Published online Jul 26, 2020. doi: 10.4252/wjsc.v12.i7.527
Peer-review started: February 26, 2020
First decision: April 26, 2020
Revised: May 9, 2020
Accepted: May 28, 2020
Article in press: May 28, 2020
Published online: July 26, 2020
Processing time: 150 Days and 20.5 Hours
Induced pluripotent stem (iPS) cells present a seminal discovery in cell biology and promise to support innovative treatments of so far incurable diseases. To translate iPS technology into clinical trials, the safety and stability of these reprogrammed cells needs to be shown. In recent years, different non-viral transposon systems have been developed for the induction of cellular pluripotency, and for the directed differentiation into desired cell types. In this review, we summarize the current state of the art of different transposon systems in iPS-based cell therapies.
Core tip: The seminal discovery of induced pluripotent stem (iPS) cells has opened up the possibility of converting most somatic cell types into a pluripotent state. The iPS cells possess most of the advantages of embryonic stem cells without the ethical stigma associated with derivation of the latter. This procedure has had a large impact on the generation of custom-made pluripotent cells, ideal for cell-type specific differentiation and regenerative medicine with or without genetic correction. In this review, we focus on updated information of transposon system-mediated cellular reprogramming to iPS cells and their application in cellular therapy.
- Citation: Kumar D, Anand T, Talluri TR, Kues WA. Potential of transposon-mediated cellular reprogramming towards cell-based therapies. World J Stem Cells 2020; 12(7): 527-544
- URL: https://www.wjgnet.com/1948-0210/full/v12/i7/527.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v12.i7.527
Transposon systems currently provide a promising toolbox for cell therapy, disease modeling, and drug discovery[1-4]. Importantly, the non-viral transposon systems can be an important alternative to viral vectors, which are commonly used for cellular reprogramming for transfection of somatic cells with exogenous Oct4, Sox2, Klf4, and c-Myc genes to induce cellular pluripotency and establish induced pluripotent stem (iPS) cells[5-8]. However, the limited cargo size of retro and lenti viral vectors of about 7 kb pairs hampers transfer of larger therapeutic genes[9]. In addition, the construction of viral vectors is cumbersome, expensive and requires living cells for their scale up, which further complicates the quality control and downstream processing[10].
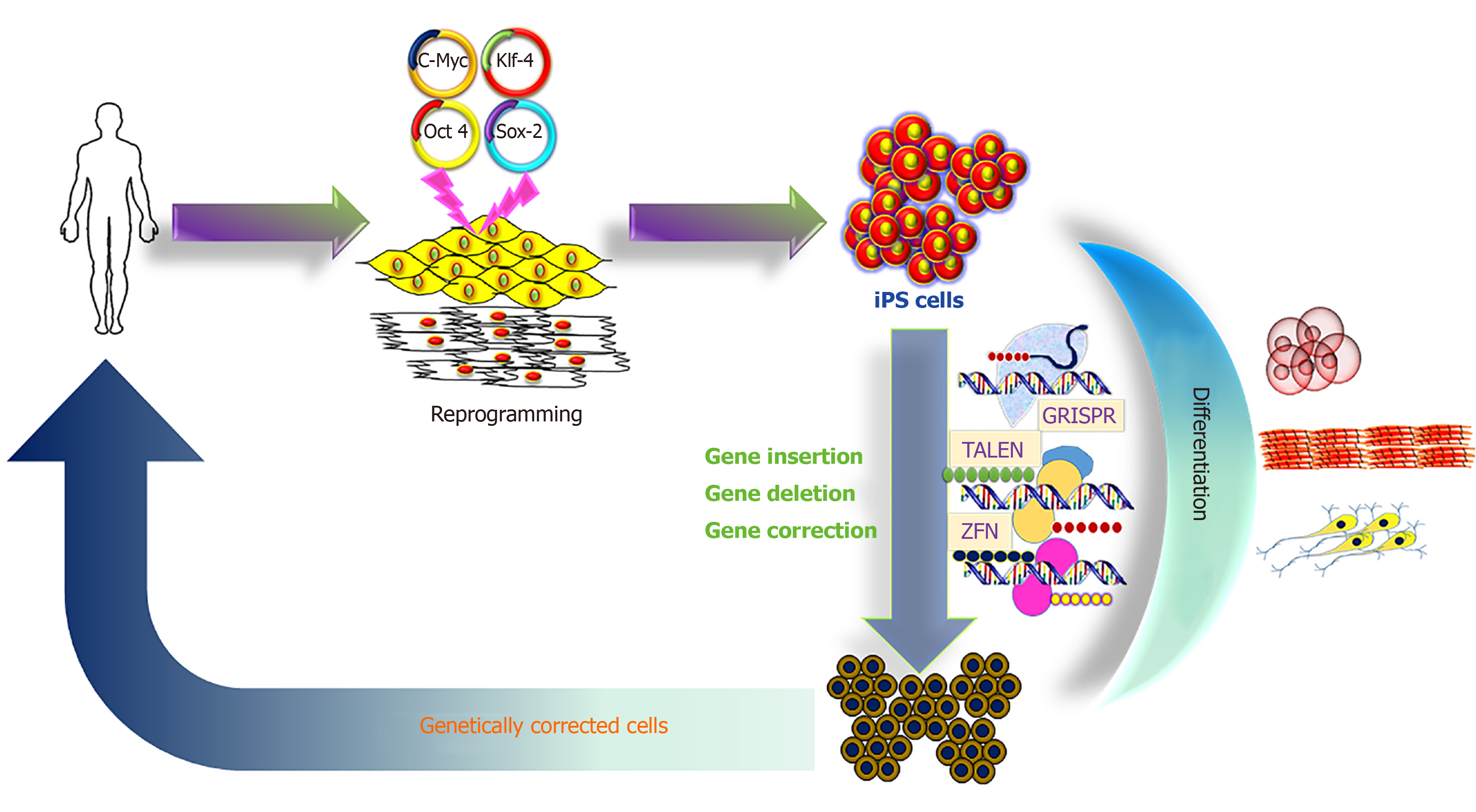
The iPS cell technology promises to provide an unlimited source of cells for innovative therapies, and to treat so far incurable diseases[11-13]. A hypothetical schedule would require a small tissue sample from the patient, to reprogram the somatic cells to iPS cells with unlimited proliferative capacity, to perform gene correction in the iPS cells, then to direct differentiation into the desired precursor cells, which are finally transplanted into the patient (Figure 1).
