Published online Mar 26, 2022. doi: 10.4252/wjsc.v14.i3.231
Peer-review started: August 23, 2021
First decision: October 3, 2021
Revised: October 11, 2021
Accepted: February 15, 2022
Article in press: February 15, 2022
Published online: March 26, 2022
Processing time: 213 Days and 19.5 Hours
The generation of induced pluripotent stem cells (iPSC) has been a game-changer in translational and regenerative medicine; however, their large-scale applicability is still hampered by the scarcity of accessible, safe, and reproducible protocols. The porcine model is a large biomedical model that enables translational applications, including gene editing, long term in vivo and offspring analysis; therefore, suitable for both medicine and animal production.
To reprogramme in vitro into pluripotency, and herein urine-derived cells (UDCs) were isolated from porcine urine.
The UDCs were reprogrammed in vitro using human or murine octamer-binding transcription factor 4 (OCT4), SRY-box2 (SOX2), Kruppel-like factor 4 (KLF4), and C-MYC, and cultured with basic fibroblast growth factor (bFGF) supplementation. To characterize the putative porcine iPSCs three clonal lineages were submitted to immunocytochemistry for alkaline phosphatase (AP), OCT4, SOX2, NANOG, TRA1 81 and SSEA 1 detection. Endogenous transcripts related to the plu
The UDCs were isolated from swine urine samples and when at passage 2 submitted to in vitro reprogramming. Colonies of putative iPSCs were obtained only from UDCs transduced with the murine factors (mOSKM), but not from human factors (hOSKM). Three clonal lineages were isolated and further cultured for at least 28 passages, all the lineages were positive for AP detection, the OCT4, SOX2, NANOG markers, albeit the immunocytochemical analysis also revealed heterogeneous phenotypic profiles among lineages and passages for NANOG and SSEA1, similar results were observed in the abundance of the endogenous transcripts related to pluripotent state. All the clonal lineages when cultured in suspension without bFGF were able to form EBs expressing ectoderm and mesoderm layers transcripts.
For the first time UDCs were isolated in the swine model and reprogrammed into a pluripotent-like state, enabling new numerous applications in both human or veterinary regenerative medicine.
Core Tip: The porcine induced pluripotent stem cells (piPSCs) derived from urine derived cells (UDCs) may facilitate their routine and large-scale use by avoiding injury or stress during collection for autologous purposes. However, the precise reprogramming process and characterization is not fully elucidated in other species than murine or human. The generation of piPSCs from UDCs can contribute as a biomedical model for regenerative and translational medicine, as well as for animal production and to elucidate the reprogramming process in porcine, a large animal model.
- Citation: Recchia K, Machado LS, Botigelli RC, Pieri NCG, Barbosa G, de Castro RVG, Marques MG, Pessôa LVF, Fantinato Neto P, Meirelles FV, Souza AF, Martins SMMK, Bressan FF. In vitro induced pluripotency from urine-derived cells in porcine. World J Stem Cells 2022; 14(3): 231-244
- URL: https://www.wjgnet.com/1948-0210/full/v14/i3/231.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v14.i3.231
The generation of pluripotent cells in vitro has been reported in numerous studies; however, pluripotent cell generation protocols and their characterization are not as robust in animal models as they are in humans and mice. The generation of induced pluripotent stem cells (iPSCs), unlike embryonic stem cells, creates the possibility of autologous therapies and circumvents ethical barriers. iPSCs applications range from basic to applied research, for example, from regenerative medicine to the enhancement of animal production to generate functional gametes or even iPSCs-derived embryos[1-3]. For wild and domestic animal models, the establishment of pluripotent cells and their maintenance in vitro may enable diverse translational, clinical and reproductive applications. A robust approach, along with a well-known understanding of the pluripotency pathways for each species, is still to be reported, as previously discussed and reviewed[4-6].
The reprogramming of porcine cells into a pluripotent state can significantly contribute not only to applications in veterinary medicine and animal production, but also, the porcine as a large biomedical model is greatly acknowledged for their physiological and immunological similarities to humans, being suitable to preclinical and translational studies, in special when compared to the murine model[3,5,7-11]. Cells used for the in vitro reprogramming into iPSCs are mostly from invasive collection procedures, such as from embryos and interrupted gestations (embryonic and foetal cells), or biopsies (adult fibroblasts and mesenchymal cells)[1,12,13]. The derivation of cultured cells from embryos or foetuses impedes their development, and consequently is considered an unethical practice in humans. The isolation of adult fibroblasts and other tissue-derived cells through biopsies is usual, especially when autologous therapies or in vitro modelling of specific genomes is needed. Biopsies, however, usually demand minimally invasive procedures performed by health professionals. Post procedure care may lead to complications such as scars, inflammation, and infection. In particular, the ability of iPSCs to model in vitro syndromes or diseases from patients with affected cognitive, neurological, and muscular-skeletal functions may be impaired by such procedures, often requiring special attention and ethics approval. Therefore, using cells from a noninvasive source for the generation of iPSCs would facilitate their use in regenerative and translational human or veterinary medicine, aiming for its large scale use without resulting in injuries or stress[14,15].
Urine-derived cells (UDCs) have been recently reported in humans, and the in vitro modelling of diseases using these cells or iPSCs derived from them is increasingly being explored[16,17]. Studies on the in vitro differentiation of human UDCs into cardiomyocytes[16] and hepatocyte-like cells[18], the generation of patient-specific iPSC lineages for multiple sclerosis[19], X-linked retinoschisis[20], heart failure[21], phenylketonuria[22], glaucoma[23], and retinitis pigmentosa[24], and recently, the derivation of iPSCs from UDCs in nonhuman primates[25] reinforce the importance of this recent in vitro modelling tool.
Noninvasive cell isolation in domestic animals has also been recently reported from milk[26], an exclusive female possibility, and from urine in the rabbit and canine models[27,28]; however, no pluripotent cells have been derived from these models aiming at its use in regenerative medicine so far. In this context, porcine are nonprimate large animals widely known to present physiological and immunological similarities with humans, as well as they are considered an important species for animal production, with standardized management with pathogen-free conditions[29,30], and consequently, their use as a biomedical model is advantageous compared to nonhuman primates. The fully reprogramming, consistent and robust characterization of porcine iPSCs (piPSCs) are not frequently reported; however, in vitro differentiation of these cells into other cell types, and importantly, the generation of chimeras has been presented and discussed, endorsing their use for in vitro disease modeling or even for cell therapy[14,18,19,31,32].
Herein, we describe urine collection, cellular isolation, and in vitro reprogramming of a noninvasive cell source used for iPSC generation in a large domestic animal, the porcine model. Three clonal lineages were evaluated throughout the passages. Porcine iPSCs derived from UDCs are important not only for agricultural traits, for example, for enabling the in vitro generation of gametes and embryos and contributing to future genetic improvement, but also as an excellent platform for the in vitro and in vivo modelling of several diseases.
All procedures were performed following the National Council for Control of Animal Experimentation (CONCEA) rules and were approved by the Ethics Committee on Animal Experimentation of the Faculty of Animal Science and Food Engineering and Faculty of Veterinary Medicine and Animal Sciences, University of São Paulo (protocols 6372070119 and 7051150717).
Swine urine samples (approximately 250 mL) were collected from three females at reproductive age (2 year) after spontaneous urination. The samples were identified as UDC1, UDC2, and UDC3, and processed following the protocol previously described for human samples[33]. Briefly, the urine was aliquoted into conical tubes and centrifuged at 400 × g and 25 °C for 10 min; the supernatant was removed, leaving approximately 1 mL in each tube, washed with 45 mL of D-polybutylene succinate (PBS) (Life Technologies) containing 1% penicillin/streptomycin (Life Technologies), and centrifuged at 200 × g and 25 °C for 10 min. The supernatant was discarded, and the pellet was resuspended in 12 mL of previously prepared medium containing 22.5 mL DMEM high glucose (Life Technologies), 2.5 mL FBS (HyClone), 0.25 mL penicillin/streptomycin (Life Technologies), 0.25 mL 100 × GlutaMAX supplement (Life Technologies), 0.25 mL 100 × nonessential amino acid solution (Life Technologies), 25 mL REBM medium (Renal Epithelial Basal Medium, Lonza) and REGM supplements: 5 μL/mL FBS, hEGF, insulin, hydrocortisone, GA-1000, transferrin, triiodothyronine, epinephrine (all 0.5 μL/mL, Lonza), and basic fibroblast growth factor (bFGF) (2.5 ng/mL, PeproTech).
