Published online Dec 26, 2021. doi: 10.4252/wjsc.v13.i12.1918
Peer-review started: June 18, 2021
First decision: July 17, 2021
Revised: August 11, 2021
Accepted: December 10, 2021
Article in press: December 10, 2021
Published online: December 26, 2021
Processing time: 189 Days and 18.4 Hours
One of the most challenging tasks of modern biology concerns the real-time tracking and quantification of mRNA expression in living cells. On this matter, a novel platform called SmartFlareTM has taken advantage of fluorophore-linked nanoconstructs for targeting RNA transcripts. Although fluorescence emission does not account for the spatial mRNA distribution, NanoFlare technology has grown a range of theranostic applications starting from detecting biomarkers related to diseases, such as cancer, neurodegenerative pathologies or embryonic developmental disorders.
To investigate the potential of SmartFlareTM in determining time-dependent mRNA expression of prominin 1 (CD133) and octamer-binding transcription factor 4 (OCT4) in single living cells through differentiation.
Brain fragments from the striatum of aborted human fetuses aged 8 wk postconception were processed to obtain neurospheres. For the in vitro differentiation, neurospheres were gently dissociated with Accutase solution. Single cells were resuspended in a basic medium enriched with fetal bovine serum, plated on poly-L-lysine-coated glass coverslips, and grown in a lapse of time from 1 to 4 wk. Live cell mRNA detection was performed using SmartFlareTM probes (CD133, Oct4, Actin, and Scramble). All the samples were incubated at 37 °C for 24 h. For nuclear staining, Hoechst 33342 was added. SmartFlareTM CD133- and OCT4-specific fluorescence signal was assessed using a semiquantitative visual approach, taking into account the fluorescence intensity and the number of labeled cells.
In agreement with previous PCR experiments, a unique expression trend was observed for CD133 and OCT4 genes until 7 d in vitro (DIV). Fluorescence resulted in a mixture of diffuse cytoplasmic and spotted-like pattern, also detectable in the contacting neural branches. From 15 to 30 DIV, only few cells showed a scattered fluorescent pattern, in line with the differentiation progression and coherent with mRNA downregulation of these stemness-related genes.
SmartFlareTM appears to be a reliable, easy-to-handle tool for investigating CD133 and OCT4 expression in a neural stem cell model, preserving cell biological properties in anticipation of downstream experiments.
Core Tip: The detection of RNA transcripts in living cells is a challenge in embryonic development and cancer related studies. In the last decade, a straightforward and noninvasive approach has emerged, exploiting the combination of nanotechnology and the physiological behavior of stem cells. Although SmartFlareTM technology is far from providing an unambiguous localization of specific mRNAs, it might help in elucidating the time-dependent dynamics of RNA expression at single-cell level, where results are coherent with those coming from both qRT-PCR and fluorescence in situ hybridization (FISH), the gold standards for mRNA analysis.
- Citation: Diana A, Setzu MD, Kokaia Z, Nat R, Maxia C, Murtas D. SmartFlareTM is a reliable method for assessing mRNA expression in single neural stem cells. World J Stem Cells 2021; 13(12): 1918-1927
- URL: https://www.wjgnet.com/1948-0210/full/v13/i12/1918.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i12.1918
During the last decades, the proper signature of neural stem cells and their derivatives has been accomplished by tracking both proteins and mRNAs. Thus, the experimental setting is a real challenge when the production of certain proteins is scarce and the sensitivity threshold of the laboratory methods is inadequate. Furthermore, a snapshot of this phenomenon does not account for the pathway dynamics, such as axonal transport, fast secretion, and developmental mechanisms orchestrated by molecular gradients.
Historically, simultaneous detection at single-cell level by means of immunochemical and FISH techniques can provide ultimate confirmation for the presence of a variety of signaling molecules. Nevertheless, the real-time monitoring of specific RNA transcripts and downstream proteins is limited by cell fixation and permeabilization dictated by the above techniques and the required lysis of tissues to extract RNA for qRT-PCR. This last molecular option provides information about gene expression levels, in heterogeneous populations, hiding the small but relevant differences and changes taking place in individual cells. Ultimately, the aforementioned methodologies make incompatible further analysis (e.g., cell sorting and collection) particularly meaningful for addressing developmental issues. Within this context, an affordable and reproducible method aiming at encompassing both the kinetics and quantification of endogenous RNAs at cellular level has been brought by a group of researchers[1,2]. SmartFlareTM technology combines the high sensitivity of oligonucleotide-linked nanoparticles with natural receptor-mediated endocytosis to uptake the same nanoconstructs. In particular, target RNA-specific complementary single stranded RNA (capture strand) is hybridized with a complementary “reporter” sequence bound to a fluorophore (Cy3 or Cy5) at its 5' -end that, for vicinity to the central gold particle, is permanently quenched. Only upon pairing with the target RNA sequence, the reporter strand can be released and consequently gain the feature to flare with fluorescent emission at the proper wavelength and intensity, consistent with the expression level of the target RNA. Since the introduction of the SmartFlareTM concept[2-4], this molecular procedure has been successfully exploited for the identification and assessment of both tumor and immune cell subsets[5-8]. Interestingly, the SmartFlareTM technique could provide a wide spectrum of research applications, as identifying RNAs into mammalian conceptuses at different developmental stages has already been used as a proper model[9]. Indeed, SmartFlareTM allows the detection of RNAs specific for hereditary diseases, sex determination, performance and con
To answer to some developmental issues related to the expression of the transcription factor Octamer-Binding Transcription Factor 4 (OCT4), involved in the differentiation process of human neurospheres in a time-dependent fashion[15,16], the mRNA pattern of OCT4 at single-cell level was analyzed from 3 to 30 d in vitro (DIV) using specific SmartFlareTM probes to assess a possible downregulation strictly linked to cellular maturation from stem/progenitor to neural phenotype. In parallel, a SmartFlareTM probe for Prominin 1 (CD133), encoding for a transmembrane gly
Our findings suggest that SmartFlareTM technology is a straightforward tool for discriminating gene transcripts specifically related to some neural stem cell markers.