In this respect, Sleeping Beauty (SB) and piggyBac (PB) transposon systems appear as attractive tools for somatic cell reprogramming due to their efficient gene delivery and their ability to be excised from the cells after reprogramming, which helps overcome the limitations of viral-based reprogramming technologies. Transposon systems have a number of additional advantages, such as (1) Cargo capacity of up to 100 kb[14,15]; (2) No bias to integrate in expressed genes or promoter regions; (3) Possibility of seamless removal of the transposon[16,17]; (4) Cost-effective production of the basic plasmids; (5) Reduced innate immunogenicity; and (6) No requirement for a specialized biosafety facility.
The translation of this iPS cell-based therapy into clinical testing needs authorization approval to initiate safety and efficacy studies, and to exclude risks of insertional oncogenesis or immunogenicity[18,19]. SB and PB transposon systems have been successfully used to obtain reprogrammed iPS cells from human somatic cells[16,20], but also somatic cells from the murine model[21-24], and cells from large model species, such as pig[25], horse[26], bat[27], monkey[28], rat[29], cattle[30,31] and buffalo[32]. Here, we review the potential of transposon-mediated cellular reprogramming and its clinical applications in cell-based therapy and the associated risks.
DNA transposons, also known as Class II elements or mobile genetic elements, were first described as “jumping genes” by McClintock[33] and were found to be responsible for color mosaicism of maize cob kernels. DNA transposons have been divided into two major groups: (1) Cut-and-paste; and (2) Rolling-circle transposons[34]. In vertebrates, commonly cut-and-paste group of transposons are found, which include the Tc1/mariner, hATs, PB and SB families, all of which are characterized by inverted terminal repeats of 10 to 1000 bp flanking their transposase gene[35]. Transposons are discrete DNA segments which can move from one site to another within a genome, and sometimes between genomes catalyzed by the transposase[36,37]. Transposons are species-specific, found in the genomes of all prokaryotes and eukaryotes, whereas in humans approximately 46% of the genome is derived from retro- (RNA) and DNA transposons[38,39].
Transposons are important sources of genome structures that are actively used to regulate the multicellular embryonic development. These structures include binding sites with transcription factors, enhancers and silencers, promoters, insulators, alternative splicing sites, and non-coding RNA. Moreover, transposons are involved in the emergence and evolution of new protein-coding genes through exonization, domestication, and the formation of retrogenes. The activation of transposons is needed to regulate the differentiation and reproduction of cells in the body; however, in terminally differentiated cells, upon reaching predetermined sizes of organs, molecular systems are activated that block a further cascade of transposon activation[40,41]. Due to the wide distribution and diversity of transposons, they contribute significantly to genomic variation and as such, they are powerful drivers of genome evolution[36,42-45].
For this purpose, SB and PB transposon systems are identified as efficient vectors for cellular reprogramming. The SB originated from salmonid fish species, where it existed as an inactive element[46]; from this a synthetic transposon system was constructed using a reverse engineering approach to eliminate the accumulated mutations[46]. PB was derived from an active element discovered in the moth Trichoplusia ni[47]. These transposons have no orthologous elements in mammalian species, which prevents the re-mobilization of transposons by potential endogenous transposases. This has been experimentally verified in transgenic mice and pigs[48,49]. Presently, the hyperactive versions of SB (SB100X) or PB (hypPB) seem to be the most active transposon systems. They possess comparable activity levels in mammalian cells, and are independent of cellular co-factors[50,51]. Both of these transposons have been employed for stable expression of reprogramming factors and are suitable for the derivation of iPS cells as proven in various studies[16,22,23,25,26,30-32,52]. Other transposons namely: Frog Prince, Mos1, Tol2 and Passport are also active in mammalian cells, but they are still under-investigated in iPS cell generation[53].
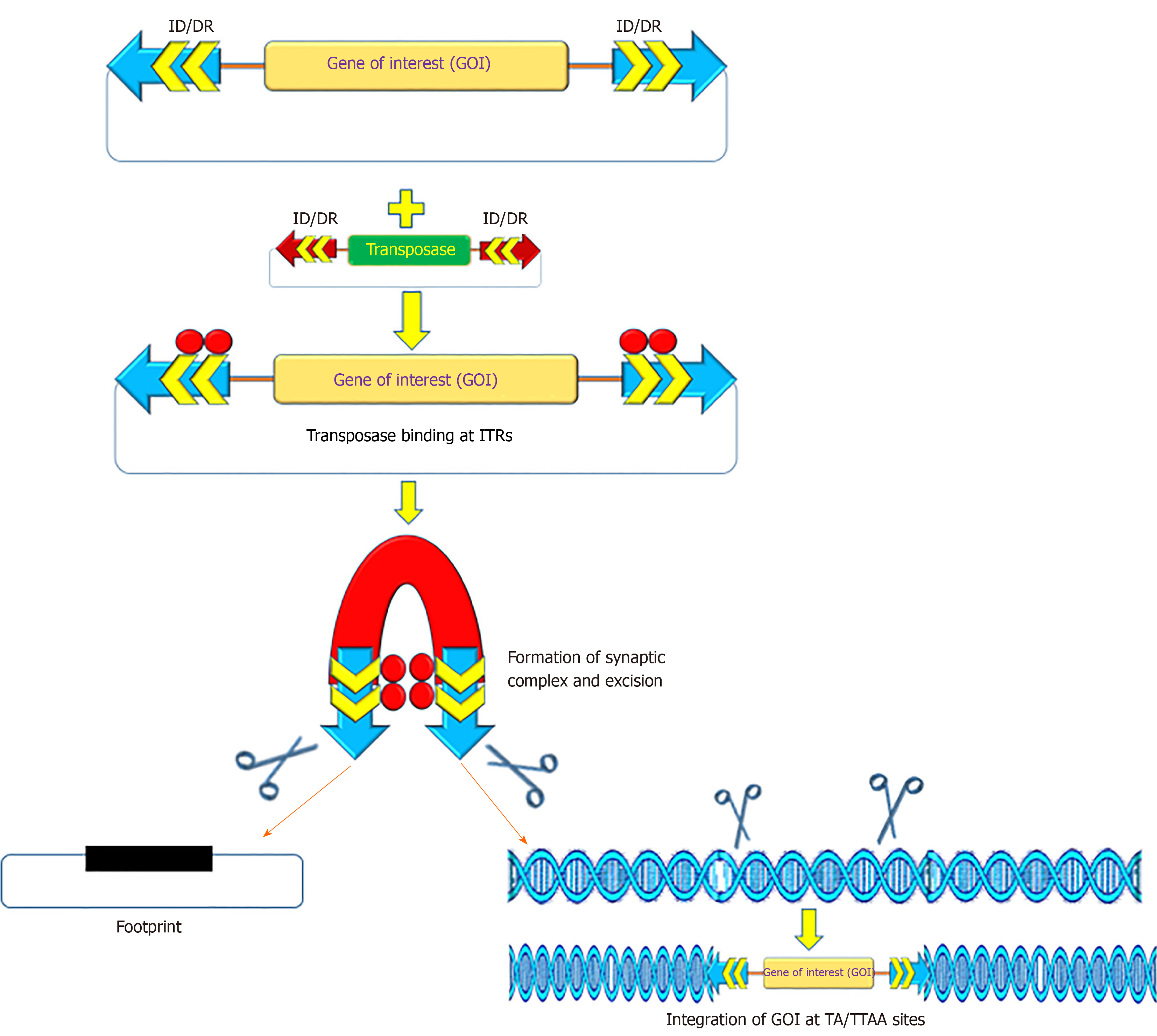
The recombinant PB and SB systems mobilize or transfer gene(s) of interest through a “cut and paste” mechanism (Figure 2)[2,54,55]. For most applications, recombinant transposon systems encompass a donor plasmid that carries one or more genes flanked by the inverted terminal repeats (ITRs) sequences essential for transposition[2,56,57]. The transposase gene can be positioned on a separate plasmid (trans) or in the same plasmid (cis). Once the transposase protein is expressed, it binds to the ITR sequences, which catalyzes the removal of the gene of interest (cut) and integrates (paste) the transposon sequence into the genome of a host cell[57]. The SB transposase catalyzes integrations at consensus TA-dinucleotides[46], whereas the PB requires TTAA-tetranucleotide sequences[58-60]. The efficiency of transposition of these transposon systems has been further increased due to generation of highly active and efficient transposases, namely hyp(er) PB (hypPB) and hyperactive SB 100X (hySB100X)[50,51,61,62]. The hySB100X showed a 30% higher transposition rate compared with SB100X. hySB100X was obtained by mutation in short hydrophilic residues in the catalytic domain of the SB100X transposase molecule, which required direct DNA contact to increase the DNA binding affinity of the transposon[62]. Furthermore, the transposition rate of these transposons is affected by topological conformations, chromatin condensation and CpG-methylation patterns of the target DNA[63,64]. Genomic insertion for SB100X prefers target regions with higher AT content, in a palindromic core unit[65,66]; whereas PB transposase integration requires a TTAA recognition sequence and exhibits a bias toward insertions in genes[67].