Cells were plated onto 0.1% gelatine (Sigma-Aldrich)-coated 24-well plates. The medium was replaced at D3 (3rd day after plating) and then partially refreshed every day. The UDC1 cell lineage was further used in the cellular reprogramming protocol, and clonal iPSC lineages were used for statistical analyses.
In vitro reprogramming was performed by transducing UDCs with polycistronic lentiviral vectors harboring either murine or human transcription factors OCT4, SOX2, KLF-4 and C-MYC (mOSKM or hOSKM, STEMCCA, Millipore), as previously reported[34,35]. Briefly, for the production of lentiviral particles, the lipofection protocol (Lipofectamine 3000, Life Technologies) was performed using OSKM and auxiliary vectors TAT, REV, Hgpm2, VSVG, in 293 FT cells (Life Technologies) as previously described. UDCs, at a concentration of 2 × 104 per well, were transduced with viral particles and incubated overnight at 38.5 °C, 5% CO2, and maximum humidity for 12-16 h, when media were refreshed.
After 5-6 d, the transduced cells were replaced onto a 6-well plate coated with a monolayer of mitomycin C (M4287 Sigma-Aldrich)-inactivated MEFs and cultured in iPSC medium composed of DMEM/F12 knockout medium supplemented with 20% KSR, 1% glutamine, 3.85 μM β-mercaptoethanol, 1% nonessential amino acids, 1% penicillin/streptomycin (all from Life Technologies), and 10 ng/mL bFGF (PeproTech) and incubated at 38.5 °C, 5% CO2 and maximum humidity. After approximately 1 wk, colonies were manually picked at the first passage, and further on, clonal lineages (putative iPSCs, or iPSC-like cells) were dissociated for passaging (TrypLe Express, Life Technologies). Three clonal lineages (C1, C2, and C3) were further analysed throughout passaging. Cryopreservation (10% DMSO), and therefore a freeze-and-thaw cycle, was performed at approximately passage 18 and again at approximately passage 30.
The reprogramming efficiency was assessed by analysing the ratio of morphologically typical and alkaline phosphatase (AP)-positive iPSCs colonies per the number of transduced cells initially plated (2 × 104 cells per well of a 6-well plate). The AP detection protocol was performed using the Alkaline Phosphatase Detection Kit (86R, Sigma-Aldrich) according to the manufacturer’s instructions.
Immunocytochemistry was used to detect OCT4, SOX2, NANOG, SSEA1, and TRA1 81 in two different passage windows for the three lineages: p16, p15, and p9 for C1, C2, and C3, respectively, and again after p20 (p23, p22, and p22, respectively). The cultured putative piPSCs were fixed in paraformaldehyde for 10 min and washed in PBS. The pluripotency-related markers test was performed as previously described[36]. Briefly, the antibodies were used to detect OCT4 (1:100, cat# SC8628, Santa Cruz), SOX2 (1:500, cat# ab97959; Abcam), NANOG (1:100, cat# ab77095, Abcam), SSEA1 (1:50, cat# MAB4301, Millipore) and TRA1 81 (1:50, cat# MAB4381, Millipore), and the respective secondary antibodies were used (donkey anti-goat 594, cat# A11058, donkey anti-rabbit 488, A21206, 1:500, donkey anti-goat 488, cat# A11055, Invitrogen, 1:500 goat anti-mouse 594, cat# A21044, Invitrogen). When necessary, the cells underwent permeabilization and blocking following previously described methods[37]. At the end of each protocol, the cell nuclei were labelled with Hoechst 33342 (1:1000) and analysed using the EVOS™ photodocumentation system.
RNA extraction and reverse transcription: The specific expression of endogenous factors OCT4, SOX2, NANOG, and exogenous reprogramming factors (mOSKM) was evaluated in UDCs and reprogrammed cells. Additionally, porcine embryos were collected on day 5 after insemination and cultured in vitro for 24 h to obtain blastocysts[38]. A pool of 20 porcine blastocysts was used as a positive technical control for pluripotency-related gene expression.
UDCs and iPSCs were recovered from culture plates and centrifuged in microtubes. The pellets were resuspended in linear acrylamide (0.05 mg/mL, Ambion) and UltraPure™ DNase/RNase-Free Distilled Water (Invitrogen), and RNA was extracted using TRIzol Reagent (Invitrogen) following the manufacturer’s instructions. The RNA samples were analyzed regarding quantity and quality using a spectrophotometer (Nanodrop 2000). Reverse transcription of the extracted RNA was performed using the commercial High-Capacity cDNA Reverse Transcription Kit (QIAGEN) according to the manufacturer’s instructions.
Gene expression quantification: The three reprogrammed clonal lineages (C1, C2, and C3) were analysed for the expression of the endogenous factors OCT4, SOX2, and NANOG as well as exogenous reprogramming factors (mOSKM) at different time points of in vitro culture: Early passages (EP: 15 to 18), intermediate passages (IP: 20 to 24), and late passages (LP: 29 to 32). To quantitatively evaluate expression, primers were designed using Primer-BLAST software (NCBI) with GenBank sequences (Supplementary Table 1). Polymerase chain reaction (PCR) products were sequenced for specificity analysis. The reference genes were glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and beta-actin 3 (β-ACTIN-3) and normalization was performed based on their geometric means. The primers for endogenous pluripotency gene expression were designed to detect porcine and not murine transcripts, whereas exogenous expression was detected using the mOSKM primers.
Relative expression of candidate genes was quantified by SYBR Green PCR Master Mix (Life Technologies) using the QuantStudio 5 PCR System (Thermo Fisher). Cycling conditions for amplification were 95 °C for 15 min; 40 cycles of 95 °C for 15 s, 60 °C for 5 s, and 72 °C for 30 s; and 72 °C for 2 min; the melting curve was analysed up to 90 cycles starting at 50 °C with a 0.5 °C increase. The three clonal lineages were considered biological triplicates when compared to UDCs, whereas different passages from the same lineage were considered biological triplicates when these were compared, and all reactions were performed in technical duplicates. The relative gene abundance was performed by 2ΔCT[39].
The piPSCs from the three lineages at passages 15-16 and also at passages 24-25 were replated into a 6-well plate previously treated with 0.6% agarose and cultured in bFGF-free iPSC medium for 48 to 60 h. The embryoid bodies (EBs) were collected and centrifuged at 900 × g for 5 min, and RNA extraction was performed as described before. Reverse transcription was performed using the commercial High-Capacity cDNA Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions to evaluate the expression of endodermal (AFP), mesodermal (VIMENTIN and BMP4), and ectodermal (β-TUBULIN III) genes by reverse transcription quantitative real-time PCR (RT-qPCR), as described before (Supplementary Table 1).
Data obtained from the experimental procedures were analyzed using the statistical program Statistical Analysis System (SAS University Edition), with previous verification of the normality of the residues by the Shapiro-Wilk test (PROC UNIVARIATE). The variables that did not meet the statistical assumptions were submitted to a logarithmic transformation [Log (X + 1)]. The original or transformed data, when necessary, were submitted to analysis of variance. When significant with the variance analysis, the data related to the different cell lineages were submitted to the Bonferroni test. A significance level of 5% was considered for all statistical analyses.