Forebrain tissues were obtained from aborted human fetuses aged 8 wk postconception (Lund and Malmö University Hospitals) in accordance with guidelines approved by the Lund/Malmö Ethical Committee (ethical permit No. Dnr 6.1.8-2887/2017). Brain fragments from the striatum were subjected to microdissection under a stereomicroscope (Leica, Germany), incubated for 30 min in an expansion medium at 37 °C, and then mechanically dissociated in order to obtain a single-cell suspension. Expansion medium DMEM/F-12 (1:1; InVitrogen, Life Technologies, United States), 2.92 g/100 mL L-glutamine, 23.8 mg/100 mL HEPES, 7.5% NaHCO3, 0.6% glucose, and 2% heparin (all from Sigma-Aldrich, United States) contained B27 supplement (1%; InVitrogen), human Leukemia Inhibitory Factor (LIF; 10 ng/mL; Sigma-Aldrich), Epidermal Growth Factor (EGF; R&D Systems, United States), and Fibroblast Growth Factor (20 ng/mL and 10 ng/mL, respectively; R&D Systems, United States). Live cells were thereafter counted by the Trypan Blue dye exclusion method before plating in culture flasks at the fixed density of 50.000 cells/mL, at 37 °C in a humidified atmosphere with 5% CO2. After several weeks, neurospheres were developed and supplied by the Laboratory of Stem Cells and Restorative Neurology (Lund). To determine the capacity of cells to form secondary spheres, single neu
For in vitro differentiation, pelleted neurospheres were incubated with Accutase solution for gentle dissociation for 10 min at room temperature (RT), followed by DMEM/F-12 addition for halting the enzymatic activity. After centrifugation, single cells were resuspended in 500 μL basic medium (without growth factors and heparin) containing 1% fetal bovine serum (FBS; differentiation medium) and plated on poly-L-lysine-coated glass coverslips (5000-10000 cells/cm2)[16,19]. During the differentiation period (1-4 wk), the specific medium was refreshed every third day.
Live-cell mRNA detection was performed using SmartFlareTM probes, according to the manufacturer’s protocol (Merck Millipore, Temecula, CA, United States). Briefly, all the used probes were rehydrated by 50 μL of sterile nuclease-free double-distilled water to each vial and kept in the dark until needed. Immediately before the use, the stock solutions were diluted 1:20 in sterile phosphate-buffered saline. Four μL of the same solutions were added to 200 μL of the medium for each tested probe. For each experiment, performed in triplicate, two control samples were run in parallel: a negative one made of a scramble construct that, therefore, does not recognize any cellular sequence and used to quantify the unspecific background (Scramble SmartFlareTM Probe); a positive control (uptake SmartFlareTM Probe) that permanently emits fluorescence supplying the information that the SmartFlareTM particles are uptaken by the target cell type. The following reagents were used: CD133 Hu-Cy3 SmartFlareTM RNA Probe (SF-958), Oct4 Hu-Cy3 SmartFlareTM RNA Probe (SF-438), Actin-Cy3 SmartFlareTM RNA Probe (SF-145), Scramble-Cy3 SmartFlareTM RNA Probe (SF-103), and uptake-Cy3 SmartFlareTM RNA Probe (SF-114), all provided by Merck Millipore. All samples were incubated at 37 °C in a humidified atmosphere with 5% CO2 for 24 h, since in previous experiments the suggested 16 h incubation was evaluated not sufficient for the complete probe internalization. For nuclear staining, 10 μg/mL Hoechst 33342 (InVitrogen) was added 5 min before evaluation. Observations were made using an inverted microscope (IX 71; Olympus, Tokyo, Japan) with a x40 planapochromatic objective (PlanApo series; Olympus), taking care to grab all images with the same exposure time and filter set. Images (12-bit) were taken with a cooled monochrome CCD camera (Moticam Pro285D, Motic, China) with a 1360 × 1024 pixel chip. Image processing and analysis were performed using the Image-Pro Plus software (Media Cybernetics, United States).
SmartFlareTM CD133- and OCT4-specific fluorescence signal was assessed using a semiquantitative visual approach by three observers in a blinded fashion. This evaluation took into account both the fluorescence intensity and the number of labeled cells.