For cellular reprogramming, the transfection of the transcription factors into somatic cells using the transposon system is relatively straightforward. The transposons-mediated cellular reprogramming leads to an overall efficiency of approximately 0.02%[20,22,23,30], which nears the initially obtained reprogramming efficiencies by viral vectors. The obtained reprogramming efficiency from transposons is higher than other reported non-integrative delivery systems including either replicating episomal vectors or minicircles[68,69], although lower than Sendai viral vectors or synthetic mRNA[70,71]. Transposons-mediated transposition is a self-regulated activity via overproduction inhibition, a mechanism by which transposition activity is down-regulated when the transposase is over concentrated in cells[72]. Ideally, the transposase is expressed only for a short period, which prevents continuous transposon re-mobilization. However, it is also important to minimize the number of vector copies per cell as it poses an increased risk of insertional oncogenesis[73].
Transposon systems are widely used for gene delivery applications[58,74-76]. However, like the lenti viruses, transposon vectors are mutagenic, because of their random integration. Recently, clustered regularly interspaced short palindromic repeats (CRISPR) and Cas9 nucleases have emerged as excellent tools for site-specific mutation of genomes[77]. This system is an attractive candidate for targeting through extensive base pairing with the target[78]. In contrast, most DNA binding proteins remain bound to their target sites only for a matter of seconds or minutes. However, double-stranded breaks induced by CRISPR-Cas9 nucleases showed undesirable outcomes in terms of large deletions extending over many kilobases at high frequency and complex genomic rearrangements[79]. To overcome the challenges of nuclease-based gene delivery, various research groups have attempted to use site-specific DNA binding proteins such as SB, PB, Mos1, and ISY100-fused with zinc finger protein, transcription activator like effector (TALE) and/or Gal4 to target specific loci[80-82]. Owens et al[83] fused a TALE DNA-binding domain (DBD) with PB to direct the transposase to stimulate insertional activity of PB at the intended target sequence. This approach allowed the isolation of clones harboring single-copy insertions at the CCR5 locus. Subsequently, attempts were made using catalytically dead Cas9 (dCas9) for targeting PB insertions to the human endogenous hypoxanthine phosphoribosyl transferase (HPRT) locus[82]. Surprisingly, the dCas9-PB chimera protected it from insertions instead of targeting the HPRT locus. Although, PB is considered to be the most efficient system for gene delivery in vivo[84,85], it impedes the development of advanced applications such as direct delivery of transposons[86]. To resolve this difficulty, Chen and Wang described a Cas-Transposon (CasTn) system for genomic insertions which uses a Himar1 transposase fused with a dCas9 nuclease to mediate programmable, site-directed transposition[87]. They demonstrated that the Himar–dCas9 fusion protein improved the frequency of transposon insertion at a single targeted TA dinucleotide by > 300-fold compared to the un-fused transposase. This work highlights CasTn as a new modality for host-independent, programmable and site-directed DNA insertions[87].
More recently, Hew et al[88] tested a group of RNA-guided transposase vectors comprising mutations in the native PB DBD for their ability to target a single sequence in the CCR5 gene. This RNA-guided transposition in human cells might be a framework for improved targeting vectors with potential applications in gene therapy and genome editing research[88]. Similarly, Stecker et al[89] found that the CRISPR-associated transposase derived from Scytonema hofmanni (ShCAST), catalyzes the site-specific RNA-directed unidirectional integration and is located a fixed distance to one side of the targeted DNA site. These sequence-specific integrations offer significant advantages over traditional virus-based integrating vectors by avoiding insertion into unwanted regions[90-93]. Another approach applied to generate “transient transgenesis” by mutation at position 248 in the SB transposase to gain further insight into the transposition mechanism and for the generation of reprogramming factor-free iPS cells[17]. The amino acid present at position 248 of the SB transposase is involved in an interaction with target DNA, and because of the absence of integration activity, transposon removal by these transposase mutants results in extra-chromosomal circles, thereby terminating the transposition reaction[17,94]. This indicates that by the switching of a single amino acid, the SB transposase has into efficient unidirectional removal ability with utility in cellular reprogramming. In addition, soluble variants of the SB protein have been developed by genetic engineering, which allows for more control over the exposure time[95]. These underlying genome engineering procedures will reduce costs and improve the safety of genome modifications.
