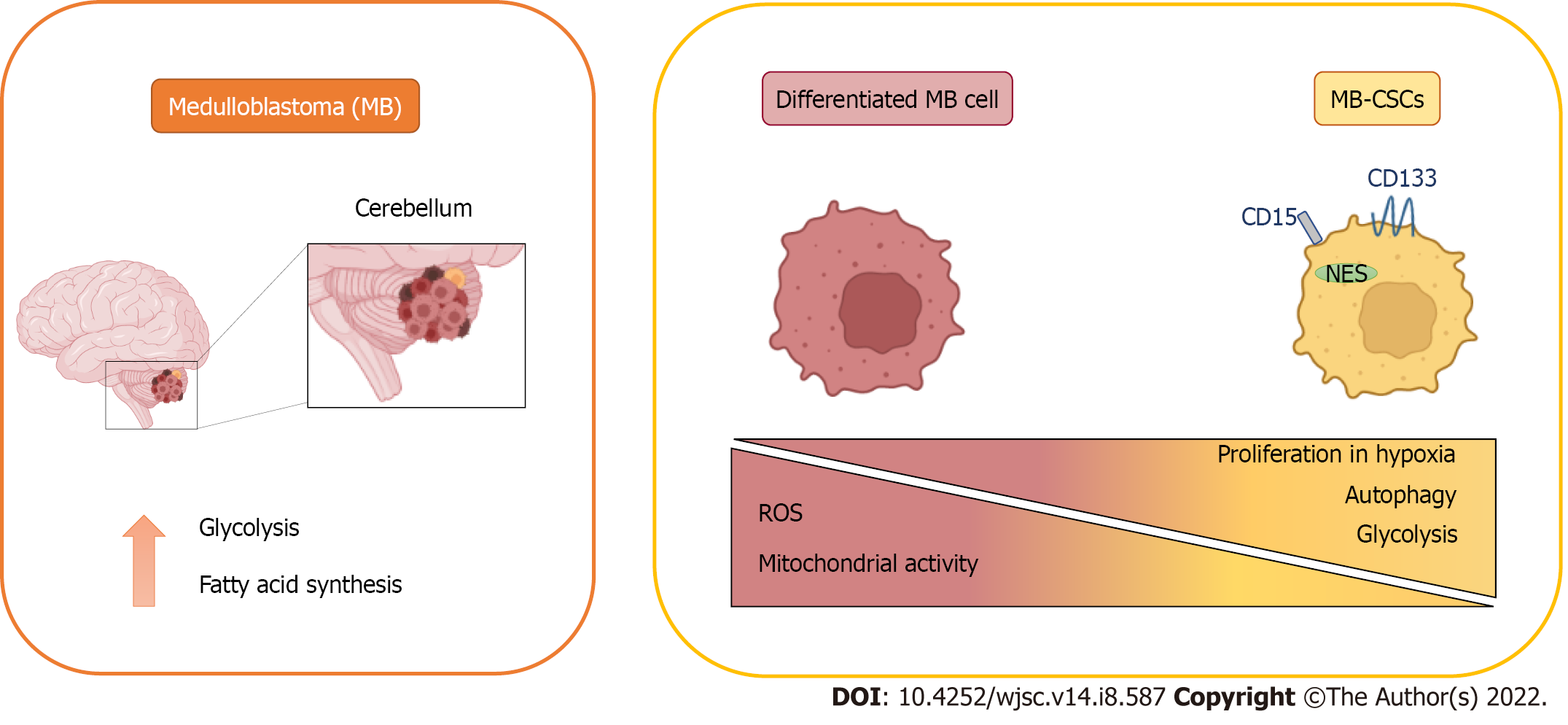
The original identification of brain CSCs in MB and other tumours involved the expression of the neural stem cell markers cluster of differentiation 133 (CD133) and Nestin[20] (Figure 1), enriching for a subpopulation of cells with enhanced neurosphere formation ability. These results were further developed in a subsequent study demonstrating that injecting as little as 100 CD133+ MB cells produced a tumour resembling the patient’s original tumour[8], definitively linking CD133 expression with CSCs in MB. In further studies, CD133+ MB cells have been characterised by their radiation[25] and chemo
Besides the well-established role of CD133, the surface marker cluster of differentiation 15 (CD15, SSEA-1) was found as a marker of MB-CSCs in two independent studies using the Patched mutant mouse model of SHH MB (Figure 1)[30]. In both studies, CD15+ cells showed high tumorigenicity in transplantation experiments, giving rise to tumours histologically similar to the original ones. However, these studies contrasted in the definition of CD15+ cells as progenitor or stem cell-like tumour cells, considering their CD133 expression and their ability to form neurospheres composed by different cellular subpopulations. Interestingly, SRY-box transcription factor 2-positive (Sox2+) cells form a quiescent subpopulation within CD15+ cells with enhanced tumour-initiating abilities[31] and both Sox2+ and CD15+ signatures were associated with poor prognosis and lower survival rates in patients[30,31]. Noteworthy, CD15+ signature was not restricted to any specific MB subgroup, suggesting that this marker predicts poor survival in MB patients, in general[30].
Interestingly, the specific expression pattern of CSC markers can distinguish the functional heterogeneity of MB-CSCs in less aggressive subtypes such as SHH[28]. Indeed, isolation of diverse clones from the well-studied SHH cell line DAOY revealed that highly self-renewing cells expressed CD271 [p75 neurotrophin receptor (p75NTR)], while those with enhanced migratory capacity expressed CD133. However, CD271 expression is much lower in aggressive tumours from Groups 3 and 4, and does not allow for functional classification of their MB-CSCs.
Highly proliferative ki67+ CSCs have been also linked to worse prognosis, recurrence, and metastasis in MB patients[32]. Although the percentage of cells expressing CD133 was increased only slightly in relapsed tumours, authors found that the most significantly altered parameter in recurring tumours was the proliferative index of CSCs, calculated as the percentage of ki67+ CSCs, especially in distant metastases.
Additionally, cells with CSC-like properties have been identified in MB patient samples using the side population (SP) technique, which entails the evaluation of the Hoechst 33342 dye exclusion by flow cytometry. Isolated MB-SP cells showed an increased neurosphere formation ability, as well as an overexpression of the stem cell markers Nestin, Notch1, and ABCG2[33].