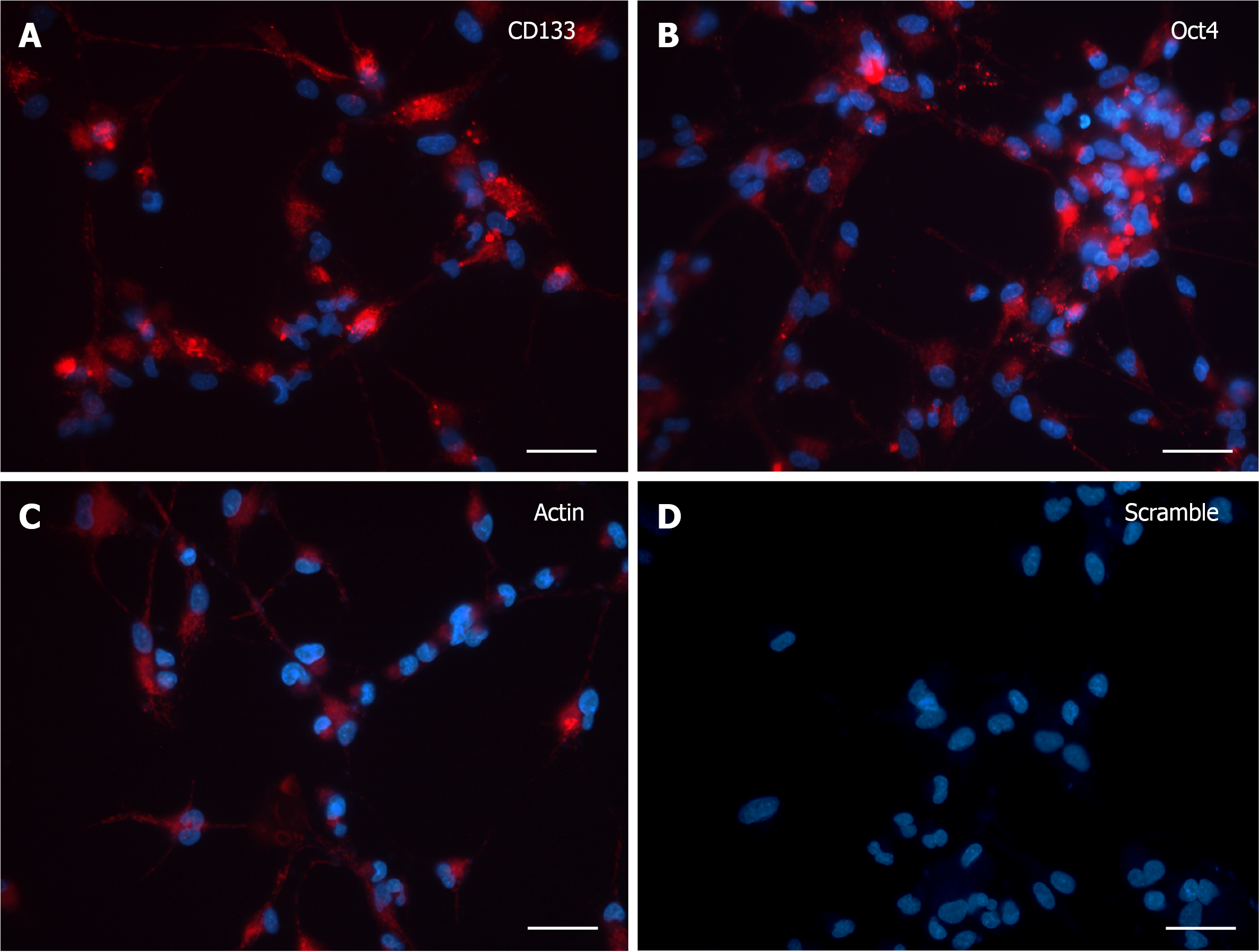
CD133 and OCT4 gene expression was analyzed by SmartFlareTM technology in dissociated human neurospheres upon differentiation commitment, accomplished by switching to growth factor withdrawn media along one-month time frame (from 3 to 30 DIV), with 3 DIV as the minimum time needed by cells both to adhere to the substrate and to grow cytoplasmic area and processes. At 3 DIV, after incubation with specific SmartFlareTM probes, the morphological expression pattern for CD133 and OCT4 mRNAs (Figure 1A and B) was consistent with the Actin-positive cells (Figure 1C). Remarkably, when Hoechst-stained cells were not massively clustered but discernible as single elements, it was possible to evaluate that all cells displayed a diffuse but strong fluorescent signal, sometimes visible as converging single dots filling the thin cytoplasmic processes too. Similarly, the fluorescence of the CD133 reporter probe was as intense as that of Oct4. The Actin housekeeping probe was clearly internalized as a fluorescent patch distributed from the perinuclear area to the peripheral branches, where it appeared as a granular content connecting distant cells (Figure 1C). Fluorescence detection in those living cells was considered a specific marker for mRNAs presence when compared to scramble experiments (Figure 1D), where any background was undetectable in most cells.