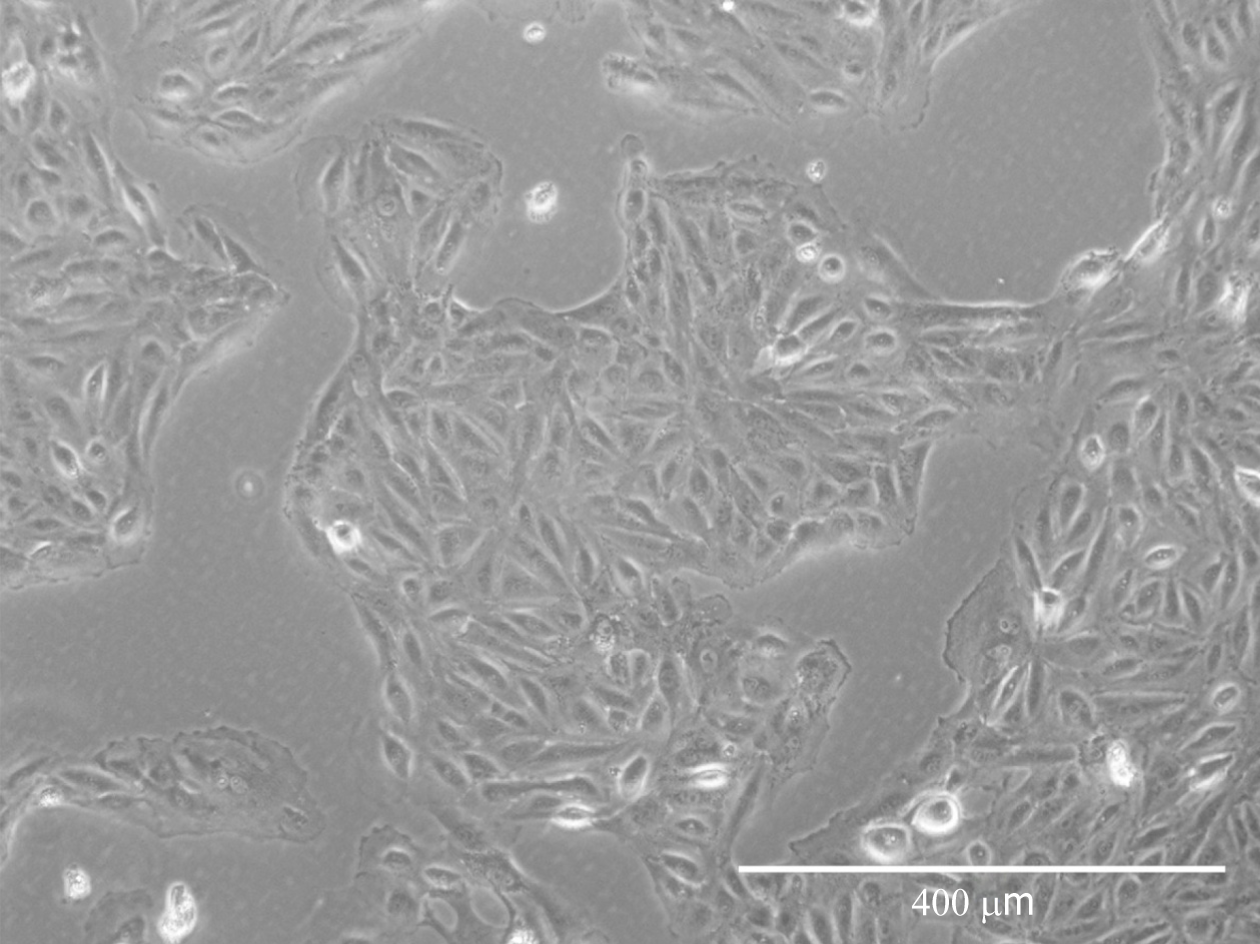
Cells isolated from urine first appeared resembling epithelial-like colonies at 3 to 5 d post isolation and then acquired fibroblastic morphology after passaging (Figure 1).
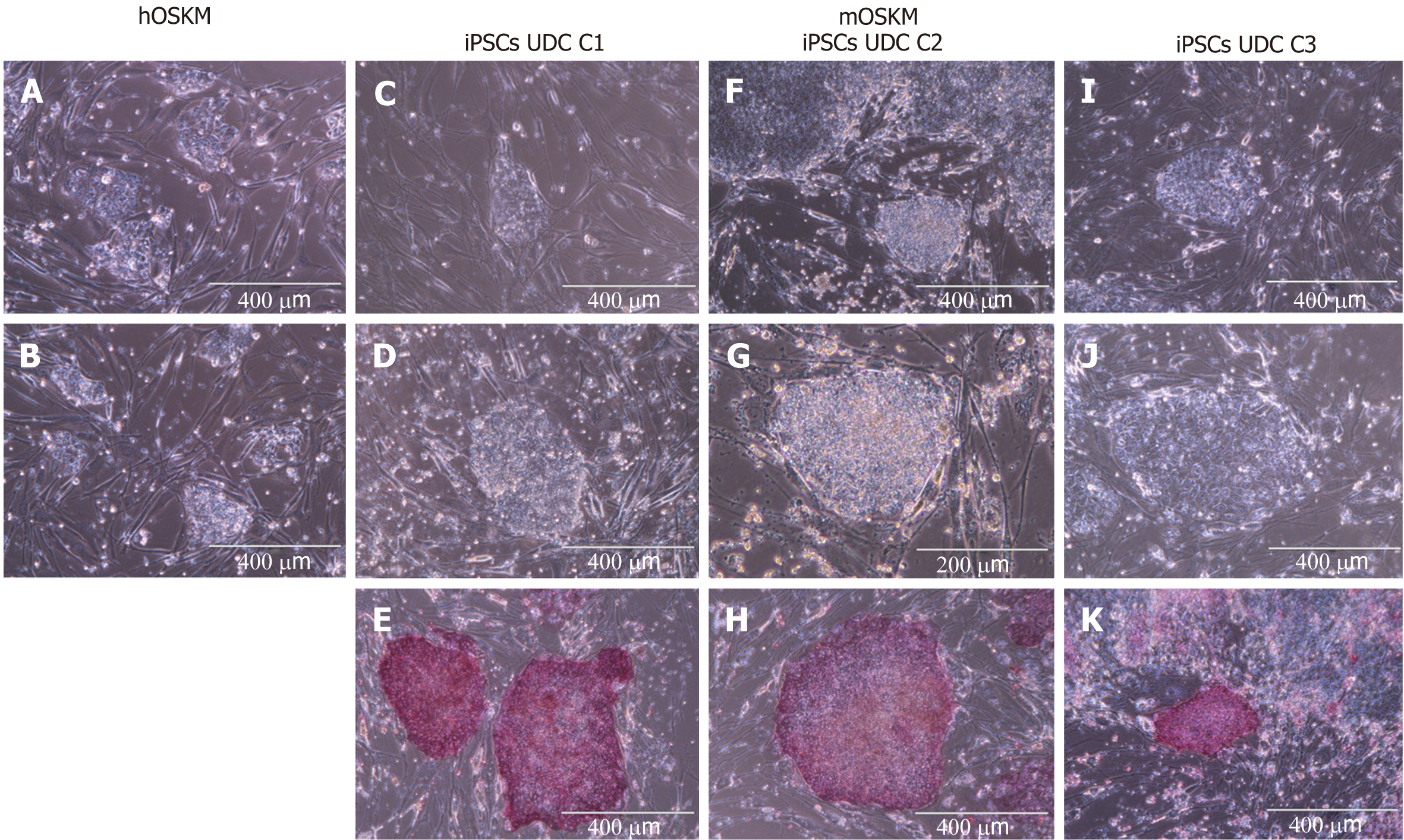
Cellular reprogramming was performed using murine (mOSKM) or human (hOSKM) polycistronic lentiviral vectors. The transduced cells were evaluated for morphological alterations, and twelve days after transduction, typical colonies were observed and tested for AP presence.
Reprogramming efficiency was assessed by analysing the ratio of typical AP-positive iPSC colonies per number of cells initially plated for transduction (Figure 2 and Supplementary Table 2). Repro
Eight colonies were chosen, manually picked, and replated onto new MEFs to obtain clonal lineages. Three clonal lineages designated as C1, C2, and C3 were remained in the culture at least 28 passages and were positive for AP, however, C2 colonies spontaneously differentiated after 28 passages, and the colonies C1 and C3 were further remained in culture for at least 30 passages.