MB tumour cells must be provided not only with the essential metabolites to cover their energy requirements, but also with the macromolecules demanded for tumour growth. Consistent with this, increased lipogenesis and aerobic glycolysis have been detected in MB[34]. However, MB metabolic profile is not uniform among the different subtypes (Figure 1)[35].
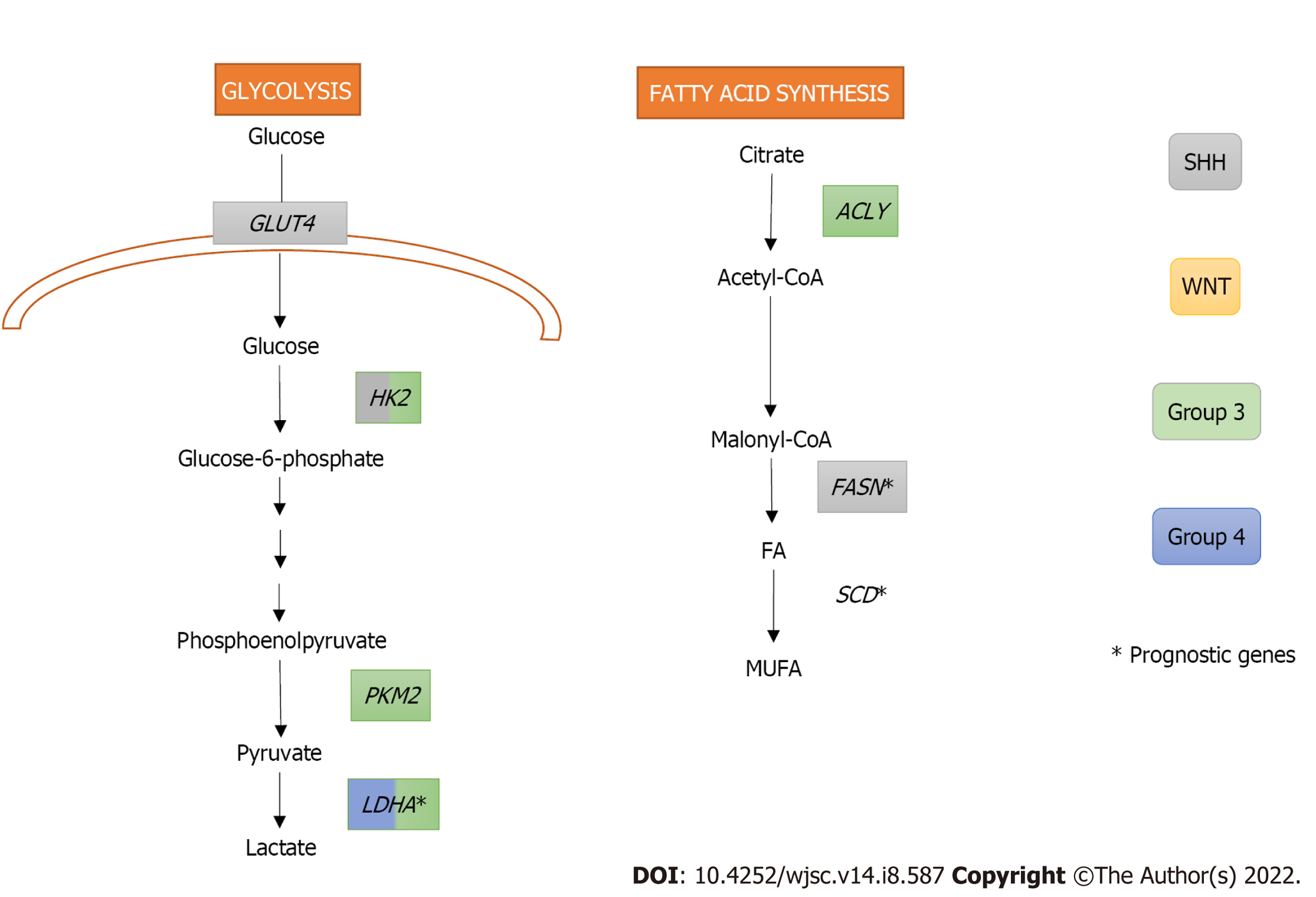
SHH oncogenic signalling inhibits fatty acid oxidation while increasing fatty acid synthesis, an early critical step of lipogenesis, to promote tumour grow. Indeed, extensive lipid accumulation and elevated levels of the lipogenic enzyme fatty acid synthase (FASN) were detected in SHH-driven MBs from transgenic mice. In fact, the Rb/E2 factor (E2F) tumour suppressor complex acted downstream of SHH, controlling FASN expression, and promoting lipogenesis and tumour growth[36]. Furthermore, FASN and stearoyl-CoA desaturase (SCD), both with crucial roles in the fatty acid synthesis pathway, were identified as prognostic genes in the SHH subgroup, Group 3, and Group 4 of MB (Figure 2)[37].
In many cancers, energy metabolism shifts from oxidative phosphorylation to aerobic glycolysis for ATP production, phenomenon known as the Warburg effect[38]. Several studies indicate that this effect also occurs in MB[39-41].
High expression levels of the oncogene c-MYC, which is a downstream target of the WNT signalling pathway and is overexpressed in Group 3 MB, has been related with increased glycolysis in MB[41-43]. Indeed, MBs from Groups 3 and WNT express significantly higher levels of the glycolytic enzyme lactate dehydrogenase A (LDHA) downstream c-MYC than non-neoplastic cerebellum (Figure 2). Interestingly, LDHA levels have been linked with poor prognosis in Group 3 MBs[43]. Besides LDHA, other glycolytic genes, such as hexokinase 2 (HK2) and pyruvate kinase M2 (PKM2), are upregulated both in murine MYC-amplified and human Group 3 MBs. PKM2 upregulation is also associated with a subsequent downregulation of the PKM1 isoform, which promotes oxidative phosphorylation in MYC-amplified MB tumour cells[42].