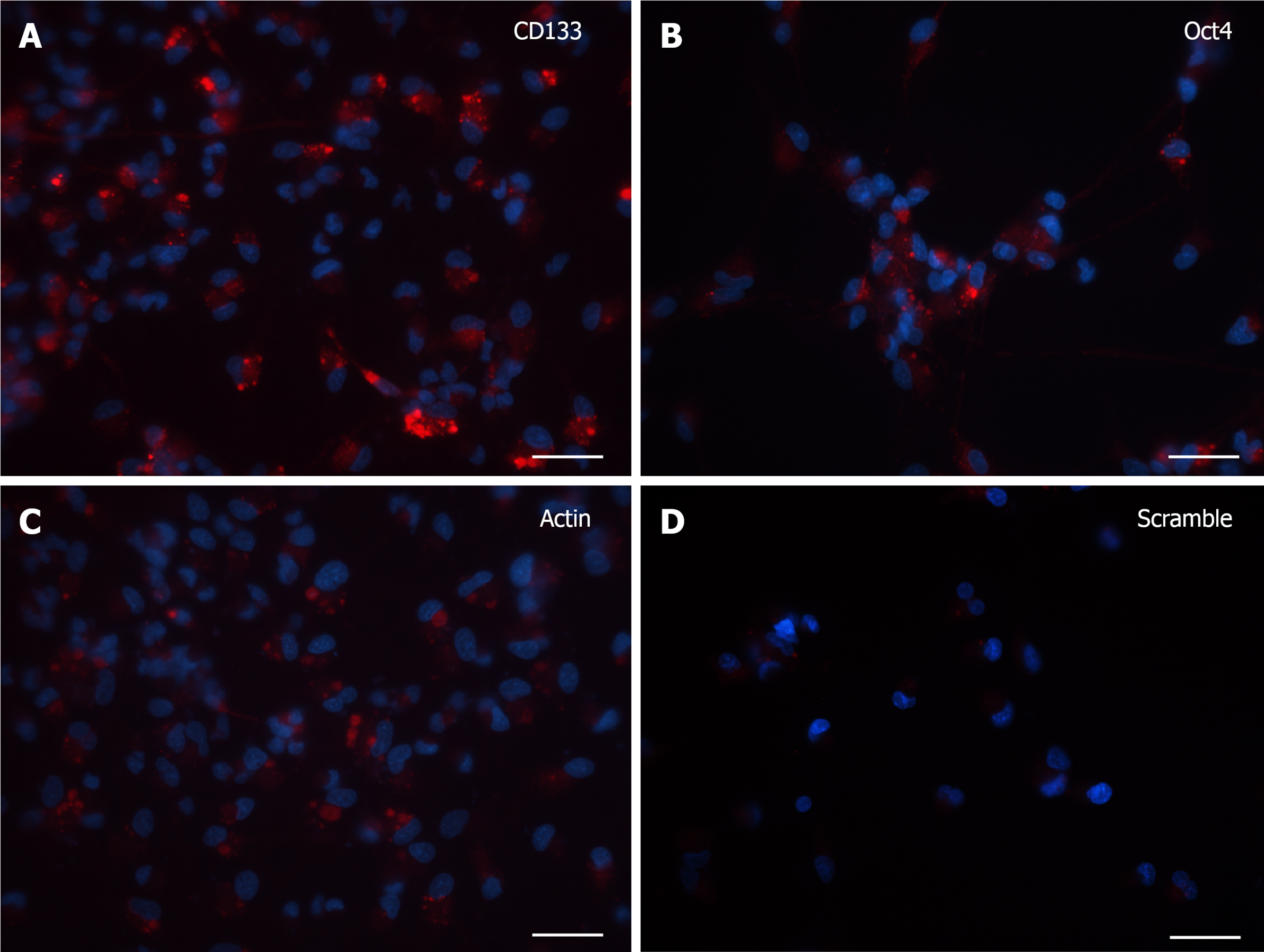
At 7 DIV, microscopic images exhibited a clear fluorescence both with CD133 and Oct4 probes (Figure 2A and B). Although the robust arborization network was still detected, in visible branches of very few cells it was observed the presence of fluorescent dots, representative of the molecular beacon-associated mRNAs. The reliability of the results was confirmed by the positive and negative controls (Figure 2C and D).
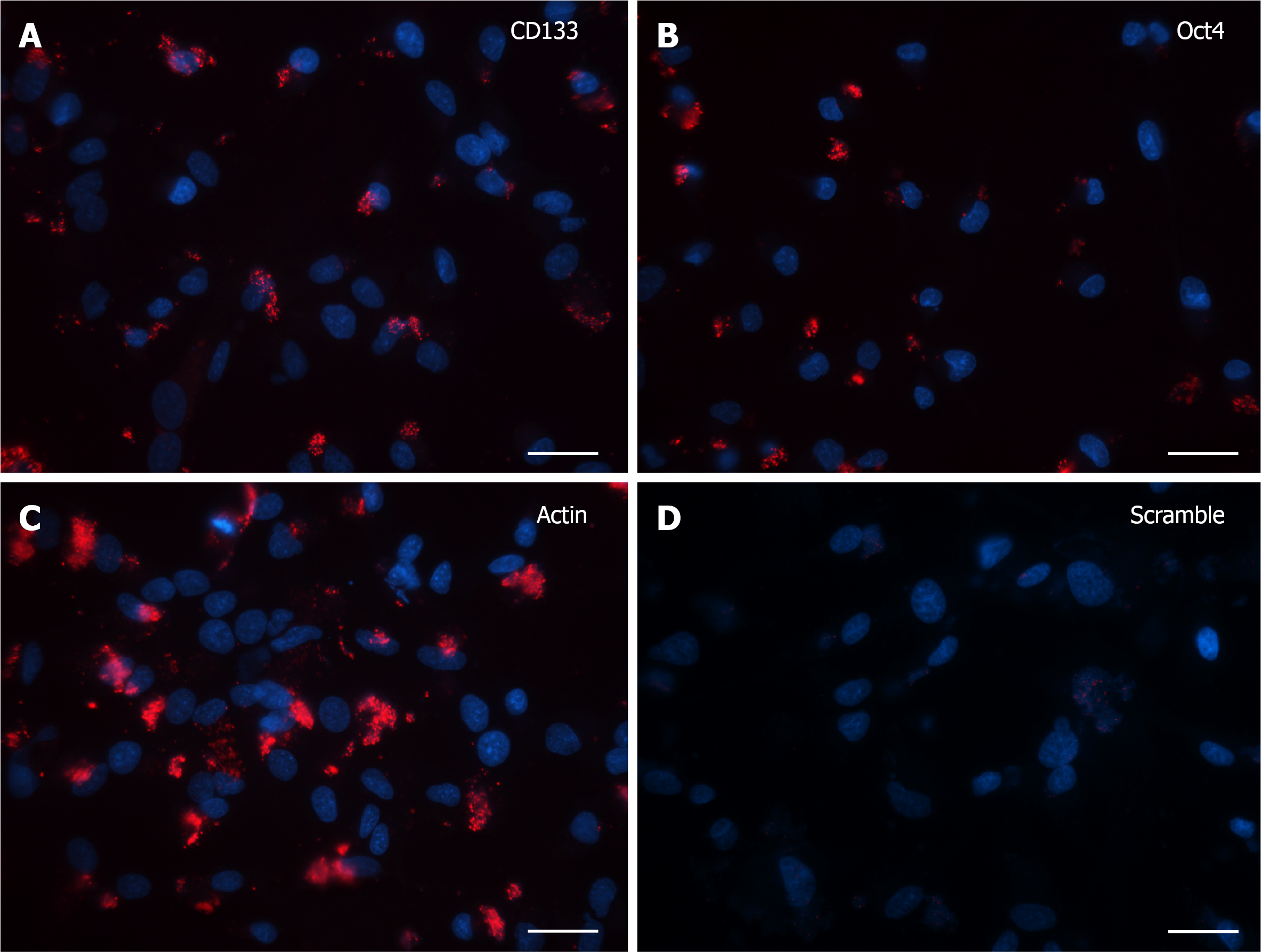