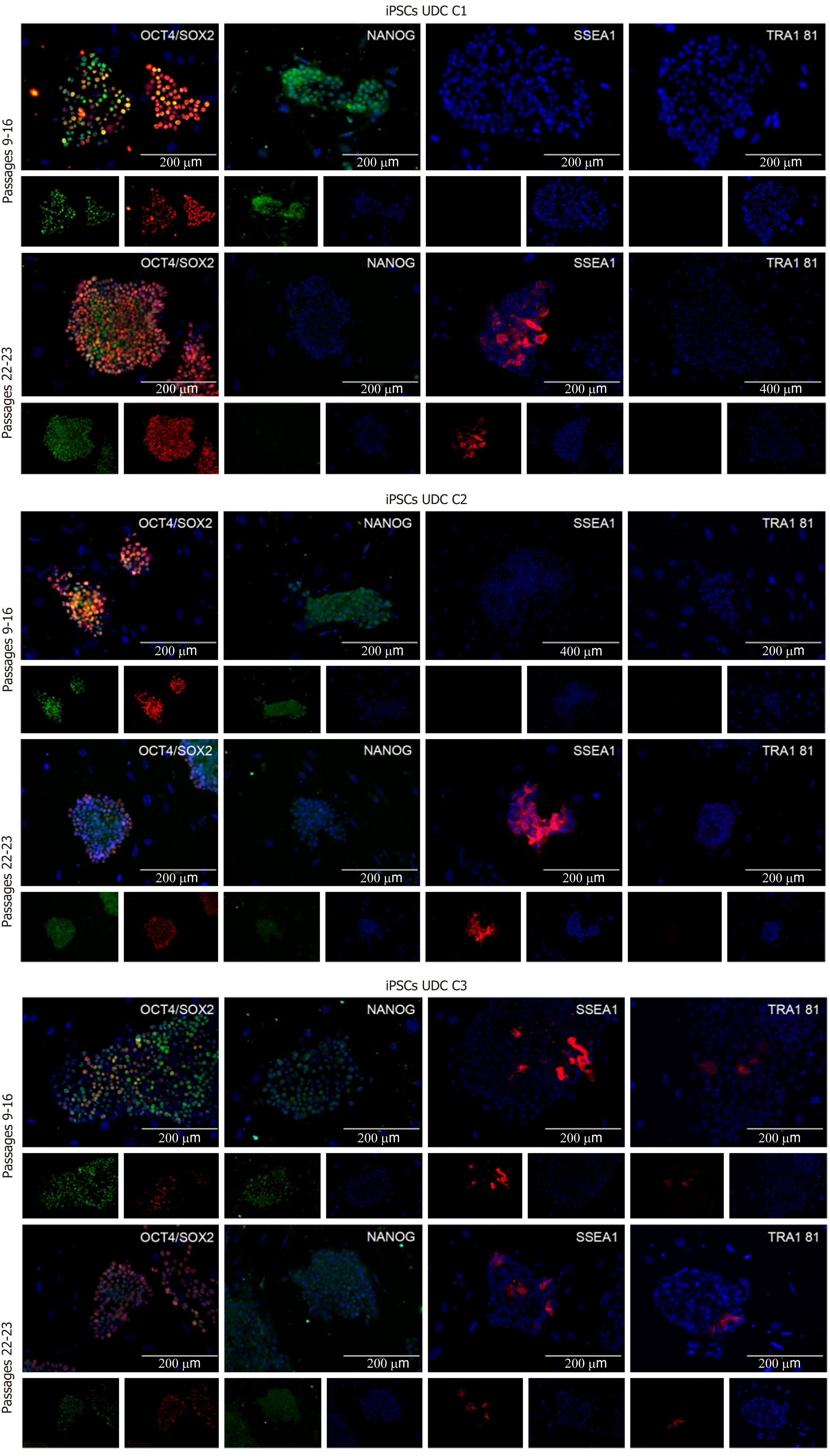
The clonal lineages were tested in two distinct passaging windows: Between p9 and p16 (p16, p15, and p9, respectively, for C1, C2 and C3) and after p20 (p23, p22, and p22, respectively), enabling analysis among colonies and between culture periods. Cell lineages at p9-16 were positive for OCT4, SOX2, and NANOG and generally negative for SSEA1 and TRA1 81. The C3 (p9) clonal lineage presented some cells positive for SSEA1 and TRA1 81 (Figure 3 and Supplementary Table 3).
In passages > p20, detection of OCT4 and SOX2 was observed, and some cells were also positive for SSEA1. C1 and C2 were negative for NANOG and TRA1 81; however, C3 cells presented mild positivity for both NANOG and TRA1 81 (Figure 3). The results are summarized in Supplementary Table 3.
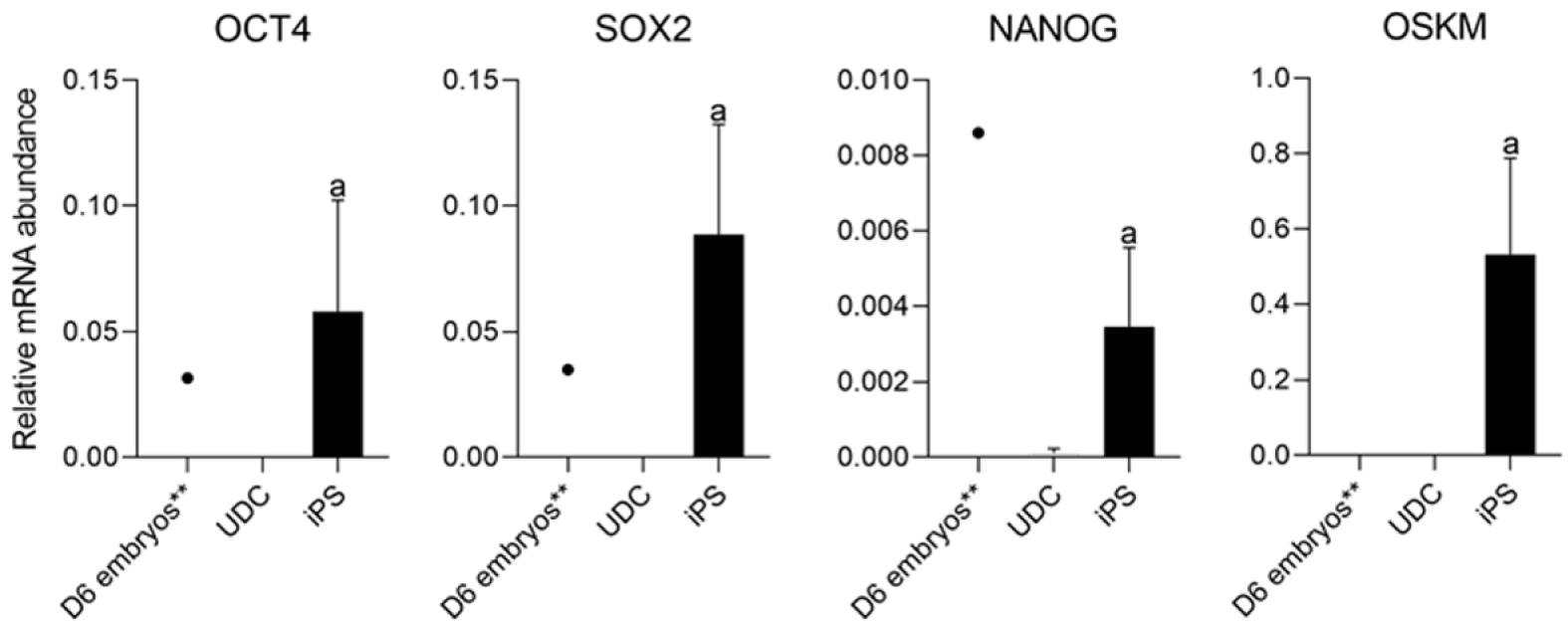
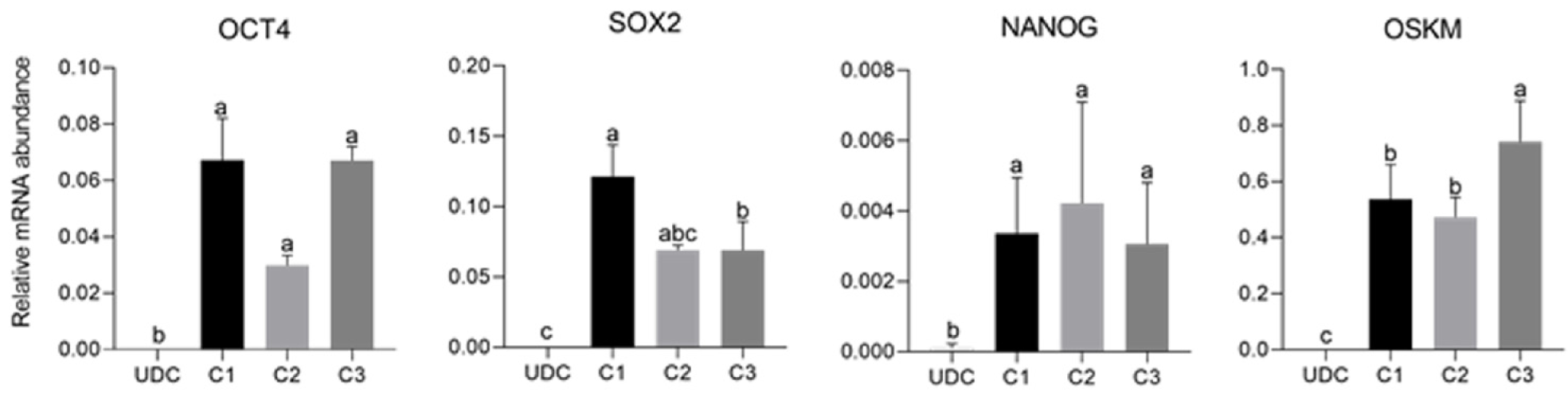
As expected, mOSKM was not amplified in UDCs or blastocysts; and endogenous genes were expressed in blastocysts (Figure 4). Then, reprogrammed lineages were compared to each other and the analysis of the expression of endogenous OCT4, SOX2, NANOG, and mOSKM in the different lineages (C1, C2, and C3) revealed that exogenous reprogramming factors were still detected in later passages of iPSCLCs. C3 showed higher expression of the exogenous vector (P < 0.0001) and lower expression of SOX2 than lineage C1 (P = 0.0099). OCT4 and NANOG expression did not differ among lineages (Figure 5 and Supplementary Table 4).
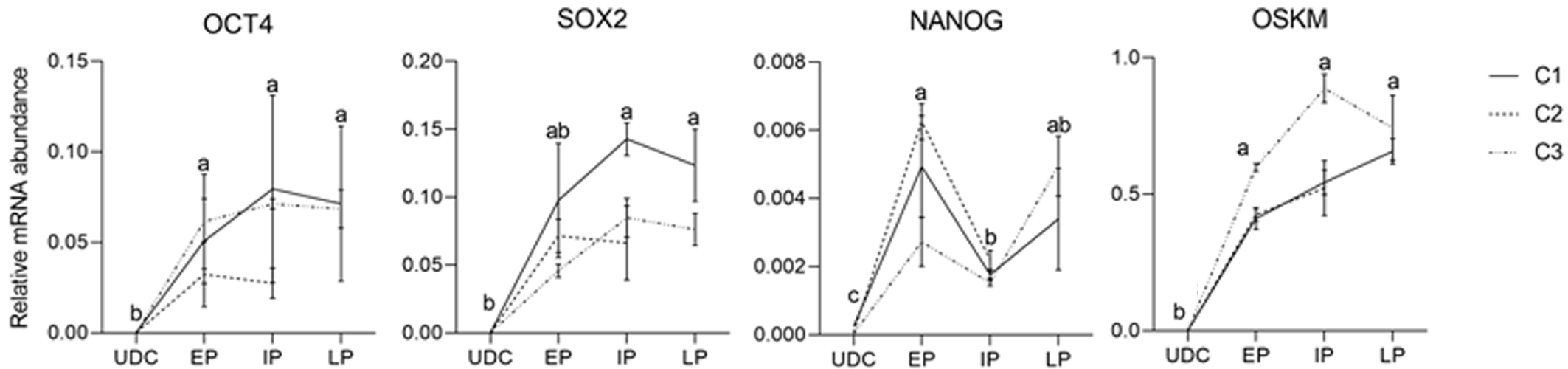
The analysis of the effect of time in culture (passaging) of the endogenous gene expression in UDCs and the iPSCLCs different groups (EP: 15 to 18; IP: 20 to 24; and LP: 29 to 32) revealed that IP and LP presented higher expression of SOX2, augmenting during culture period; and OCT4 levels were detected in all periods, differing from UDCs. The expression of the exogenous vector did not differ among passages. NANOG expression, however, decreased in intermediate passages, possibly due to a freeze-thaw cycle between EP and IP in vitro. At LP, NANOG was again slightly increased. The LP group of the C2 Lineage was not shown once these cells underwent spontaneous differentiation at passage 28 (Supplementary Table 5 and Figure 6).
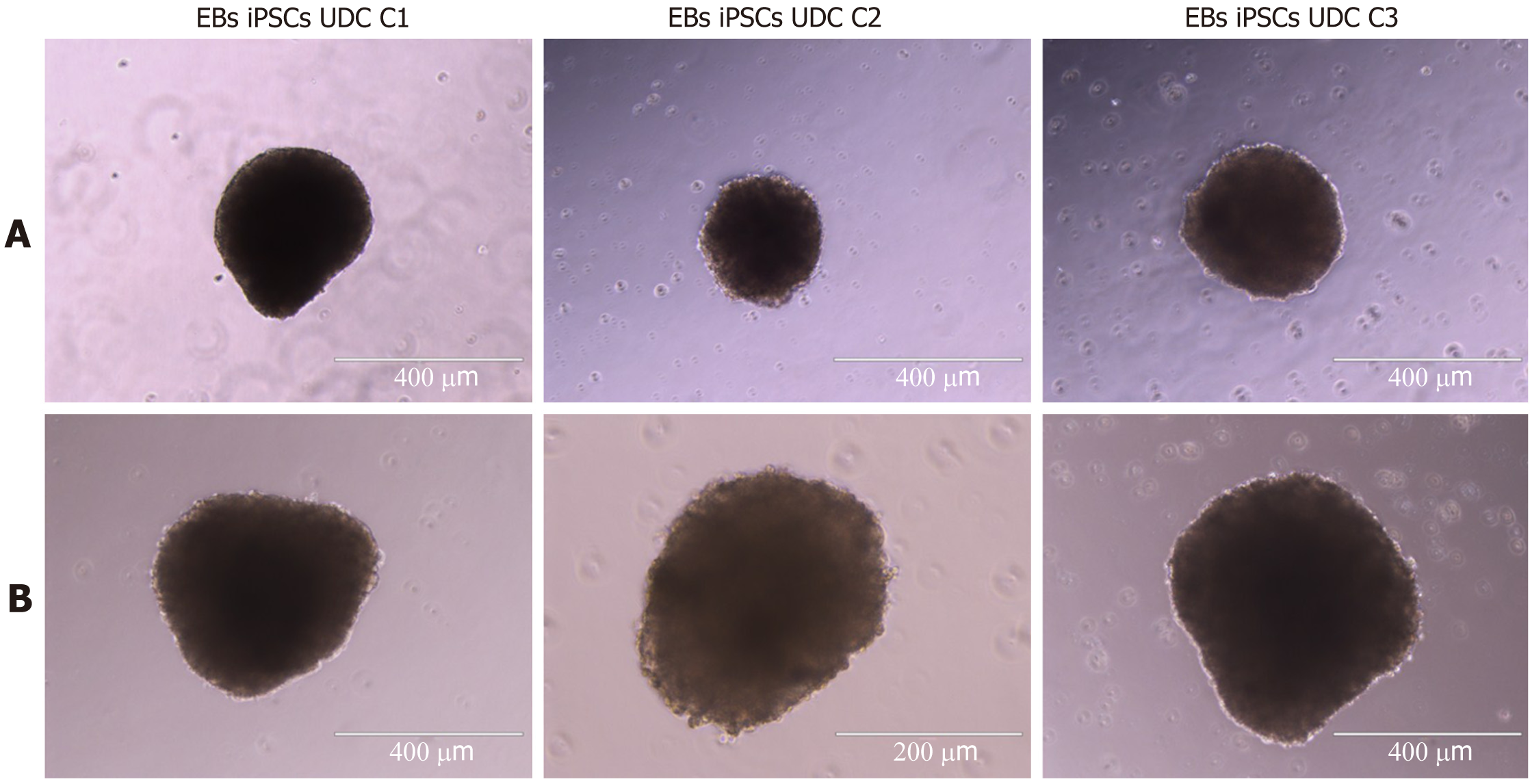
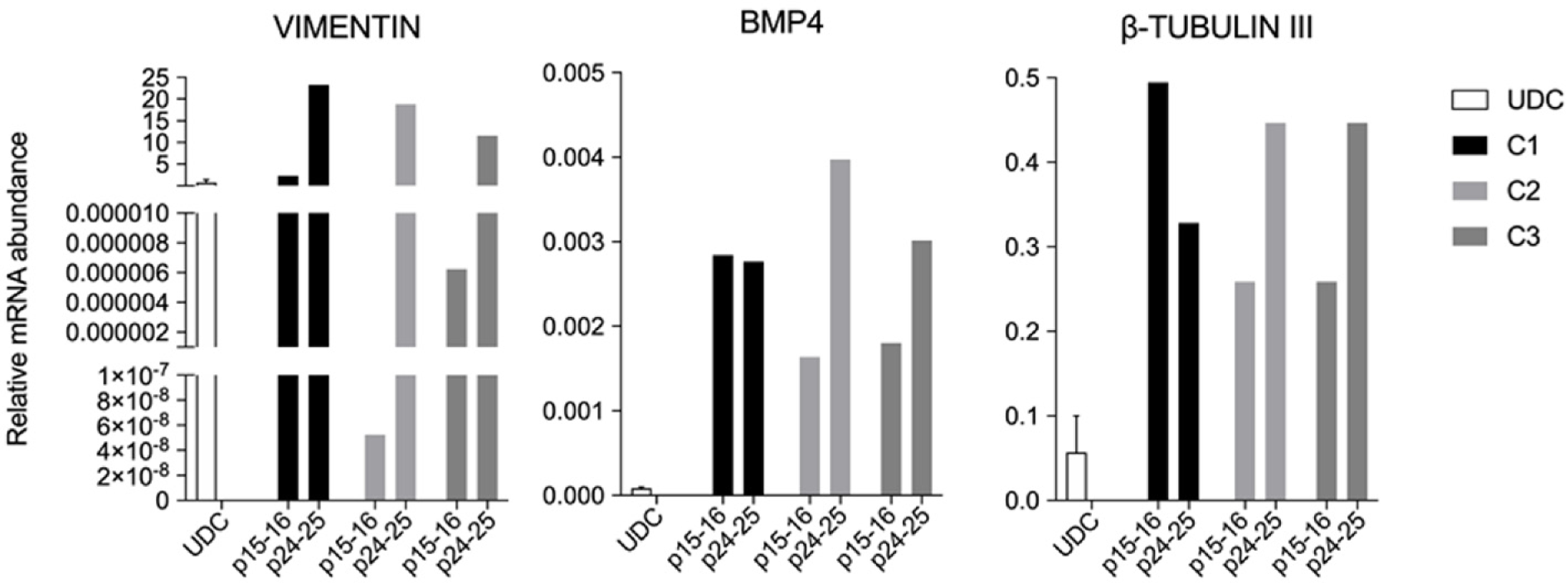
All clonal lineages were replated as single cells onto a nonadherent plate without bFGF supplementation, and these cells formed EBs with typical morphology at different passages (Figure 7). The expression of VIMENTIN, BMP4 and β-TUBULIN-III was detected in the EBs (Figure 8), and AFP was not detected in our conditions (data not shown).