Induction of glycolysis in SHH MBs requires activating both SHH and insulin/insulin growth factor (IGF)/phosphoinositide 3-kinase (PI3K) pathways[40]. In addition, downstream activation of Myc-Max effector complex is also necessary for HK2 upregulation. Interestingly, rather than LDHA upregulation, SHH subgroup showed higher levels of LDHB than the other groups[41] suggesting that LDHB could be performing the expected role of LDHA[40]. Peroxisome proliferator-activated receptor gamma (PPARγ), a key regulator of fatty acid and glucose metabolism, is also upregulated in SHH-induced mouse MBs. This transcription factor, controlled by E2F1, induces the expression of key glycolytic enzymes including HK2, PKM2, and glucose transporter type 4 (GLUT4)[39].
Although increased uptake of 18F-fluoro-2-deoxyglucose, a readout of aerobic glycolysis, has been detected in human MB tumours[44] and tested in clinical trials for MB detection and follow-up (i.e. NCT00381797), some evidence questions the universality of the Warburg phenotype in MB. On the one hand, an increased expression of genes associated with the mitochondrial respiratory chain and oxidative phosphorylation was detected in human SHH MBs, but not in mouse SHH-MB models. This suggests that murine SHH-MB models do not recapitulate the metabolic features of human MBs[45] faithfully. On the other hand, a recent publication also questions the Warburg effect in MYC-amplified MBs: higher glucose incorporation into the tricarboxylic acid (TCA) cycle was detected in orthotopic tumours compared with normal brain, indicating that tumour cells use glycolysis and oxidative phosphorylation simultaneously. In addition, significant differences in glucose and glutamine metabolism were detected in high MYC-amplified MBs comparing in vitro, flank xenografts, and orthotopic xenograft MB models, evidencing the importance of using orthotopic models for metabolic analysis in these tumours[46].
Group 3 MB tumours not only display an enrichment of the glycolytic pathway but also the upregulation of proteins involved in other metabolic pathways such as gluconeogenesis (PCK2, PC), glutamine anabolism (GLUL), calcium signalling (RYR3, CAMK2D), fatty acid synthesis (ACLY), glutathione-mediated antioxidant pathway (PTGDS, GSTZ1), and the drug metabolism pathway (IMPDH1, GUSB)[47]. In addition, the upregulation of the TCA cycle, synthesis of nucleotides, hexosamines, amino acids, and glutathione was also detected in MYC-amplified orthotopic xenograft MBs[46]. This highly metabolic profile could be responsible for the aggressiveness and chemoresistance of Group 3 and MYC-amplified tumours.
Importantly, several metabolic pathways with prognostic value have been identified in MB including alanine, aspartate, and glutamate metabolism in SHH subgroup, pentose phosphate pathway and TCA cycle in Group 3, and one carbon pool by folate in Group 4[37].
Thanks to recent advances, we now know that cellular metabolism and stemness are highly interconnected in normal development and cancer. Indeed, CSCs from different cancer types show distinct metabolic features when compared with their more differentiated progenies, though their dominant metabolic phenotype varies across tumour entities, patients and even subclones within a tumour[48]. In fact, it has been shown that, depending on the type of cancer, CSCs may be either highly glycolytic or oxidative phosphorylation-dependent[49]. Moreover, recent evidence from both pluripotent embryonic and adult stem cell studies suggests that reactive oxygen species (ROS) in synchrony with metabolism mediate the maintenance of CSC[50]. A major role for the metabolic regulator mTOR in stemness maintenance has also been described in MB-CSCs[51,52]. Surprisingly, although MB stem cells are thought to be responsible for chemoresistance and recurrence in this cancer[53], their metabolic properties have barely been studied.
A decreased oxidative phosphorylation, mitochondrial mass and endogenous ROS production have been detected in established radioresistant MB stem-like clones (rMSLCs). These clones showed a higher pyruvate kinase activity and lactate production than parental cells. Mass spectrometry also detected an increased concentration of glycolysis intermediates (3-phosphoglyceric acid, 2-phosphoglyceric acid, phosphoenolpyruvic acid) and amino acids (arginine, glycine, histidine, leucine, lysine, serine) in rMSLCs, as well as decreased levels of NADH and ATP, and dysregulation of TCA cycle intermediates. Additionally, they showed an increased rate of conversion of pyruvic acid into lactic acid, and a lower rate of conversion of pyruvic acid into acetyl-CoA, which indicated an increased glycolysis[54].
MB cells expressing high levels of BMI1 (a key regulator of neural stem cells) and low chromodomain-helicase-DNA-binding protein 7 (CHD7) (an ATP-dependent chromatin remodeller) showed a decreased mitochondrial function concomitant with higher glycolytic activity. Moreover, enhanced expression of glycolytic genes [HK2, PFKP, enolase 4 (ENO4), pyruvate dehydrogenase kinase 1(PDK1) and LDHB] and increased levels of valine and leucine were detected in BMI1high CHD7low Group 3 MB tissues. Since both markers have been previously related to hypoxia, it was proposed that cells with this signature might present advantages in hypoxic conditions thanks to these metabolic adaptations (Figure 1)[55].
Indeed, oxygen restriction in non-vascularised regions may support cell cycle entry and self-renewal via Notch1 signalling, since hypoxia is one of the main expansion inducers of the CD133+ population[25,56]. In contrast, MB-CSCs associated to a more quiescent state have been reported to reside in a perivascular niche[57], in close interaction with endothelial cells that secrete factors to maintain their stemness.
Finally, it has been demonstrated that autophagy positively regulates stemness in MB cells. Indeed, the expression of the pro-autophagy factor AMBRA1 depends on c-MYC levels, and it is strongly upregulated in Group 3 MB-CSCs[58].
In general, all these reports suggest that the metabolic phenotype harboured by MB-CSCs is mainly glycolytic, which is favoured by hypoxia. Both glycolysis and hypoxia generate the antioxidant redox status needed to maintain their stemness. Moreover, the elevated availability of macromolecules needed to support their enhanced proliferation may be sustained by biosynthesis derived from glycolysis intermediates and an increased autophagy in these cells.