Cells grown for 15 DIV presented a marked decrease in the SmartFlareTM flu
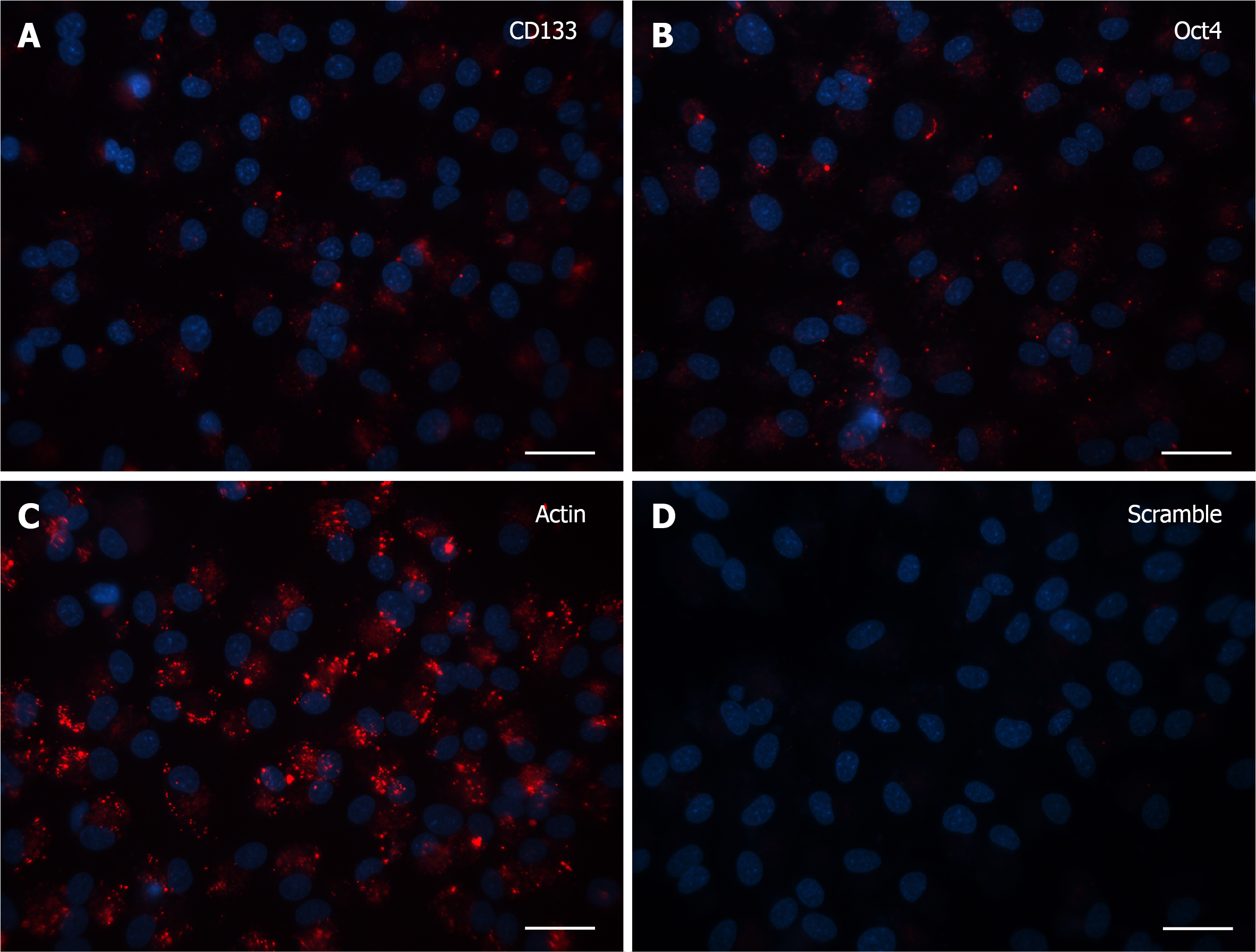
Finally, after 30 DIV, even fewer positive cells with specific signal were noticed and again the only morphological feature consisted of single dot-like elements, both in CD133 and Oct4 probe-treated cells (Figure 4A and B). Accordingly, in the last two experiments (15 and 30 DIV), Actin (Figure 3C and 4C, respectively) and Scramble (Figure 3D and 4D, respectively) signals were representative of the specificity of the resulting fluorescence.