Commonly, somatic cells were reprogrammed to pluripotency by the exogenous introduction of transcription factors (Oct3/4, Sox2, Klf4 and c-Myc). The resulting iPS cells demonstrate the features of embryonic stem (ES) cells, including the ability to form chimeras and contribute to the germ line[5]. Thereafter, iPS cells were generated either by the protein transduction approach[96], or in combination with small chemical molecules[97] without genetic modification. These reprogramming approaches suffer from low efficiency and require complicated and prolonged cell culture conditions[96,97]. Furthermore, these approaches need either extraction of crude cell lysates of cells expressing defined reprogramming factors or preparation of a large amount of recombinant reprogramming transcription factors from bacteria, which may be contaminated with unknown detrimental genetic materials. Thus, the use of a suitable gene-delivery reprogramming approach is a critical step in the generation of iPS cells for basic and clinical research.
More recently, DNA transposons appeared as alternative tools for cellular reprogramming in a wide range of cell types, including fibroblasts using cocktails of transcription factors. This technique is straightforward, less time consuming and easy to handle as compared to viral vectors (Figure 3). In general, PB and SB systems have been used for iPS cells generation in a broad range of domesticated and farm animal species[16,20,22,23,25,30,32,98-101], in addition to human cells[102-105]. The generation of iPS cells from domesticated and companion animal species such as cattle, pig, horse and buffalo is critically important for the establishment of disease models and economically valuable for the production of medically useful substances, e.g., enzymes and growth hormones, which are either absent or inadequate in patients suffering from specific genetic diseases. More importantly, either iPS cells or differentiated cells from iPS cells could be directly used for cellular therapies, drug screening, and disease modeling thus significantly decreasing the extent to which animals are used for research purposes[4,106-110].
In this direction, cellular reprogramming through transposon systems represents one of the unique features of the excision of gene expression cassettes from the iPS cell genome through re-expression of integration-deficient transposase variants. Alternatively, excision can be achieved by either clustered regularly interspaced short palindromic repeats/CRISPR-associated protein-9 (CRISPR/Cas9) or Cre/loxP recombination technology[22,94]. Using these technologies enable the production of “transgene-free” iPS cells, which could be beneficial in minimizing the risk of reactivation of reprogramming factors leading to oncogenic potential[94]. Similarly, Woltjen et al[111] showed that PB-mediated transgene excision does not leave a genetic trace in the host genome, thus providing the feasibility of seamless modification for “genetically unmodified iPS cells” production.
Currently iPS cells are considered a valuable resource for studying medicine and regenerative biology due to their tremendous differentiation capacity into almost all cell types of the body. In principle, the differentiated cells derived from iPS cells should behave in the same way as their in vivo counterparts in terms of both molecular and functional aspects, but it remains a challenge to direct cell fate decisions under in vitro conditions towards specific cell types[112]. In general, differentiation comprises the conversion of an iPS cell to a more specialized cell type, involving a transition from proliferation to specialization. This involves successive alterations in cell morphology, membrane potential, metabolic activity and responsiveness to specific signals. Differentiation leads to acquiring specific functions of differentiated cells depending upon the tissue in which they will finally reside[113].
The transposon-mediated iPS cells can be differentiated in vitro in the absence of appropriate growth factor (LIF/bFGF) or feeder cells. Under the appropriate conditions, such as suspension culture, embryoid bodies (EBs) can be formed from iPS cells of almost all species, such as human[20], mouse[21,23], bat[27], monkey[28], prairie vole[114], horse[26], bovine[31], rat[29] and buffalo[32] with expression of lineage specific for endoderm, mesoderm and ectoderm (Table 1). Pluripotency is one of the defining features of iPS cells. Perhaps the most definitive test of pluripotency is the blastocyst complementation assay. The contribution of iPS cells to the resulting chimeras has been assessed to determine the differentiation capacity and germline contribution. True pluripotent murine iPS cells were generated using PB[115] and SB[21]. To the best of our knowledge, there is no report on the successful transposon-derived iPS cell-mediated germline contribution in large domestic animals.