Herein, cells derived from urine sample (UDCs) were in vitro reprogrammed in a large domestic animal model, the swine. Previous studies on porcine have mostly derived iPSCs from foetal or adult fibroblasts, and fewer with multipotent adult cells[1]. UDC-derived piPSCs are highly advantageous for veterinary and regenerative medicine due to the simple collection procedure, avoiding stress or injuries, and in addition, is an inexpensive procedure unlike surgeries aiming biopsies, also important for the feasibility of large-scale sample collection[14,15,17,27,40].
Raab et al[40] reported a higher reprogramming efficiency of human UDCs when compared with other somatic cell types. Indeed, several studies have reported cell heterogeneity from urine-derive cells in human, including renal tubular cells[41], urine-stem cells (renal progenitor cells)[42], and urine-derived epithelial cells[16]. It is already known the cells’ origin can influence the reprogramming process, and a more complete characterization and sorting for each cell type prior to reprogramming may be essential to understand the contribution of each cell population to the generation of iPSCs[35,43].
In recent years, it has been showed by several reports the establishment of pluripotency, or at least a state similar to embryonic pluripotency, in several species other than human and mice, and although the main molecular mechanisms involved in pluripotency acquisition in vitro are considered rather conserved between species, there are notable differences between species turning the generation of bonafide iPSCs challenging, however still extremely promising.
Indeed, the same human/mouse protocols for iPSCs generation are not extendable to other species[1,3]. Herein we used a previous strategy already reported for large animals reprogramming[34-36,43-45], and widely used to reprogram porcine somatic cells[1]. In the conditions described, the results showed that UDCs transduced with human factors failed to be maintained in culture for more than 5 passages due to early differentiation of the cells cultured in vitro, and similar results was described by Pieri et al[36] when reprogramming of porcine foetal fibroblasts. Conversely, cells transduced with murine factors were maintained in culture for at least 28 passages, showing typical morphology, positive AP detection and endogenous pluripotency-related gene expression through the different passages. Next steps to improve in vitro reprogramming must consider possible epigenetic modulation or even the identification of species-specific pluripotency pathways to improve the nonintegrative reprogramming.