In this study, we carried out a simple and noninvasive RNA-based approach to monitor intracellular gene expression in living cells by fluorescent SmartFlareTM probes. In detail, this study focused on human neurospheres as neural stem cell reservoir, as this is a well-established model to study the progression of differentiation events giving rise to both neuronal and glial lineages. This is a very interesting topic to address, since it involves OCT4, one of the key genes implicated in encoding transcription factors prone to convert somatic cells into iPSCs and, therefore, necessary for the commitment of embryological events[20]. The rationale behind the present investigation dates back to a previous study, where the immunohistochemical presence of Oct4 protein was observed in neural stem cells during the first week of differentiation but disappeared after 4 wk. Coherently, in the same research, RT-PCR experiments supported OCT4 mRNA downregulation, as illustrated by the blurred bands of the electrophoretic assay[16]. Therefore, the advantage of SmartFlareTM probe uptake has emerged for challenging the quantification of mRNA gradient in specific and individual cells. Moreover, the same technique could be useful for identifying neural stem/progenitor cells eventually sorted for further characterization, avoiding any minimal alteration of morpho-functional and biochemical properties. With regard to Oct4, there are some further but possibly misinterpreting studies describing cytoplasmic staining due to splicing variants that make it critical to distinguish transcriptional products[21-24]. For this reason, this study conceived the experimental design of choosing the cell surface antigen CD133 as an alternative positive marker of neural stem cells[25]. The localization of OCT4 mRNA within cells has already been addressed by some researchers[26] using molecular beacon transfection in differentiated human mesencephalic-derived neurospheres. However, after dissociation, adherent differentiated monolayers resulted lacking OCT4 expression. Interestingly, monolayered cells grown from neurospheres revealed the complete absence of mRNA expression just before the first week of differentiation, as further confirmed by immunocytochemistry. Indeed, the initial enthusiasm of the scientific community was damped by some studies reporting “A total lack of correlation between fluorescence intensities of SmartFlare probes and the level of corresponding RNAs assessed by RT-qPCR”[27]. Recent data might explain the resulting different amounts of mRNA detected by SmartFlareTM and qRT-PCR, due to cytoplasmic stress granules where mRNA can be sequestered and made unavailable to be processed for translation[28].
Mason et al[29] argued about the SmartFlareTM probes sequestration by the lysosomal machinery. However, by specific matching Lysosome-associated Membrane Protein 1 (LAMP-1) SmartFlareTM, these authors found a very low overlap (mean Manders’ coefficient 0.26), concluding that the unspecific SmartFlareTM fluorescence localized in lysosomes could be negligible compared to cytoplasmic staining. Our findings agree with the heterogeneity of SmartFlareTM expression, either diffuse cytoplasmic or spotted from the perinuclear site to peripheral processes (dendrites and axons). Moreover, ultrastructural evidence of gold nanoparticles, encapsulated within endosomal/lysosomal compartments, does not explain the spotted fluorescent pattern, unless enzyme digestion would degrade and remove the nanostructure links, ultimately quenching the fluorescence signal. So far, there is still no experimental evidence for that degradative machinery, and, on the other hand, it cannot be ruled out whether there are some alternative routes either passively or actively driven by cells.
By means of a qualitative analysis, the strength of the SmartFlareTM technology would not be affected by the decrease of the fluorescence intensity as a reflection of a reduced lysosomal activity, which occurs during cell differentiation[30]. Actually, as shown by our results, it is unlikely to detect all the cells in the same stage of replication or differentiation within single timepoints.
Although FISH is a well-established and reliable qualitative molecular method, the advantages of SmartFlareTM technology could reside in the opportunity of analyzing unfixed single living cells, retaining their viability, morpho-functional and biochemical properties and allowing downstream experiments[31]. In particular, this approach could help to detect and count stem/progenitor living cells, expressing markers of stemness, in terms of differential expression of the relative mRNAs, as well as microRNAs, which could find application in the profiling of tumor cell heterogeneity[32,33]. Moreover, from an empirical perspective, the SmartFlareTM could be a quicker, easier and less expensive method than techniques involving RNA isolation. Thus, in agreement with the findings by Mason et al[29], our results might validate the SmartFlareTM technology as a reliable and easy-to-handle tool, at least in the qualitative analysis framework, although, in some cases, as usually happens, the possibility of an artifact detection may arise.
In the prospect of controversial negative results, it should be considered that FBS supplementation in the culture medium could dramatically play a crucial role in the interpretation of target mRNA detection by SmartFlareTM technology, in terms of cytoplasmic distribution and localization. This methodological issue could partially explain the documentation failure by many research groups[19].
Despite the above-described unsolved criticism, some recent data on molecules and cells involved in immunological and inflammation response against cancer have renewed the interest in an innovative and effective platform to investigate some mRNA functions[34-36]. Besides, it cannot be denied that SmartFlareTM probe detection is not indicative of the real localization of single mRNA molecules. Nevertheless, those NanoFlare probes have paved the way to inspire a novel theranostic wave arising some new sticky-flares for in situ monitoring of human telomerase RNA[37], adopting photoactivation to detect mRNA in specific cells[38].
In conclusion, this new age of NanoFlare compounds has opened up or, at least, broadened biomedical applications, paying attention to preserving the physiological integrity of cellular systems with an excellent grade of selectivity and specificity[39].
Although mRNA analysis is still conventionally achieved by fluorescence in situ hybridization and qRT-PCR, there is a strong need for real-time monitoring of specific RNA transcripts in living cells, both for a qualitative and quantitative assessment. Within this context, SmartFlareTM technology is a reliable tool for evaluating the presence and the upregulation/downregulation of mRNAs in individual living cells. In addition, this nanotechnology offers the advantages of retaining cell viability, morpho-functional and biochemical properties and allowing downstream experiments.