| Species | Cell type | Transposon system | Reprogramming factors | Differentiation | Characterization of lineage specific differentiated cells through | Chimera | Germline contribution | Ref. | ||
| In vitro | In vivo | Histology | Expression of gene/protein | |||||||
| Bat | Fetal fibroblasts | PB | Human OKSMNL + NR5A2, and bat-specific miR302/367 | EBs | Teratoma | Yes | Yes | NA | NA | Mo et al[27], 2014 |
| Buffalo | Fetal fibroblasts | PB | Human OSKMNL | EBs | NA | NA | Yes (ectoderm- NF-68 and Cytokeratin 8; mesoderm- MSX1 and endoderm- GATA4) | NA | NA | Kumar et al[32], 2019 |
| Cattle | Fetal fibroblasts | PB SB | Human OSKMNL Murine OSKM | EBs | Teratoma | Yes | Yes (Ectoderm- β III-Tubulin, Nestin; Mesoderm- Vimentin and Endoderm- AFP, GATA, PAX6 | NA | NA | Talluri et al[30], 2015 |
| Fetal fibroblasts | PB | Bovine OSKM | EBs | Teratoma | Yes | Yes (Ectoderm- β III-Tubulin; Mesoderm-α-SMA and endoderm- AFP) | NA | NA | Zhao et al[31], 2017 | |
| Horse | Fetal fibroblasts | PB | Murine OSKM | EBs | Teratoma | Yes | NA | NA | NA | Nagy et al[26], 2011 |
| Human | Skin fibroblasts | PB | OSKML | EBs, Keratinocyte | Teratoma | NA | Yes (ectoderm- K14; mesoderm- desmin and endoderm- AFP) | Ethically not allowed | Ethically not allowed | Igawa et al[116], 2014 |
| Fetal fibroblasts | SB | Murine OSKM | EBs | NA | NA | Yes (Ectoderm- β III-Tubulin, Nestin; Mesoderm- Vimentin and Endoderm- AFP | Ethically not allowed | Ethically not allowed | Davis et al[20], 2013 | |
| Monkey | Skin fibroblasts | PB | Monkey OSKMNL | EBs | Teratoma | Yes | Yes (Ectoderm- β III-Tubulin; Mesoderm- SMA and Endoderm- AFP | NA | NA | Debowski et al[28], 2015 |
| Mouse | Fetal fibroblasts | PB | Murine OSKM | EBs | Teratoma | Yes | NA | Yes | Yes | Yusa et al[115], 2009 |
| Fetal fibroblasts | SB | Murine OSKM | EBs | Teratoma | NA | Yes (cardiac-cTnT and desmin and neuronal- nestin and Tuj1) | Yes | Yes | Muenthaisong et al[21], 2012 | |
| Pig | Fetal fibroblasts | SB | Murine OSKM | Neuronal lineage | Teratoma | Yes | Yes | NA | NA | Kues et al[25], 2013 |
| Prairie vole | Fetal fibroblasts | PB | Murine OSKMNL | EBs | Teratoma | Yes | NA | NA | NA | Katayama et al[114], 2016 |
| Rat | Fetal fibroblasts | PB | OSKM | EBs, chondrocyte | NA | NA | NA | NA | NA | Ye et al[29], 2015 |
The iPS cells may be directed into the lineage of interest by supplementing various growth factors into the culture media. These growth factors or stimulating agents allow directed differentiation of iPS cells towards a particular cell lineage or cell type. The differentiated cells can be identified with the help of various markers, which are highly expressed in these cells. Very few markers are specific for one cell type, and as such, a panel of markers needs to be used in order to characterize the differentiation status. In this direction, EBs derived from SB-mediated mouse iPS cells were differentiated into cardiac cells with a beat frequency[21,23]. Davis et al[20] observed that SB-mediated human iPS cells differentiated into EBs which contained hemoglobinized erythroid cells as well as spontaneously contracting cells, indicating that iPS cells could be differentiated into hematopoietic cell types and cardiomyocytes.
EBs generated from PB-mediated rat iPS cells showed numerous Alcian blue-stained regions, indicating the presence of acidic proteoglycans[29]. These acidic proteoglycans were suggestive of cartilaginous tissue, which was further confirmed by the production of collagen II. Transgene-free human iPS cells derived from PB reprogramming were successfully differentiated into epidermal keratinocytes, which were found to be similar in morphological, functional, and molecular analysis of single-cell gene expression to normal human keratinocytes[116]. The protocol for differentiation of human iPS cells into keratinocytes employed either retinoic acid or bone morphogenetic protein 4 (BMP4)[117]. Igawa et al[116] used a modified protocol in which neither BMP4 nor retinoic acid were used. Around 5 weeks of initiation of differentiation, they reported obtaining keratinocyte-like cells. These cells were propagated through successive passaging at least five times in serum-free keratinocyte medium without feeder cells. Upon characterization, these cells were positive for K5/K14, suggesting successful differentiation of keratinocytes from human iPS cells, and they called these cells induced keratinocytes[116]. These results indicate that iPS cell lines could be selected for therapeutic purposes.
Our group presented a novel approach for the differentiation of murine iPS cells derived through PB-mediated reprogramming into lentoid bodies[118]. We established a co-culture system using human NTERA-2, a committed neuronal precursor cell line[119] and P19, a murine embryonic carcinoma cell line[120] to provide a suitable niche for differentiation of the iPSs into the ectodermal lineage. The developing lentoid bodies were identified by a lens lineage-specific reporter, but also showed changed light refraction in the bright-field view. The existing data support the notion that the specific cell type reporter approach is instrumental for the optimization, development and validation of differentiation protocols for murine iPS cells. We speculate that the gained knowledge can be translated to optimize the differentiation of lens cells from human iPS cells and thus to advance the progress of patient-specific lentoid bodies as a pipeline for in vitro drug testing. It is likely that the specific cell type reporter approach is also adaptable for in vitro tracking of other cell lineages.
Cell-based therapy aims to treat diseases which cannot be addressed adequately by existing pharmaceutical interventions. The technology utilizes the cells with the ability to differentiate into specific lineages that are subsequently administered to a patient for therapeutic treatment. For this purpose, stem cells are considered ideal to restore tissue repair, or to replenish cells in the background of a genetic disease. The iPS cells can be expanded indefinitely and they are capable of differentiating in all the derivatives of the three germ layers. The generation of iPS cells is without the ethical stigma associated with ES cells, and iPS cells are able to result in personalized stem cells created from patient-specific cells. Although viral vectors are one of the most used methods for cellular reprogramming, their inherent limitations do not favor their clinical application due to hurdles in large-scale vector production and require careful biosafety characterization, which majorly impacts the costs of clinical-grade production of reprogrammed cells.
In recent years, non-viral DNA transposon based-systems have emerged as a potential tool to overcome some of the above-mentioned limitations. In transposon-mediated genetic manipulation, gene(s) of interest such as therapeutic gene rendering stable phenotypic correction, can be introduced and the resulting stem cells can be expanded in vitro and then subjected to differentiation into particular cell lineages according to the therapeutic need. The iPS cells generated through transposon-mediated cellular reprogramming are capable of differentiation into EBs in vitro and readily form teratomas in vivo. Teratoma formation confirmed that the reprogrammed iPS cells had the developmental potential to produce tissues of all three primary germ layers, i.e., ectoderm, mesoderm and endoderm[23,27,28,30,31]. However, the gold standard of the iPS cells pluripotency is determined by their ability to form germline-competent chimeras. Woltjen et al[16] demonstrated the formation of murine chimeras from transposon-reprogrammed iPS cells. However, most of the currently used transposon-mediated iPS cell lines carry constructs driven by a strong promoter, which constitutively promotes the reprogramming factors that will prevent the contribution to a normal ontogenesis[25,26,30]. Thus, the transposon-mediated iPS cell lines in several species have not yet been tested for their capability to generate chimera and mediate germline transmission. The recent progress achieved in the area of integration-deficient, but excision-competent transposase variants[61] will further simplify the transposon removal after complete reprogramming and the achievement of autonomous stemness.