Current standard treatments for MB include surgical resection of the tumour, craniospinal radiotherapy and systemic chemotherapy. Unfortunately, only 70% of patients survive. In addition, radiotherapy causes neurocognitive, neuroendocrine and psychosocial deficits, as well as the development of secondary malignancies[59,60]. Consequently, new treatment strategies for MB are needed, such as targeted therapies that are less toxic and more effective in reducing the resulting long-term side effects[61].
Targeted therapies for WNT and SHH MB entail inhibiting the major pathways implicated in their tumourigenesis[62]. As an example, the use of Smo inhibitors in the SHH MB group has been linked to tumour suppression in mouse models, and tumour regression in MB patients[62,63]. On the other hand, research on WNT MBs has focused on nuclear β-catenin inhibitors, leading to reduced cell migration and invasion[63,64].
In Groups 3 and 4 there are no major advances in targeted therapy as they have a low somatic mutation rate[61,63]. Group 3 MB is characterised by MYC oncogene amplification, so some epigenetic MYC inhibitors, such as histone deacetylases (HDACs) and bromodomain inhibitors, have been tested[65,66]. In addition, PI3K inhibitors can act in synergy with HDAC modulators, preventing tumour proliferation and prolonging survival in Group 3 MB mice[67].
Apart from subgroup-specific therapies, other molecular approaches linked to the general population of MB patients are being explored nowadays. Selective inhibition of cell division protein kinase (CDK) 4/6 arrests the cell cycle potently and is therefore a favourable target for cancer treatment[68]. Another strategy is related to immunotherapy, as it may be effective against MB innate immunosuppressive properties and thus it may extend patient survival[69].
Thoroughly investigated in the context of other cancer types, metabolic reprogramming might be also the source of putative targets for the design of new treatments against MB (Table 2). For example, specifically in lipid metabolism, direct inhibition of FASN by C75 reduced tumour proliferation in SHH tumours and increased survival in vivo, associated to reduced lipogenesis. In addition, C75 may act in synergy with the CDK inhibitor roscovitine[36].
| MB group | MB | MB-CSCs | ||
| General drug (target) | Group specific drug (target) | General drug (target) | Group specific drug (target) | |
| WNT[2] | C75 (FASN)[36], Oxamate (LDH)[41], DFMO (ODC)1, Isotretinoin (RAR, RXR)1 | DCA (PDK)[54]; Curcumin (pleiotropic)[73]; IP6 (mTOR)[55] | ||
| SHH[2] | GW9662 (PPARγ)[39] | |||
| Group 3[2] | GSK2837808a, FX11 (LDHA)[43]; TAK228, Sirolimus, Temsirolimus, Everolimus (mTOR)[71]; JHU395 (Glutamine)[72] | WP1066 (STAT3) & Chloroquine (autophagy)[58] | ||
| Group 4[2] | ||||
Since glycolysis seems crucial for the metabolic requirements of MB, it may be important to target glycolytic enzymes. In that regard, LDH tetramers inhibition by oxamate attenuated glycolysis, proliferation, and motility significantly in MB cell lines[41]. In addition, LDHA antagonists GSK2837808a and FX11 reduced the growth of Group 3 MB cells[43]. Conditional suppression of HK2 altered both energy homeostasis and the balance between proliferation and differentiation markedly, thereby reducing tumour growth. Similarly, blockade of IL6 by bazedoxifene showed glycolysis downregulation, and impaired viability and proliferation of MB cells[70]. Finally, it is important to mention that pharmacological inhibition of PPARγ by GW9662 increases SHH-driven MB cell death and mouse survival in vivo through glycolysis inhibition[39].
Another interesting therapeutic strategy for MB is based on the inhibition of the metabolic regulator mTOR. Indeed, the TORC1/2 kinase inhibitor TAK228 improved the survival of orthotopic MYC-amplified Group 3 MB tumours through glutathione depletion[71]. In addition, the dual inhibition of PI3Kα and mTOR blocked the sphere-forming ability of SHH-driven MB cells, and repressed tumour growth in vivo[52]. In fact, several clinical trials are currently testing different conventional treatments in combination with mTOR inhibitors, such as Sirolimus (NCT02574728), Temsirolimus (NCT00784914) or Everolimus (NCT03387020) against MB and other brain tumours.
The elevated MYC expression characteristic of Group 3 MB is related to an increased glutamine transport and utilisation. For this reason, the glutamine antagonist 6-diazo-5-oxo-l-norleucine has been tested as therapy against Group 3 MB, although with little success in clinical trials due to its poor pharmacodynamic properties. Still, the prodrug isopropyl 6-diazo-5-oxo-2-[phenyl (pivoxyloxy) methoxy-carbonyl] hexanoate (JHU395) has shown an improved cell penetration, enhanced growth reduction and increased apoptosis of Group 3 MB cells in vitro, as well as longer survival of mice in vivo[72].
Different metabolic targeting approaches related to the toxic effect of ROS accumulation and redox imbalance have also been tested in clinical trials. On the one side, two clinical trials have studied the antitumoral properties of Photodynamic Therapy, which uses light and photosensitizing drugs to kill tumour cells via ROS accumulation, in MB (NCT00002647, NCT01682746). Unfortunately, no results have been posted to date. On the other hand, the antiprotozoal compound Nifurtimox, with pro-oxidant properties, has also been approved by the Food and Drug Administration for its use in MB. In the Phase II trial NCT00601003, the combination of Nifurtimox with cyclophosphamide and topotecan is being tested as treatment for relapsed or refractory neuroblastoma and MB.
There are other alternative therapeutic approaches targeting MB metabolism currently being tested in clinical trials. On the one hand, the polyamine synthesis inhibitor difluoromethylornithine (DFMO) is being tested in a phase II clinical trial as maintenance therapy for high molecular risk or very high risk and relapsed or refractory MB (NCT04696029). On the other hand, the combination of Vorinostat and Isotretinoin (13-cis-retinoic acid, vitamin A derivative) has been tested in a phase I clinical trial targeting young patients with recurrent or refractory lymphoma, leukemia, or solid tumours such as MB (NCT00217412), although no results have been posted to this day.