SmartFlareTM technology is a devoted and straightforward method for the spati
To study the dynamics of differentiation-related RNA transcripts in human neural stem cells.
The presence of CD133 and OCT4 mRNA-linked nanoprobes in neurosphere-derived cells (from 3 to 30 DIV) was investigated by SmartFlareTM as a reliable insight into neural stem cell differentiation.
Until 7 DIV, all the cells displayed a strong SmartFlareTM fluorescent signal indicative of CD133 and OCT4 mRNA expression, as single dots encompassing both the cytoplasmic domain and the related processes. Upon 15 DIV, cells showed a marked decrease in the fluorescence, both for CD133 and Oct4 probes. In cells grown for 30 DIV, the CD133 and Oct4 probe uptake was very scant but still consisted of single dot-like elements, representative of a downregulation of the same genes.
Our findings propose the SmartFlareTM technology as a reliable and straightforward tool in the context of a qualitative expression analysis applied to a broad panel of mRNAs in single living stem cells.
The NanoFlare technology, such as SmartFlareTM, could enhance the scenario of biomedical applications in the field of marker identification mirroring both normal and pathological conditions, with the advantage of ensuring the physiological integrity of cellular systems.
The authors would like to thank Dr. Emanuela Monni (Laboratory of Stem Cells & Restorative Neurology, Lund Stem Cell Center, Lund University, Sweden), for kindly supplying the neurospheres from the human forebrain tissues.
| 1. | Kallijärvi J, Hämäläinen RH, Karlberg N, Sainio K, Lehesjoki AE. Tissue expression of the mulibrey nanism-associated Trim37 protein in embryonic and adult mouse tissues. Histochem Cell Biol. 2006;126:325-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 2. | Seferos DS, Giljohann DA, Hill HD, Prigodich AE, Mirkin CA. Nano-flares: probes for transfection and mRNA detection in living cells. J Am Chem Soc. 2007;129:15477-15479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 564] [Cited by in RCA: 568] [Article Influence: 29.9] [Reference Citation Analysis (0)] |
| 3. | Prigodich AE, Randeria PS, Briley WE, Kim NJ, Daniel WL, Giljohann DA, Mirkin CA. Multiplexed nanoflares: mRNA detection in live cells. Anal Chem. 2012;84:2062-2066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 233] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 4. | Briley WE, Bondy MH, Randeria PS, Dupper TJ, Mirkin CA. Quantification and real-time tracking of RNA in live cells using Sticky-flares. Proc Natl Acad Sci U S A. 2015;112:9591-9595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 98] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 5. | McClellan S, Slamecka J, Howze P, Thompson L, Finan M, Rocconi R, Owen L. mRNA detection in living cells: A next generation cancer stem cell identification technique. Methods. 2015;82:47-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 6. | Halo TL, McMahon KM, Angeloni NL, Xu Y, Wang W, Chinen AB, Malin D, Strekalova E, Cryns VL, Cheng C, Mirkin CA, Thaxton CS. NanoFlares for the detection, isolation, and culture of live tumor cells from human blood. Proc Natl Acad Sci U S A. 2014;111:17104-17109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 172] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 7. | Seftor EA, Seftor REB, Weldon D, Kirsammer GT, Margaryan NV, Gilgur A, Hendrix MJC. Melanoma tumor cell heterogeneity: a molecular approach to study subpopulations expressing the embryonic morphogen nodal. Semin Oncol. 2014;41:259-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 8. | Toptygina AP, Zakirov RS, Kapitanova KS, Semikina EL. Detection of Small Subsets of CD4+ Lymphocytes with SmartFlare Nanoprobes. Bull Exp Biol Med. 2019;168:270-274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 9. | Budik S, Tschulenk W, Kummer S, Walter I, Aurich C. Evaluation of SmartFlare probe applicability for verification of RNAs in early equine conceptuses, equine dermal fibroblast cells and trophoblastic vesicles. Reprod Fertil Dev. 2017;29:2157-2167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 10. | Choi YH, Gustafson-Seabury A, Velez IC, Hartman DL, Bliss S, Riera FL, Roldán JE, Chowdhary B, Hinrichs K. Viability of equine embryos after puncture of the capsule and biopsy for preimplantation genetic diagnosis. Reproduction. 2010;140:893-902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 11. | Beckelmann J, Budik S, Bartel C, Aurich C. Evaluation of Xist expression in preattachment equine embryos. Theriogenology. 2012;78:1429-1436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 12. | Beckelmann J, Budik S, Helmreich M, Palm F, Walter I, Aurich C. Sex-dependent insulin like growth factor-1 expression in preattachment equine embryos. Theriogenology. 2013;79:193-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Hamilton CK, Combe A, Caudle J, Ashkar FA, Macaulay AD, Blondin P, King WA. A novel approach to sexing bovine blastocysts using male-specific gene expression. Theriogenology. 2012;77:1587-1596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 14. | Lahm H, Doppler S, Dreßen M, Werner A, Adamczyk K, Schrambke D, Brade T, Laugwitz KL, Deutsch MA, Schiemann M, Lange R, Moretti A, Krane M. Live fluorescent RNA-based detection of pluripotency gene expression in embryonic and induced pluripotent stem cells of different species. Stem Cells. 2015;33:392-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 15. | Monni E, Congiu T, Massa D, Nat R, Diana A. Human neurospheres: From stained sections to three-dimensional assembly. Translat Neurosci. 2011;2:43-48. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 16. | Massa D, Pillai R, Monni E, Kokaia Z, Diana A. Expression analysis of pluripotency-associated genes in human fetal cortical and striatal neural stem cells during differentiation. Translat Neurosci. 