Several advantages of transposon systems have encouraged investigators to carry out a clinical trial for the treatment of B-cell malignancies using SB-modified T-cell therapy[121]. The results published in 2016 showed that the use of SB-modified chimeric antigen receptor (CAR) T-cells is safe when infused after allogeneic or autologous hematopoietic stem cell transplantation as an adjuvant therapy. Modified cells survived for an average of 51 or 201 d in the allogeneic or autologous setting, respectively, and patients showed progression-free survival rates that were improved when compared to historical data[122]. Thereafter, iPS cell-based clinical trials have been initiated to treat Parkinson’s disease, heart disease and macular degeneration, highlighting the rapid progress that continues to be made in this area[123,124]. To treat Duchenne muscular dystrophy, Filareto et al[125] showed that SB-mediated ectopic expression of micro-utrophin in dystrophic iPS-derived skeletal muscle progenitors restored the muscle pathology by contributing to dystrophin–glycoprotein complex formation, which resulted in improved muscle contraction strength. PB-mediated expression of drug-inducible MYOD1 gene in human iPS cells lead to more efficient differentiation into myocytes[102]. Similarly, SB-mediated overexpression of PAX3 in iPS cells induced differentiation into MYOD positive myogenic progenitors and produced multinucleated myofibers[126]. Transposon-mediated iPS cells derived from patients suffering from either sickle cell disease caused by a β-globin gene mutation or Huntington’s disease caused by trinucleotide repeat expansions in the Huntingtin gene were successfully used for gene editing[127-129]. The most commonly used transposons PB and SB were successfully used to generate human iPS cells from patient-derived cells with a disease-causing genetic background[16,22,130]. These studies indicated that transposons are capable of introducing functional gene copies in patient-derived iPS cells containing defective genes. Recent evidence showed that transposon-mediated gene transfer was demonstrated in several types of cells such as ES cells, iPS cells, CD34+ hematopoietic stem cells or myoblasts[131].
Transposon-based gene delivery could also be used in combination with designer nucleases in iPS cells to correct gene defects. Yusa[132] reported that the endonuclease-based gene targeting efficiency increased using the PB transposon and it occurred due to the possibility of seamless removal of the drug marker enabled by re-transfection of the transposase. More recently, a transposon system was used in combination with CRISPR/Cas9 for the generation of iPS cells from Huntington disease patients to correct mutations in the Huntingtin gene and corrected cells were then differentiated successfully into excitable, synaptically active forebrain neurons[129]. Similarly, Wang et al[94] demonstrated that PB in combination with CRISPR/Cas9 for genome editing in iPS cells, in which the transposon delivered Cas9 gene followed by delivery of sgRNA caused modification. Subsequent transient transposase expression of inducible Cas9 cassette was removed and yielded genome-edited iPS cells with seamless transgene removal.
The treatment of several human diseases often involves genetic manipulation of iPS cells prior to transplantation, which may further threaten their genomic stability. Overall, genomic aberrations can affect differentiation capability, identity and tumorigenicity of iPS cells. In the promising era of iPS cell research and therapy, the genomic stability of iPS cells and their safety, efficiency, and specificity remains one of the highest concerns prior to clinical translation[133]. Hence preclinical trials in mice and other animal models are necessary in the future to confirm the in vivo therapeutic potential of reprogrammed cells. Challenges for reprogrammed cells are that they not only contain the in vivo delivery and dosage, but also their stability and potential off-target effects[4]. These challenges are currently hindering the progress to translate this potentially promising approach to clinical applications, but they appear to be solvable due to rapidly evolving advances in cellular reprogramming.
The use of SB systems appears to be safe in human cells with respect to off-target effects, as they originate from fish genomes, and the mammalian genome does not contain sufficient transposons to allow them to be cleavage by the transposase[50,73]. Hence, the SB transposon exhibits the least deviation in genome-wide distribution and no apparent bias was observed for either the heterochromatic or euchromatic region and weak correlation with transcriptional status of targeted genes was detected[134]. In addition, the ITRs region have negligible promoter/enhancer activity, and therefore they are unable to initiate transcription of genes that flank the integration site[135]. This system is highly efficient in transfecting even those cell types which are hard to transfect. On the other hand, PB systems have a wide target site that favor integration into genes and near chromatin marks characteristic of active transcription units[73,134,136,137]. These observations indicate that transposons (SB and PB) might be safe for therapeutic gene delivery in clinical trials.
After delivery of the transposon system, the transposition may undergo multiple rounds of remobilization[138,139], which should be minimized by carefully controlling the transposase dose[136]. In mouse embryonic stem cells approximately 95% of genomic transposon excision was reported to be precise and 5% of the transpositions showed genomic alterations[138]. It was also observed that frequent transposition into unknown sites could result in micro-deletions, footprint mutations as well as chromosomal rearrangements in the genome, which makes it labor intensive to identify integration-free iPS cells with intact genomes[138,140]. As a consequence, the transposase expression window should be tightly controlled to achieve traceless excision without inducing any genomic alterations and cytotoxicity[141].
Due to its non-viral nature and integration capacity, some of the transposon systems were adapted for use in gene therapy practices. To achieve efficient and safe use, the transposon systems were split into two plasmids, one containing the sequence encoding the transposase enzyme and the other comprising an expression cassette flanked by ITRs. However, in spite of these advantages, DNA transposon based vectors are essentially gene-inserting tools that still need assistance for efficient cellular uptake. Therefore, its activity depends on cell type, transfection method, and plasmid size. Moreover, it is important to note that these vectors have been largely used in the preclinical setting, and clinical trials are in progress to evaluate their efficacy, safety and presumed advantages.
Transposon-based gene transfer followed by cellular reprogramming might be associated with the important risk factor of genotoxicity. The genotoxicity could be induced either by interaction of the transposase with endogenous DNA sequences, or the genome-wide insertion profile of the transposon vector. To increase the efficacy and safety of cellular reprogramming, many efforts have been made to obtain potential molecules that can improve reprogramming efficiency or replace some of the vital transcription factors[142]. In this direction, various small molecules such as histone deacetylase inhibitors, DNA methyltransferase inhibitors, methylases, and demethylase inhibitors, Rho-associated protein kinase, and Wnt pathway regulators have been recognized to be effective in inducing reprogramming of terminally differentiated cells[143-146]. Huangfu et al[147] showed that valproic acid, a histone deacetylase inhibitor, increased the efficiency of transcription factor-mediated cellular reprogramming from 0.50% to 11.8%, indicating chromatin modification is one of the major rate-determining steps during cellular reprogramming. In addition to these, other molecules have also been tested to improve cellular reprogramming efficiency, including RepSOX2, E-616452 (2-[3-(6-methyl-2-pyridinyl)-1H-pyrazol-4-yl]-1,5-naphthyridine), and OAC1 (Oct4-activating compound 1), which facilitate the mesenchymal-epithelial transition (MET), and activate the stemness-associated promoter regions of mature fibroblasts[148,149]. Nowadays, the use of these small molecules is more trustworthy for introducing transcription factors into cells, but it remains a challenge to break through the efficiency threshold due to inadequate gene delivery and limitations in cellular uptake[150].