Lineages at p9-16 were positive for SSEA1 and weakly positive for NANOG. These results correlate with a decrease in NANOG expression at IPs, and it might be an effect associated with the freeze-and-thaw process, which was performed in the lineages between EP and IP in this study. Interestingly, the abundance of NANOG transcription increased between the IP and LP. Li et al[12] reported that the staining for NANOG, SOX2 and OCT4, increased at passage 20 when compared to p10, indicating a stabilization of the pluripotency phenotype of intermediate type piPSCs. In addition, an elegant discussion was provided by Yamanaka[46] on the heterogeneous profile of each iPSCs lineage, leading to different phenotypes. Furthermore, in our conditions, we infer that a longer time in culture without the freeze-and-thaw process may lead to better reprogramming, as observed by the late acquisition of the SSEA1+ phenotype, a reported marker for human naïve stem cells[47].
All lineages formed EBs that expressed VIMENTIN, BMP4 and b-TUBULIN-III, known markers of mesodermal and ectodermal lineages, respectively. However, none of the EBs presented AFP transcripts, a marker of the endodermal lineage. Rodríguez et al[41] has shown that EBs differentiated from piPSCs cultured in different conditions have shown mesoderm, endoderm and ectoderm markers after 15 d of undirected differentiation, and moreover, some markers not or mildly found at D15, were shown after D30 of differentiation[41]. Hence, further markers and other periods during spontaneous differentiation should be tested for complete characterization and discussion.
Overall, the results presented describe novel ways to derive in vitro reprogrammed cells in an important biomedical model, the porcine model. The isolation of UDCs is also relevant for other reproductive technologies, for example, for the conservation of many mammal species through nuclear reprogramming, or even to produce in vitro viable gametes, which could decrease the interval between generations for the acquisition of a genetically superior herd. Although the scenario of complete and robust in vitro cellular reprogramming is still under discussion in the porcine model; the advances described herein, in our conditions, are valuable for both translational studies and animal production, hence these putative piPSCs can be used to enable future autologous therapies, to the creation of gene-edited or not in vitro and in vivo biomedical models, to the study of the mechanism of cell differentiation, and also to future generation of gamete- or embryos-derived from iPSCs, contributing to the conservation and propagation of genetic material.