2012;3:242-248. [RCA] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 17. | Barraud P, Stott S, Møllgård K, Parmar M, Björklund A. In vitro characterization of a human neural progenitor cell coexpressing SSEA4 and CD133. J Neurosci Res. 2007;85:250-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 67] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 18. | Peh GS, Lang RJ, Pera MF, Hawes SM. CD133 expression by neural progenitors derived from human embryonic stem cells and its use for their prospective isolation. Stem Cells Dev. 2009;18:269-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 55] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 19. | Golab K, Krzystyniak A, Langa P, Pikuła M, Kunovac S, Borek P, Trzonkowski P, Millis JM, Fung J, Witkowski P. Effect of serum on SmartFlare™ RNA Probes uptake and detection in cultured human cells. Biomed J Sci Tech Res. 2020;28:21788-21793. [PubMed] |
| 20. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14327] [Cited by in RCA: 14573] [Article Influence: 809.6] [Reference Citation Analysis (0)] |
| 21. | Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, Andrews PW. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells. 2008;26:3068-3074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 224] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 22. | Wang X, Dai J. Concise review: isoforms of OCT4 contribute to the confusing diversity in stem cell biology. Stem Cells. 2010;28:885-893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 166] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 23. | Lee SH, Jeyapalan JN, Appleby V, Mohamed Noor DA, Sottile V, Scotting PJ. Dynamic methylation and expression of Oct4 in early neural stem cells. J Anat. 2010;217:203-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 24. | Alexander RE, Cheng L, Grignon DJ, Idrees MT. Cytoplasmic OCT4 staining is a sensitive marker of neuroendocrine differentiation. Hum Pathol. 2014;45:27-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 25. | Mizrak D, Brittan M, Alison M. CD133: molecule of the moment. J Pathol. 2008;214:3-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 396] [Cited by in RCA: 406] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 26. | Ilieva M, Dufva M. SOX2 and OCT4 mRNA-expressing cells, detected by molecular beacons, localize to the center of neurospheres during differentiation. PLoS One. 2013;8:e73669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 27. | Czarnek M, Bereta J. SmartFlares fail to reflect their target transcripts levels. Sci Rep. 2017;7:11682. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 28. | Wolczyk M, Podszywalow-Bartnicka P, Bugajski L, Piwocka K. Stress granules assembly affects detection of mRNA in living cells by the NanoFlares; an important aspect of the technology. Biochim Biophys Acta Gen Subj. 2017;1861:1024-1035. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 29. | Mason D, Carolan G, Held M, Comenge J, Cowman S, Levy R. The spherical nucleic acids mRNA detection paradox. Science Open Res. 2015;. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 30. | Kobayashi T, Piao W, Takamura T, Kori H, Miyachi H, Kitano S, Iwamoto Y, Yamada M, Imayoshi I, Shioda S, Ballabio A, Kageyama R. Enhanced lysosomal degradation maintains the quiescent state of neural stem cells. Nat Commun. 2019;10:5446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 31. | Villani V, Thornton ME, Zook HN, Crook CJ, Grubbs BH, Orlando G, De Filippo R, Ku HT, Perin L. SOX9+/PTF1A+ Cells Define the Tip Progenitor Cells of the Human Fetal Pancreas of the Second Trimester. Stem Cells Transl Med. 2019;8:1249-1264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 32. | Diana A, Gaido G, Murtas D. MicroRNA Signature in Human Normal and Tumoral Neural Stem Cells. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 33. | Diana A, Gaido G, Maxia C, Murtas D. MicroRNAs at the Crossroad of the Dichotomic Pathway Cell Death vs. Stemness in Neural Somatic and Cancer Stem Cells: Implications and Therapeutic Strategies. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 34. | Zakiryanova GK, Kustova E, Urazalieva NT, Amirbekov A, Baimuchametov ET, Nakisbekov NN, Shurin MR. Alterations of oncogenes expression in NK cells in patients with cancer. Immun Inflamm Dis. 2017;5:493-502. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 35. | Zakiryanova GK, Kustova E, Urazalieva NT, Baimuchametov ET, Nakisbekov NN, Shurin MR. Abnormal Expression of c-Myc Oncogene in NK Cells in Patients with Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 36. | Banerjee H, Joyner J, Stevenson M, Kaha W, Krauss C, Hodges S, Santos E, Worthington M, Rousch J, Payne G, Manglik V, Banerjee N, Morris B, Bell D, Mandal S. Short Communication: Studying the Role of Smart Flare Gold Nano Particles in Studying Micro RNA and Oncogene Differential Expression in Prostate Cancer Cell Lines. J Can Res Updates. 2017;6:25-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 37. | Wu Q, Liu Z, Su L, Han G, Liu R, Zhao J, Zhao T, Jiang C, Zhang Z. Sticky-flares for in situ monitoring of human telomerase RNA in living cells. Nanoscale. 2018;10:9386-9392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 38. | Lin M, Yi X, Huang F, Ma X, Zuo X, Xia F. Photoactivated Nanoflares for mRNA Detection in Single Living Cells. Anal Chem. 2019;91:2021-2027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 39. | Chenab KK, Eivazzadeh-Keihan R, Maleki A, Pashazadeh-Panahi P, Hamblin MR, Mokhtarzadeh A. Biomedical applications of nanoflares: Targeted intracellular fluorescence probes. Nanomedicine. 2019;17:342-358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Neurosciences
Country/Territory of origin: Italy
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Liao Z S-Editor: Wu YXJ L-Editor: A P-Editor: Wu YXJ