As compared to integration of retrovirus[151] and lentivirus[152], the integration profile of PB[137] and SB are safe, and are currently being tested for several clinical trials of T cell immunotherapy. Furthermore, to exclude the possibility of remobilization, the transposase could be transfected in the form of RNA, which seems to be less toxic to the cells[153].
Recently, transposon systems have been developed as attractive tools for somatic cell reprogramming, which has significant potential in speeding up patient-specific cell based therapies, as they can overcome some of the limitations of viral-based reprogramming technologies[125,154]. Furthermore, transposon systems have unique features for excising the exogenous reprogramming cassette from the iPS genome through re-expression of the transposase. Transposon systems eventually gives rise to “transgene-free” iPS cells, which is valuable in minimizing the risk of reactivation of reprogramming factors with oncogenic potential[94]. In addition to gene delivery, gene correction can also be achieved with a combination of transposons and designer endonucleases including ZFN, TALEN or CRISPR/Cas9. The introduction of a site-specific DNA double-strand break by endonuclease activity allows homologous recombination at target genes, followed by traceless removal of selectable gene cassettes by the transposase. This strategy has been used in SCD patient-derived iPS cells without any detectable off-target activity and undesirable chromosomal alterations[127]. More recently, the practice of mRNA encoding transposases to prevent continued mobilization of transposons and modification of ITRs, and the generation of hyperactive and codon-optimized transposase variants enhanced the overall transposition efficiencies[88]. This broadens the spectrum of possible therapeutic alternatives for gene therapy in particular, and gene correction in iPS cells. A number of preclinical studies performed as disease models that simulate the cognate human disorders have highlighted the potential of transposons for gene therapy[154]. Thus, iPS cell biology will continue to play a major role not only in the advancement of medical sciences, but also in improving the understanding of basic sciences. Looking forward, the continued advancement and refinement of transposon based-technologies and the steps toward their clinical translation will likely herald an exciting era in gene therapy.
| 1. | Sánchez Alvarado A, Yamanaka S. Rethinking differentiation: stem cells, regeneration, and plasticity. Cell. 2014;157:110-119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 207] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 2. | Kumar D, Talluri TR, Anand T, Kues WA. Transposon-based reprogramming to induced pluripotency. Histol Histopathol. 2015;30:1397-1409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 3. | Fidan K, Ebrahimi A, Çağlayan ÖH, Özçimen B, Önder TT. Transgene-Free Disease-Specific iPSC Generation from Fibroblasts and Peripheral Blood Mononuclear Cells. Methods Mol Biol. 2016;1353:215-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 4. | Kumar D, Anand T, Kues WA. Clinical potential of human-induced pluripotent stem cells: Perspectives of induced pluripotent stem cells. Cell Biol Toxicol. 2017;33:99-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 5. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17989] [Cited by in RCA: 18599] [Article Influence: 930.0] [Reference Citation Analysis (1)] |
| 6. | Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3332] [Cited by in RCA: 3108] [Article Influence: 163.6] [Reference Citation Analysis (12)] |
| 7. | Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861-872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14327] [Cited by in RCA: 14573] [Article Influence: 809.6] [Reference Citation Analysis (0)] |
| 8. | Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917-1920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7589] [Cited by in RCA: 7335] [Article Influence: 386.1] [Reference Citation Analysis (0)] |
| 9. | Lundstrom K. Viral Vectors in Gene Therapy. Diseases. 2018;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 252] [Cited by in RCA: 343] [Article Influence: 42.9] [Reference Citation Analysis (0)] |
| 10. | van der Loo JC, Wright JF. Progress and challenges in viral vector manufacturing. Hum Mol Genet. 2016;25:R42-R52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 182] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 11. | Chang EA, Jin SW, Nam MH, Kim SD. Human Induced Pluripotent Stem Cells : Clinical Significance and Applications in Neurologic Diseases. J Korean Neurosurg Soc. 2019;62:493-501. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 12. | Wattanapanitch M. Recent Updates on Induced Pluripotent Stem Cells in Hematological Disorders. Stem Cells Int. 2019;2019:5171032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 13. | Zakrzewski W, Dobrzyński M, Szymonowicz M, Rybak Z. Stem cells: past, present, and future. Stem Cell Res Ther. 2019;10:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 937] [Cited by in RCA: 1051] [Article Influence: 150.1] [Reference Citation Analysis (35)] |
| 14. | Skipper KA, Andersen PR, Sharma N, Mikkelsen JG. DNA transposon-based gene vehicles - scenes from an evolutionary drive. J Biomed Sci. 2013;20:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 15. | Rostovskaya M, Fu J, Obst M, Baer I, Weidlich S, Wang H, Smith AJ, Anastassiadis K, Stewart AF. Transposon-mediated BAC transgenesis in human ES cells. Nucleic Acids Res. 2012;40:e150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 112] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 16. | Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766-770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1423] [Cited by in RCA: 1269] [Article Influence: 74.6] [Reference Citation Analysis (0)] |
| 17. | Kesselring L, Miskey C, Zuliani C, Querques I, Kapitonov V, Laukó A, Fehér A, Palazzo A, Diem T, Lustig J, Sebe A, Wang Y, Dinnyés A, Izsvák Z, Barabas O, Ivics Z. A single amino acid switch converts the Sleeping Beauty transposase into an efficient unidirectional excisionase with utility in stem cell reprogramming. Nucleic Acids Res. 2020;48:316-331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 18. | Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1459] [Cited by in RCA: 1336] [Article Influence: 74.2] [Reference Citation Analysis (0)] |