The results presented herein report, for the first time, the isolation and reprogramming of cells derived through the noninvasive collection of urine in a porcine model. Under our conditions, three putative iPSC lineages generated with murine OSKM presented typical morphology, AP and endogenous pluripotency-related gene expression, which was analyzed in three different passaging periods of the in vitro culture, and two lineages were maintained in vitro for more than 28 passages. Further studies on pluripotency induction in domestic animals are still needed to thoroughly understand and achieve full reprogramming, including more complete molecular profiles during in vitro and in vivo reprogramming processes, representing a novel tool for biomedical models of regenerative and translational medicine and animal production improvement.
Induced pluripotent stem cells (iPSCs) derived from large animal models can greatly contribute to translational medicine and also to animal production, although robust and safe protocols are still uncommon. Cellular reprogramming of urine derived cells presents great advantages for iPSCs use in regenerative medicine due to the easy collection, injury and stress free, and is herein described for the first time in large animals.
The porcine iPSCs generation is promising for both translational medicine and animal production; and iPSCs derived from a noninvasive cell source would greatly contribute to its large-scale use, especially for in vivo autologous purposes using large animal models.
Isolate cells from porcine urine and generate iPSCs through their transduction with Yamanaka’s human or murine factors.
We isolated urine-derived cells (UDCs), which were reprogrammed in vitro into pluripotent cells. The porcine induced pluripotent cells generated were investigated regarding morphology, markers and endogenous transcripts related to the pluripotency.
From the porcine urine samples we isolated the UDCs, and colonies were formed when murine factors were used in the reprogramming. Endogenous pluripotent markers were detected in all three isolated lineages, in different time points during in vitro culture, and were able to differentiate into embryoid bodies (EBs) with mesoderm and ectoderm transcripts.
In an unprecedented way, UDCs were isolated from noninvasive collection and reprogrammed into a pluripotent state using murine factors, the cells formed colonies presenting the expected characteristics, such as colonies with limited borders, transcripts and markers related to the pluripotency, and ability to differentiate into EBs.
As we reported here, iPSCs can be derived from an easy collection and noninvasive source in the porcine model, and with our methodology represents a novel tool for iPSCs production in large animals and biomedical models of regenerative or translational medicine.
The authors would like to acknowledge Professors José Eduardo Krieger, Clara Steichen, and Carlos Eduardo Ambrosio for scientific discussion.
| 1. | Pessôa LVF, Bressan FF, Freude KK. Induced pluripotent stem cells throughout the animal kingdom: Availability and applications. World J Stem Cells. 2019;11:491-505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 2. | Sierra H, Cordova M, Chen CJ, Rajadhyaksha M. Confocal imaging-guided laser ablation of basal cell carcinomas: an ex vivo study. J Invest Dermatol. 2015;135:612-615. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 597] [Cited by in RCA: 674] [Article Influence: 56.2] [Reference Citation Analysis (0)] |
| 3. | Pieri NCG, de Souza AF, Botigelli RC, Machado LS, Ambrosio CE, Dos Santos Martins D, de Andrade AFC, Meirelles FV, Hyttel P, Bressan FF. Stem cells on regenerative and reproductive science in domestic animals. Vet Res Commun. 2019;43:7-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 4. | Kitala D, Maurício AC. Novel Perspectives of Stem Cell Manufacturing and Therapies. In: Vicari de Figueiredo Pessôa L, Caroline Godoy Pieri N, Recchia K, Fernandes Bressan F. Induced Pluripotent Stem Cells from Animal Models: Applications on Translational Research. 2020. |
| 5. | Nowak-Imialek M, Kues W, Carnwath JW, Niemann H. Pluripotent stem cells and reprogrammed cells in farm animals. Microsc Microanal. 2011;17:474-497. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 6. | Bogliotti YS, Wu J, Vilarino M, Okamura D, Soto DA, Zhong C, Sakurai M, Sampaio RV, Suzuki K, Izpisua Belmonte JC, Ross PJ. Efficient derivation of stable primed pluripotent embryonic stem cells from bovine blastocysts. Proc Natl Acad Sci U S A. 2018;115:2090-2095. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 184] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 7. | Perin EC, Miller LW, Taylor DA, Willerson JT. Stem Cell and Gene Therapy for Cardiovascular Disease. In: Migliati E. Large Animal Models for Cardiac Cell Therapy. New York: Elsevier, 2015: 25-36. |
| 8. | Cong X, Zhang SM, Ellis MW, Luo J. Large Animal Models for the Clinical Application of Human Induced Pluripotent Stem Cells. Stem Cells Dev. 2019;28:1288-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 9. | Su Y, Zhu J, Salman S, Tang Y. Induced pluripotent stem cells from farm animals. J Anim Sci. 2020;98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 10. | Cooper DKC, Hara H, Iwase H, Yamamoto T, Jagdale A, Kumar V, Mannon RB, Hanaway MJ, Anderson DJ, Eckhoff DE. Clinical Pig Kidney Xenotransplantation: How Close Are We? J Am Soc Nephrol. 2020;31:12-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 50] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 11. | Ellis CE, Korbutt GS. Justifying clinical trials for porcine islet xenotransplantation. Xenotransplantation. 2015;22:336-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Li D, Secher J, Hyttel P, Ivask M, Kolko M, Hall VJ, Freude KK. Generation of transgene-free porcine intermediate type induced pluripotent stem cells. Cell Cycle. 2018;17:2547-2563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | Esteban MA, Xu J, Yang J, Peng M, Qin D, Li W, Jiang Z, Chen J, Deng K, Zhong M, Cai J, Lai L, Pei D. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J Biol Chem. 2009;284:17634-17640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 298] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 14. | Gao P, Jiang D, Liu W, Li H, Li Z. Urine-derived Stem Cells, A New Source of Seed Cells for Tissue Engineering. Curr Stem Cell Res Ther. 2016;11:547-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 15. | Ji X, Wang M, Chen F, Zhou J. Urine-Derived Stem Cells: The Present and the Future. Stem Cells Int. 2017;2017:4378947. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 16. | Jiang YF, Chen M, Zhang NN, Yang HJ, Rui Q, Zhou YF. In vitro and in vivo differentiation of induced pluripotent stem cells generated from urine-derived cells into cardiomyocytes. Biol Open. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Bento G, Shafigullina AK, Rizvanov AA, Sardão VA, Macedo MP, Oliveira PJ. Urine-Derived Stem Cells: Applications in Regenerative and Predictive Medicine. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 18. | Zhou M, Shen L, Qiao Y, Sun Z. Inducing differentiation of human urine-derived stem cells into hepatocyte-like cells by coculturing with human hepatocyte L02 cells. J Cell Biochem. 2020;121:566-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Massa MG, Gisevius B, Hirschberg S, Hinz L, Schmidt M, Gold R, Prochnow N, Haghikia A. Multiple Sclerosis Patient-Specific Primary Neurons Differentiated from Urinary Renal Epithelial Cells via Induced Pluripotent Stem Cells. PLoS One. 2016;11:e0155274. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 20. | Yan X, Guo Y, Chen J, Cui Z, Gu J, Wang Y, Mao S, Ding C, Tang S. Establishment of CSUASOi001-A, a non-integrated induced pluripotent stem cell line from urine-derived cells of a Chinese patient carrying RS1 gene mutation. Stem Cell Res. 2019;38:101466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 21. | Cao Y, Xu J, Wen J, Ma X, Liu F, Li Y, Chen W, Sun L, Wu Y, Li S, Li J, Huang G. Generation of a Urine-Derived Ips Cell Line from a Patient with a Ventricular Septal Defect and Heart Failure and the Robust Differentiation of These Cells to Cardiomyocytes via Small Molecules. Cell Physiol Biochem. 