Though much less studied in the context of MB, metabolic pathways also play a key role in cancer resistance linked to CSCs. Treatment with the PDK inhibitor DCA suppressed CSC-like phenotypes, and increased intracellular ROS levels and radiosensitivity by inhibiting glycolysis and inducing mito
Besides metabolism, we can also find other targets, such as the new SHH inhibitor Chemotype 12 that overcomes the associated SMO receptor drug resistance. It reduced the ability to form spheres in number and size, associated with a reduced expression of stemness markers [NANOG, octamer-binding transcription factor 4 (OCT4)], and thus impaired stem-like cell growth in vitro and in vivo[75]. Moreover, CDK inhibitors, such as Ribociclib, can also target CSCs[68]. Oncolytic engineered herpes simplex viruses (oHSVs) may also have the potential to eliminate MB-CSCs specifically, as they express the primary HSV-1 entry molecule nectin-1 (CD111) and are shown to be highly sensitive to clinically relevant oHSVs in vitro and in vivo[76].
Further research is urgently needed to elucidate the metabolic profile of MB tumours. Recent evidence suggests that metabolic features among MB groups and even among the different cell subpopulations within a tumour are highly heterogeneous. In addition, new data obtained in more clinically relevant models challenge most of the existing knowledge in the field, obtained from genetically modified mouse models. Moreover, the metabolic features of MB-CSCs remain absolutely unknown nowadays. The identification of metabolic vulnerabilities of these tumours and their highly aggressive CSCs could pave the way to the design of new therapeutic strategies improving the current treatment for this paediatric disease.
We want to thank Laura Sancho for proofreading the manuscript.
| 1. | Hatten ME, Roussel MF. Development and cancer of the cerebellum. Trends Neurosci. 2011;34:134-142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 138] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 2. | Northcott PA, Robinson GW, Kratz CP, Mabbott DJ, Pomeroy SL, Clifford SC, Rutkowski S, Ellison DW, Malkin D, Taylor MD, Gajjar A, Pfister SM. Medulloblastoma. Nat Rev Dis Primers. 2019;5:11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 271] [Cited by in RCA: 458] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 3. | Wang J, Garancher A, Ramaswamy V, Wechsler-Reya RJ. Medulloblastoma: From Molecular Subgroups to Molecular Targeted Therapies. Annu Rev Neurosci. 2018;41:207-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 4. | Thomas A, Noël G. Medulloblastoma: optimizing care with a multidisciplinary approach. J Multidiscip Healthc. 2019;12:335-347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 5. | Hovestadt V, Smith KS, Bihannic L, Filbin MG, Shaw ML, Baumgartner A, DeWitt JC, Groves A, Mayr L, Weisman HR, Richman AR, Shore ME, Goumnerova L, Rosencrance C, Carter RA, Phoenix TN, Hadley JL, Tong Y, Houston J, Ashmun RA, DeCuypere M, Sharma T, Flasch D, Silkov A, Ligon KL, Pomeroy SL, Rivera MN, Rozenblatt-Rosen O, Rusert JM, Wechsler-Reya RJ, Li XN, Peyrl A, Gojo J, Kirchhofer D, Lötsch D, Czech T, Dorfer C, Haberler C, Geyeregger R, Halfmann A, Gawad C, Easton J, Pfister SM, Regev A, Gajjar A, Orr BA, Slavc I, Robinson GW, Bernstein BE, Suvà ML, Northcott PA. Resolving medulloblastoma cellular architecture by single-cell genomics. Nature. 2019;572:74-79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 187] [Cited by in RCA: 297] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 6. | Dalerba P, Clarke MF. Cancer stem cells and tumor metastasis: first steps into uncharted territory. Cell Stem Cell. 2007;1:241-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 129] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 7. | Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells - what challenges do they pose? Nat Rev Drug Discov. 2014;13:497-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 660] [Cited by in RCA: 810] [Article Influence: 67.5] [Reference Citation Analysis (0)] |
| 8. | Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5422] [Cited by in RCA: 5627] [Article Influence: 255.8] [Reference Citation Analysis (0)] |
| 9. | Malanchi I, Santamaria-Martínez A, Susanto E, Peng H, Lehr HA, Delaloye JF, Huelsken J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481:85-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 943] [Cited by in RCA: 1086] [Article Influence: 72.4] [Reference Citation Analysis (0)] |
| 10. | Sancho P, Alcala S, Usachov V, Hermann PC, Sainz B Jr. The ever-changing landscape of pancreatic cancer stem cells. Pancreatology. 2016;16:489-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 11. | Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3316] [Cited by in RCA: 3436] [Article Influence: 107.4] [Reference Citation Analysis (0)] |
| 12. | Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983-3988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7830] [Cited by in RCA: 7800] [Article Influence: 339.1] [Reference Citation Analysis (0)] |
| 13. | Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030-1037. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2377] [Cited by in RCA: 2455] [Article Influence: 129.2] [Reference Citation Analysis (0)] |
| 14. | Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K, Coghlan L, Tang DG. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696-1708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 727] [Cited by in RCA: 731] [Article Influence: 36.6] [Reference Citation Analysis (10)] |
| 15. | Trosko JE. On the potential origin and characteristics of cancer stem cells. Carcinogenesis. 2021;42:905-912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 16. | Bjerkvig R, Tysnes BB, Aboody KS, Najbauer J, Terzis AJ. Opinion: the origin of the cancer stem cell: current controversies and new insights. Nat Rev Cancer. 2005;5:899-904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 426] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 17. | Cordeiro BM, Oliveira ID, Alves MT, Saba-Silva N, Capellano AM, Cavalheiro S, Dastoli P, Toledo SR. SHH, WNT, and NOTCH pathways in medulloblastoma: when cancer stem cells maintain self-renewal and differentiation properties. Childs Nerv Syst. 2014;30:1165-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | Miyata H, Ikawa E, Ohama E. Medulloblastoma in an adult suggestive of external granule cells as its origin: a histological and immunohistochemical study. Brain Tumor Pathol. 1998;15:31-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 19. | Schüller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, Huillard E, Sun T, Ligon AH, Qian Y, Ma Q, Alvarez-Buylla A, McMahon AP, Rowitch DH, Ligon KL. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123-134. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 570] [Cited by in RCA: 515] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 20. | Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821-5828. [PubMed] |
| 21. | Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, Schüller U, Machold R, Fishell G, Rowitch DH, Wainwright BJ, Wechsler-Reya RJ. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135-145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 580] [Cited by in RCA: 529] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 22. | Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, Kranenburg TA, Hogg T, Poppleton H, Martin J, Finkelstein D, Pounds S, Weiss A, Patay Z, Scoggins M, Ogg R, Pei Y, Yang ZJ, Brun S, Lee Y, Zindy F, Lindsey JC, Taketo MM, Boop FA, Sanford RA, Gajjar A, Clifford SC, Roussel MF, McKinnon PJ, Gutmann DH, Ellison DW, Wechsler-Reya R, Gilbertson RJ. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095-1099. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 681] [Cited by in RCA: 628] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 23. | Kawauchi D, Robinson G, Uziel T, Gibson P, Rehg J, Gao C, Finkelstein D, Qu C, Pounds S, Ellison DW, Gilbertson RJ, Roussel MF. A mouse model of the most aggressive subgroup of human medulloblastoma. Cancer Cell. 2012;21:168-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 237] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 24. | Pei Y, Moore CE, Wang J, Tewari AK, Eroshkin A, Cho YJ, Witt H, Korshunov A, Read TA, Sun JL, Schmitt EM, Miller CR, Buckley AF, McLendon RE, Westbrook TF, Northcott PA, Taylor MD, Pfister SM, Febbo PG, Wechsler-Reya RJ. An animal model of MYC-driven medulloblastoma. Cancer Cell. 2012;21:155-167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 251] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 25. | Blazek ER, Foutch JL, Maki G. Daoy medulloblastoma cells that express CD133 are radioresistant relative to CD133- cells, and the CD133+ sector is enlarged by hypoxia. Int J Radiat Oncol Biol Phys. 2007;67:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 198] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 26. | Garg N, Bakhshinyan D, Venugopal C, Mahendram S, Rosa DA, Vijayakumar T, Manoranjan B, Hallett R, McFarlane N, Delaney KH, Kwiecien JM, Arpin CC, Lai PS, Gómez-Biagi RF, Ali AM, de Araujo ED, Ajani OA, Hassell JA, Gunning PT, Singh SK. CD133+ brain tumor-initiating cells are dependent on STAT3 signaling to drive medulloblastoma recurrence. Oncogene. 2017;36:606-617. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 27. | Annabi B, Rojas-Sutterlin S, Laflamme C, Lachambre MP, Rolland Y, Sartelet H, Béliveau R. Tumor environment dictates medulloblastoma cancer stem cell expression and invasive phenotype. Mol Cancer Res. 2008;6:907-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 54] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 28. | Morrison LC, McClelland R, Aiken C, Bridges M, Liang L, Wang X, Di Curzio D, Del Bigio MR, Taylor MD, Werbowetski-Ogilvie TE. Deconstruction of medulloblastoma cellular heterogeneity reveals differences between the most highly invasive and self-renewing phenotypes. Neoplasia. 2013;15:384-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 29. | Raso A, Mascelli S, Biassoni R, Nozza P, Kool M, Pistorio A, Ugolotti E, Milanaccio C, Pignatelli S, Ferraro M, Pavanello M, Ravegnani M, Cama A, Garrè ML, Capra V. High levels of PROM1 (CD133) transcript are a potential predictor of poor prognosis in medulloblastoma. Neuro Oncol. 2011;13:500-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 30. | Read TA, Fogarty MP, Markant SL, McLendon RE, Wei Z, Ellison DW, Febbo PG, Wechsler-Reya RJ. Identification of CD15 as a marker for tumor-propagating cells in a mouse model of medulloblastoma. Cancer Cell. 2009;15:135-147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 287] [Cited by in RCA: 273] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 31. | Vanner RJ, Remke M, Gallo M, Selvadurai HJ, Coutinho F, Lee L, Kushida M, Head R, Morrissy S, Zhu X, Aviv T, Voisin V, Clarke ID, Li Y, Mungall AJ, Moore RA, Ma Y, Jones SJ, Marra MA, Malkin D, Northcott PA, Kool M, Pfister SM, Bader G, Hochedlinger K, Korshunov A, Taylor MD, Dirks PB. Quiescent sox2(+) cells drive hierarchical growth and relapse in sonic hedgehog subgroup medulloblastoma. Cancer Cell. 2014;26:33-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 244] [Article Influence: 20.3] [Reference Citation Analysis (1)] |
| 32. | Tang X, Yao Y, Zhu J, Jin K, Wang Y, Mao Y, Zhou L. Differential proliferative index of cancer stem-like cells in primary and recurrent medulloblastoma in human. Childs Nerv Syst. 2012;28:1869-1877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 33. | Liu J, Chi N, Zhang JY, Zhu W, Bian YS, Chen HG. Isolation and characterization of cancer stem cells from medulloblastoma. Genet Mol Res. 2015;14:3355-3361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 34. | Tech K, Gershon TR. Energy metabolism in neurodevelopment and medulloblastoma. Transl Pediatr. 2015;4:12-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 35. | Blüml S, Margol AS, Sposto R, Kennedy RJ, Robison NJ, Vali M, Hung LT, Muthugounder S, Finlay JL, Erdreich-Epstein A, Gilles FH, Judkins AR, Krieger MD, Dhall G, Nelson MD, Asgharzadeh S. Molecular subgroups of medulloblastoma identification using noninvasive magnetic resonance spectroscopy. Neuro Oncol. 