2018;50:538-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 22. | Qi Z, Cui Y, Shi L, Luan J, Zhou X, Han J. Generation of urine-derived induced pluripotent stem cells from a patient with phenylketonuria. Intractable Rare Dis Res. 2018;7:87-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 23. | Zhang J, Wu S, Hu M, Liu Q. Generation of a human induced pluripotent stem cell line from urinary cells of a patient with primary congenital glaucoma using integration free Sendai technology. Stem Cell Res. 2018;29:162-165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 24. | Guo Y, Zeng Q, Liu S, Yu Q, Wang P, Ma H, Shi S, Yan X, Cui Z, Xie M, Xue Y, Zha Q, Li Z, Zhang J, Tang S, Chen J. Generation of an iPS cell line via a non-integrative method using urine-derived cells from a patient with USH2A-associated retinitis pigmentosa. Stem Cell Res. 2018;29:139-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Geuder J, Wange LE, Janjic A, Radmer J, Janssen P, Bagnoli JW, Müller S, Kaul A, Ohnuki M, Enard W. A non-invasive method to generate induced pluripotent stem cells from primate urine. Sci Rep. 2021;11:3516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 26. | Pipino C, Mandatori D, Buccella F, Lanuti P, Preziuso A, Castellani F, Grotta L, Di Tomo P, Marchetti S, Di Pietro N, Cichelli A, Pandolfi A, Martino G. Identification and Characterization of a Stem Cell-Like Population in Bovine Milk: A Potential New Source for Regenerative Medicine in Veterinary. Stem Cells Dev. 2018;27:1587-1597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 27. | Yang H, Chen B, Deng J, Zhuang G, Wu S, Liu G, Deng C, Yang G, Qiu X, Wei P, Wang X, Zhang Y. Characterization of rabbit urine-derived stem cells for potential application in lower urinary tract tissue regeneration. Cell Tissue Res. 2018;374:303-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 28. | Xu Y, Zhang T, Chen Y, Shi Q, Li M, Qin T, Hu J, Lu H, Liu J, Chen C. Isolation and Characterization of Multipotent Canine Urine-Derived Stem Cells. Stem Cells Int. 2020;2020:8894449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 29. | Du X, Feng T, Yu D, Wu Y, Zou H, Ma S, Feng C, Huang Y, Ouyang H, Hu X, Pan D, Li N, Wu S. Barriers for Deriving Transgene-Free Pig iPS Cells with Episomal Vectors. Stem Cells. 2015;33:3228-3238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 30. | Perleberg C, Kind A, Schnieke A. Genetically engineered pigs as models for human disease. Dis Model Mech. 2018;11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 162] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 31. | Wang H, Xiang J, Zhang W, Li J, Wei Q, Zhong L, Ouyang H, Han J. Induction of Germ Cell-like Cells from Porcine Induced Pluripotent Stem Cells. Sci Rep. 2016;6:27256. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 32. | West FD, Terlouw SL, Kwon DJ, Mumaw JL, Dhara SK, Hasneen K, Dobrinsky JR, Stice SL. Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev. 2010;19:1211-1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 175] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 33. | Steichen C, Si-Tayeb K, Wulkan F, Crestani T, Rosas G, Dariolli R, Pereira AC, Krieger JE. Human Induced Pluripotent Stem (hiPS) Cells from Urine Samples: A Non-Integrative and Feeder-Free Reprogramming Strategy. Curr Protoc Hum Genet. 2017;92:21.7.1-21.7.22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (1)] |
| 34. | Machado LS, Pieri NCG, Botigelli RC, de Castro RVG, de Souza AF, Bridi A, Lima MA, Fantinato Neto P, Pessôa LVF, Martins SMMK, De Andrade AFC, Freude KK, Bressan FF. Generation of neural progenitor cells from porcine-induced pluripotent stem cells. J Tissue Eng Regen Med. 2020;14:1880-1891. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 35. | Bressan FF, Bassanezze V, de Figueiredo Pessôa LV, Sacramento CB, Malta TM, Kashima S, Fantinato Neto P, Strefezzi RF, Pieri NCG, Krieger JE, Covas DT, Meirelles FV. Generation of induced pluripotent stem cells from large domestic animals. Stem Cell Res Ther. 2020;11:247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 36. | Pieri NCG, de Souza AF, Botigelli RC, Pessôa LVF, Recchia K, Machado LS, Glória MH, de Castro RVG, Leal DF, Fantinato Neto P, Martins SMMK, Dos Santos Martins D, Bressan FF, de Andrade AFC. Porcine Primordial Germ Cell-Like Cells Generated from Induced Pluripotent Stem Cells Under Different Culture Conditions. Stem Cell Rev Rep. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 37. | de Souza AF, Pieri NCG, Roballo KCS, Bressan FF, Casals JB, Ambrósio CE, Perecin F, Martins DS. Dynamics of male canine germ cell development. PLoS One. 2018;13:e0193026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 38. | Marques MG, de Souza AP, Pandolfi JRC. Metodologia para coleta in vitro de embriões em suínos. 2019. |
| 39. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 138826] [Article Influence: 5553.0] [Reference Citation Analysis (2)] |
| 40. | Raab S, Klingenstein M, Liebau S, Linta L. A Comparative View on Human Somatic Cell Sources for iPSC Generation. Stem Cells Int. 2014;2014:768391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 177] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 41. | Zhou T, Benda C, Duzinger S, Huang Y, Li X, Li Y, Guo X, Cao G, Chen S, Hao L, Chan YC, Ng KM, Ho JC, Wieser M, Wu J, Redl H, Tse HF, Grillari J, Grillari-Voglauer R, Pei D, Esteban MA. Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol. 2011;22:1221-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 318] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 42. | Burdeyron P, Giraud S, Hauet T, Steichen C. Urine-derived stem/progenitor cells: A focus on their characterization and potential. World J Stem Cells. 2020;12:1080-1096. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 30] [Article Influence: 5.0] [Reference Citation Analysis (7)] |
| 43. | Pessôa LVF, Pires PRL, Del Collado M, Pieri NCG, Recchia K, Souza AF, Perecin F, da Silveira JC, de Andrade AFC, Ambrosio CE, Bressan FF, Meirelles FV. Generation and miRNA Characterization of Equine Induced Pluripotent Stem Cells Derived from Fetal and Adult Multipotent Tissues. Stem Cells Int. 2019;2019:1393791. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 44. | Bessi BW, Botigelli RC, Pieri NCG, Machado LS, Cruz JB, de Moraes P, de Souza AF, Recchia K, Barbosa G, de Castro RVG, Nogueira MFG, Bressan FF. Cattle In Vitro Induced Pluripotent Stem Cells Generated and Maintained in 5 or 20% Oxygen and Different Supplementation. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 13] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 45. | de Castro RVG, Pieri NCG, Fantinato Neto P, Grizendi BM, Dória RGS, Meirelles FV, Smith LC, Garcia JM, Bressan FF. In Vitro Induction of Pluripotency from Equine Fibroblasts in 20% or 5% Oxygen. Stem Cells Int. 2020;2020:8814989. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 46. | Yamanaka S. Pluripotent Stem Cell-Based Cell Therapy-Promise and Challenges. Cell Stem Cell. 2020;27:523-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 812] [Article Influence: 162.4] [Reference Citation Analysis (0)] |
| 47. | Inada E, Saitoh I, Kubota N, Iwase Y, Murakami T, Sawami T, Yamasaki Y, Sato M. Increased Expression of Cell Surface SSEA-1 is Closely Associated with Naïve-Like Conversion from Human Deciduous Teeth Dental Pulp Cells-Derived iPS Cells. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: Brazilian Society of Embryo Technologies; International Embryo Technology Society; Colégio Brasileiro de Reprodução Animal; Associação Brasileira de Terapia Celular e Gênica (ABTCel-Gen); International Society for Stem Cell Research; Sociedade Brasileira de Biologia Celular (SBBC).
Specialty type: Cell and tissue engineering
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Batool SN, Gao YT, Kumar D, Wahid M S-Editor: Wang JJ L-Editor: A P-Editor: Zhang YL