2016;18:126-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 60] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 36. | Bhatia B, Hsieh M, Kenney AM, Nahlé Z. Mitogenic Sonic hedgehog signaling drives E2F1-dependent lipogenesis in progenitor cells and medulloblastoma. Oncogene. 2011;30:410-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 37. | Park AK, Lee JY, Cheong H, Ramaswamy V, Park SH, Kool M, Phi JH, Choi SA, Cavalli F, Taylor MD, Kim SK. Subgroup-specific prognostic signaling and metabolic pathways in pediatric medulloblastoma. BMC Cancer. 2019;19:571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 39. | Bhatia B, Potts CR, Guldal C, Choi S, Korshunov A, Pfister S, Kenney AM, Nahlé ZA. Hedgehog-mediated regulation of PPARγ controls metabolic patterns in neural precursors and shh-driven medulloblastoma. Acta Neuropathol. 2012;123:587-600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 40. | Gershon TR, Crowther AJ, Tikunov A, Garcia I, Annis R, Yuan H, Miller CR, Macdonald J, Olson J, Deshmukh M. Hexokinase-2-mediated aerobic glycolysis is integral to cerebellar neurogenesis and pathogenesis of medulloblastoma. Cancer Metab. 2013;1:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 41. | Valvona CJ, Fillmore HL. Oxamate, but Not Selective Targeting of LDH-A, Inhibits Medulloblastoma Cell Glycolysis, Growth and Motility. Brain Sci. 2018;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 42. | Staal JA, Lau LS, Zhang H, Ingram WJ, Hallahan AR, Northcott PA, Pfister SM, Wechsler-Reya RJ, Rusert JM, Taylor MD, Cho YJ, Packer RJ, Brown KJ, Rood BR. Proteomic profiling of high risk medulloblastoma reveals functional biology. Oncotarget. 2015;6:14584-14595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 43. | Tao R, Murad N, Xu Z, Zhang P, Okonechnikov K, Kool M, Rivero-Hinojosa S, Lazarski C, Zheng P, Liu Y, Eberhart CG, Rood BR, Packer R, Pei Y. MYC Drives Group 3 Medulloblastoma through Transformation of Sox2+ Astrocyte Progenitor Cells. Cancer Res. 2019;79:1967-1980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 44] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 44. | Gururangan S, Hwang E, Herndon JE 2nd, Fuchs H, George T, Coleman RE. [18F]fluorodeoxyglucose-positron emission tomography in patients with medulloblastoma. Neurosurgery. 2004;55:1280-8; discussion 1288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 45. | Łastowska M, Karkucińska-Więckowska A, Waschek JA, Niewiadomski P. Differential Expression of Mitochondrial Biogenesis Markers in Mouse and Human SHH-Subtype Medulloblastoma. Cells. 2019;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 46. | Pham K, Hanaford AR, Poore BA, Maxwell MJ, Sweeney H, Parthasarathy A, Alt J, Rais R, Slusher BS, Eberhart CG, Raabe EH. Comprehensive Metabolic Profiling of MYC-Amplified Medulloblastoma Tumors Reveals Key Dependencies on Amino Acid, Tricarboxylic Acid and Hexosamine Pathways. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 47. | Kp M, Kumar A, Biswas D, Moiyadi A, Shetty P, Gupta T, Epari S, Shirsat N, Srivastava S. The proteomic analysis shows enrichment of RNA surveillance pathways in adult SHH and extensive metabolic reprogramming in Group 3 medulloblastomas. Brain Tumor Pathol. 2021;38:96-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 48. | Jagust P, de Luxán-Delgado B, Parejo-Alonso B, Sancho P. Metabolism-Based Therapeutic Strategies Targeting Cancer Stem Cells. Front Pharmacol. 2019;10:203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 105] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 49. | Sancho P, Barneda D, Heeschen C. Hallmarks of cancer stem cell metabolism. Br J Cancer. 2016;114:1305-1312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 282] [Cited by in RCA: 443] [Article Influence: 44.3] [Reference Citation Analysis (0)] |
| 50. | Bigarella CL, Liang R, Ghaffari S. Stem cells and the impact of ROS signaling. Development. 2014;141:4206-4218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 478] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 51. | Aldaregia J, Odriozola A, Matheu A, Garcia I. Targeting mTOR as a Therapeutic Approach in Medulloblastoma. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 52. | Eckerdt F, Clymer J, Bell JB, Beauchamp EM, Blyth GT, Goldman S, Platanias LC. Pharmacological mTOR targeting enhances the antineoplastic effects of selective PI3Kα inhibition in medulloblastoma. Sci Rep. 2019;9:12822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 26] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 53. | Bahmad HF, Poppiti RJ. Medulloblastoma cancer stem cells: molecular signatures and therapeutic targets. J Clin Pathol. 2020;73:243-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 54. | Sun L, Moritake T, Ito K, Matsumoto Y, Yasui H, Nakagawa H, Hirayama A, Inanami O, Tsuboi K. Metabolic analysis of radioresistant medulloblastoma stem-like clones and potential therapeutic targets. PLoS One. 2017;12:e0176162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 55. | Badodi S, Pomella N, Zhang X, Rosser G, Whittingham J, Niklison-Chirou MV, Lim YM, Brandner S, Morrison G, Pollard SM, Bennett CD, Clifford SC, Peet A, Basson MA, Marino S. Inositol treatment inhibits medulloblastoma through suppression of epigenetic-driven metabolic adaptation. Nat Commun. 2021;12:2148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 56. | Pistollato F, Rampazzo E, Persano L, Abbadi S, Frasson C, Denaro L, D'Avella D, Panchision DM, Della Puppa A, Scienza R, Basso G. Interaction of hypoxia-inducible factor-1α and Notch signaling regulates medulloblastoma precursor proliferation and fate. Stem Cells. 2010;28:1918-1929. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 124] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 57. | Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, Frank A, Bayazitov IT, Zakharenko SS, Gajjar A, Davidoff A, Gilbertson RJ. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11:69-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1640] [Cited by in RCA: 1583] [Article Influence: 83.3] [Reference Citation Analysis (0)] |
| 58. | Nazio F, Po A, Abballe L, Ballabio C, Diomedi Camassei F, Bordi M, Camera A, Caruso S, Caruana I, Pezzullo M, Ferraina C, Milletti G, Gianesello M, Reddel S, De Luca CD, Ceglie D, Marinelli S, Campello S, Papaleo E, Miele E, Cacchione A, Carai A, Vinci M, Velardi E, De Angelis B, Tiberi L, Quintarelli C, Mastronuzzi A, Ferretti E, Locatelli F, Cecconi F. Targeting cancer stem cells in medulloblastoma by inhibiting AMBRA1 dual function in autophagy and STAT3 signalling. Acta Neuropathol. 2021;142:537-564. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 59. | Paul MR, Zage PE. Overview and recent advances in the targeting of medulloblastoma cancer stem cells. Expert Rev Anticancer Ther. 2021;21:957-974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 60. | Wlodarski PK, Jozwiak J. Therapeutic targets for medulloblastoma. Expert Opin Ther Targets. 2008;12:449-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 61. | Maier H, Dalianis T, Kostopoulou ON. New Approaches in Targeted Therapy for Medulloblastoma in Children. Anticancer Res. 2021;41:1715-1726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 62. | Petrirena GJ, Masliah-Planchon J, Sala Q, Pourroy B, Frappaz D, Tabouret E, Graillon T, Gentet JC, Delattre O, Chinot O, Padovani L. Recurrent extraneural sonic hedgehog medulloblastoma exhibiting sustained response to vismodegib and temozolomide monotherapies and inter-metastatic molecular heterogeneity at progression. Oncotarget. 2018;9:10175-10183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 24] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 63. | Gatto L, Franceschi E, Tosoni A, Di Nunno V, Bartolini S, Brandes AA. Molecular Targeted Therapies: Time for a Paradigm Shift in Medulloblastoma Treatment? Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 64. | Bassani B, Bartolini D, Pagani A, Principi E, Zollo M, Noonan DM, Albini A, Bruno A. Fenretinide (4-HPR) Targets Caspase-9, ERK 1/2 and the Wnt3a/β-Catenin Pathway in Medulloblastoma Cells and Medulloblastoma Cell Spheroids. PLoS One. 2016;11:e0154111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 65. | Wang J, Sui Y, Li Q, Zhao Y, Dong X, Yang J, Liang Z, Han Y, Tang Y, Ma J. Effective inhibition of MYC-amplified group 3 medulloblastoma by FACT-targeted curaxin drug CBL0137. Cell Death Dis. 2020;11:1029. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 66. | Bandopadhayay P, Bergthold G, Nguyen B, Schubert S, Gholamin S, Tang Y, Bolin S, Schumacher SE, Zeid R, Masoud S, Yu F, Vue N, Gibson WJ, Paolella BR, Mitra SS, Cheshier SH, Qi J, Liu KW, Wechsler-Reya R, Weiss WA, Swartling FJ, Kieran MW, Bradner JE, Beroukhim R, Cho YJ. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res. 2014;20:912-925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 290] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 67. | Pei Y, Liu KW, Wang J, Garancher A, Tao R, Esparza LA, Maier DL, Udaka YT, Murad N, Morrissy S, Seker-Cin H, Brabetz S, Qi L, Kogiso M, Schubert S, Olson JM, Cho YJ, Li XN, Crawford JR, Levy ML, Kool M, Pfister SM, Taylor MD, Wechsler-Reya RJ. HDAC and PI3K Antagonists Cooperate to Inhibit Growth of MYC-Driven Medulloblastoma. Cancer Cell. 2016;29:311-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 223] [Article Influence: 22.3] [Reference Citation Analysis (9)] |
| 68. | Cook Sangar ML, Genovesi LA, Nakamoto MW, Davis MJ, Knobluagh SE, Ji P, Millar A, Wainwright BJ, Olson JM. Inhibition of CDK4/6 by Palbociclib Significantly Extends Survival in Medulloblastoma Patient-Derived Xenograft Mouse Models. Clin Cancer Res. 2017;23:5802-5813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 69. | Kabir TF, Kunos CA, Villano JL, Chauhan A. Immunotherapy for Medulloblastoma: Current Perspectives. Immunotargets Ther. 2020;9:57-77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 37] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 70. | Chen X, Wei J, Li C, Pierson CR, Finlay JL, Lin J. Blocking interleukin-6 signaling inhibits cell viability/proliferation, glycolysis, and colony forming activity of human medulloblastoma cells. Int J Oncol. 2018;52:571-578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 15] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 71. | Maynard RE, Poore B, Hanaford AR, Pham K, James M, Alt J, Park Y, Slusher BS, Tamayo P, Mesirov J, Archer TC, Pomeroy SL, Eberhart CG, Raabe EH. TORC1/2 kinase inhibition depletes glutathione and synergizes with carboplatin to suppress the growth of MYC-driven medulloblastoma. Cancer Lett. 2021;504:137-145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 72. | Pham K, Maxwell MJ, Sweeney H, Alt J, Rais R, Eberhart CG, Slusher BS, Raabe EH. Novel Glutamine Antagonist JHU395 Suppresses MYC-Driven Medulloblastoma Growth and Induces Apoptosis. J Neuropathol Exp Neurol. 2021;80:336-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 73. | Lim KJ, Bisht S, Bar EE, Maitra A, Eberhart CG. A polymeric nanoparticle formulation of curcumin inhibits growth, clonogenicity and stem-like fraction in malignant brain tumors. Cancer Biol Ther. 2011;11:464-473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 154] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 74. | Hambardzumyan D, Becher OJ, Rosenblum MK, Pandolfi PP, Manova-Todorova K, Holland EC. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes Dev. 2008;22:436-448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 357] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 75. | Infante P, Alfonsi R, Ingallina C, Quaglio D, Ghirga F, D'Acquarica I, Bernardi F, Di Magno L, Canettieri G, Screpanti I, Gulino A, Botta B, Mori M, Di Marcotullio L. Inhibition of Hedgehog-dependent tumors and cancer stem cells by a newly identified naturally occurring chemotype. Cell Death Dis. 2016;7:e2376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 76. | Friedman GK, Moore BP, Nan L, Kelly VM, Etminan T, Langford CP, Xu H, Han X, Markert JM, Beierle EA, Gillespie GY. Pediatric medulloblastoma xenografts including molecular subgroup 3 and CD133+ and CD15+ cells are sensitive to killing by oncolytic herpes simplex viruses. Neuro Oncol. 2016;18:227-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell biology
Country/Territory of origin: Spain
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Sun Q, China; Zhao W, China S-Editor: Chen YL L-Editor: Filipodia P-Editor: Chen YL