| 19. | Wu C, Dunbar CE. Stem cell gene therapy: the risks of insertional mutagenesis and approaches to minimize genotoxicity. Front Med. 2011;5:356-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 87] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 20. | Davis RP, Nemes C, Varga E, Freund C, Kosmidis G, Gkatzis K, de Jong D, Szuhai K, Dinnyés A, Mummery CL. Generation of induced pluripotent stem cells from human foetal fibroblasts using the Sleeping Beauty transposon gene delivery system. Differentiation. 2013;86:30-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 21. | Muenthaisong S, Ujhelly O, Polgar Z, Varga E, Ivics Z, Pirity MK, Dinnyes A. Generation of mouse induced pluripotent stem cells from different genetic backgrounds using Sleeping beauty transposon mediated gene transfer. Exp Cell Res. 2012;318:2482-2489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 22. | Grabundzija I, Wang J, Sebe A, Erdei Z, Kajdi R, Devaraj A, Steinemann D, Szuhai K, Stein U, Cantz T, Schambach A, Baum C, Izsvák Z, Sarkadi B, Ivics Z. Sleeping Beauty transposon-based system for cellular reprogramming and targeted gene insertion in induced pluripotent stem cells. Nucleic Acids Res. 2013;41:1829-1847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 23. | Talluri TR, Kumar D, Glage S, Garrels W, Ivics Z, Debowski K, Behr R, Kues WA. Non-viral reprogramming of fibroblasts into induced pluripotent stem cells by Sleeping Beauty and piggyBac transposons. Biochem Biophys Res Commun. 2014;450:581-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 24. | Tsukiyama T, Kato-Itoh M, Nakauchi H, Ohinata Y. A comprehensive system for generation and evaluation of induced pluripotent stem cells using piggyBac transposition. PLoS One. 2014;9:e92973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 25. | Kues WA, Herrmann D, Barg-Kues B, Haridoss S, Nowak-Imialek M, Buchholz T, Streeck M, Grebe A, Grabundzija I, Merkert S, Martin U, Hall VJ, Rasmussen MA, Ivics Z, Hyttel P, Niemann H. Derivation and characterization of sleeping beauty transposon-mediated porcine induced pluripotent stem cells. Stem Cells Dev. 2013;22:124-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 60] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Nagy K, Sung HK, Zhang P, Laflamme S, Vincent P, Agha-Mohammadi S, Woltjen K, Monetti C, Michael IP, Smith LC, Nagy A. Induced pluripotent stem cell lines derived from equine fibroblasts. Stem Cell Rev Rep. 2011;7:693-702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 175] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 27. | Mo X, Li N, Wu S. Generation and characterization of bat-induced pluripotent stem cells. Theriogenology. 2014;82:283-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 28. | Debowski K, Warthemann R, Lentes J, Salinas-Riester G, Dressel R, Langenstroth D, Gromoll J, Sasaki E, Behr R. Non-viral generation of marmoset monkey iPS cells by a six-factor-in-one-vector approach. PLoS One. 2015;10:e0118424. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 29. | Ye J, Hong J, Ye F. Reprogramming rat embryonic fibroblasts into induced pluripotent stem cells using transposon vectors and their chondrogenic differentiation in vitro. Mol Med Rep. 2015;11:989-994. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 30. | Talluri TR, Kumar D, Glage S, Garrels W, Ivics Z, Debowski K, Behr R, Niemann H, Kues WA. Derivation and characterization of bovine induced pluripotent stem cells by transposon-mediated reprogramming. Cell Reprogram. 2015;17:131-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 31. | Zhao L, Wang Z, Zhang J, Yang J, Gao X, Wu B, Zhao G, Bao S, Hu S, Liu P, Li X. Characterization of the single-cell derived bovine induced pluripotent stem cells. Tissue Cell. 2017;49:521-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 32. | Kumar D, Anand T, Vijayalakshmy K, Sharma P, Rajendran R, Selokar NL, Yadav PS, Kumar D. Transposon mediated reprogramming of buffalo fetal fibroblasts to induced pluripotent stem cells in feeder free culture conditions. Res Vet Sci. 2019;123:252-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 10] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 33. | McClintock B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci USA. 1950;36:344-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 975] [Cited by in RCA: 928] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 34. | Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1898] [Cited by in RCA: 2007] [Article Influence: 105.6] [Reference Citation Analysis (0)] |
| 35. | Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331-368. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1001] [Cited by in RCA: 849] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 36. | Ivancevic AM, Walsh AM, Kortschak RD, Adelson DL. Jumping the fine LINE between species: horizontal transfer of transposable elements in animals catalyses genome evolution. Bioessays. 2013;35:1071-1082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 37. | Walsh AM, Kortschak RD, Gardner MG, Bertozzi T, Adelson DL. Widespread horizontal transfer of retrotransposons. Proc Natl Acad Sci USA. 2013;110:1012-1016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 38. | Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J; International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16054] [Cited by in RCA: 15289] [Article Influence: 611.6] [Reference Citation Analysis (7)] |
| 39. | Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1100] [Cited by in RCA: 1237] [Article Influence: 72.8] [Reference Citation Analysis (0)] |
| 40. | Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013;9:e1003470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 467] [Cited by in RCA: 505] [Article Influence: 38.8] [Reference Citation Analysis (0)] |
| 41. | Mustafin RN. The role of transposable elements in the differentiation of stem cells. Mol Genet Microbiol Virol. 2019;34:67-74. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 42. | Ayarpadikannan S, Kim HS. The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics Inform. 2014;12:98-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 43. | Erwin JA, Marchetto MC, Gage FH. Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci. 2014;15:497-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 203] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 44. | Chenais B. Transposable elements in cancer and other human diseases. Curr Cancer Drug Targets. 2015;15:227-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 45. | Ayarpadikannan S, Lee HE, Han K, Kim HS. Transposable element-driven transcript diversification and its relevance to genetic disorders. Gene. 2015;558:187-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 46. | Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1031] [Cited by in RCA: 1061] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 47. | Fraser MJ, Ciszczon T, Elick T, Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol Biol. 1996;5:141-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 239] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 48. | Dupuy AJ, Clark K, Carlson CM, Fritz S, Davidson AE, Markley KM, Finley K, Fletcher CF, Ekker SC, Hackett PB, Horn S, Largaespada DA. Mammalian germ-line transgenesis by transposition. Proc Natl Acad Sci USA. 2002;99:4495-4499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 153] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 49. | Garrels W, Holler S, Cleve N, Niemann H, Ivics Z, Kues WA. Assessment of fecundity and germ line transmission in two transgenic pig lines produced by sleeping beauty transposition. Genes (Basel). 2012;3:615-633. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 50. | Mátés L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, Ma L, Samara-Kuko E, Gysemans C, Pryputniewicz D, Miskey C, Fletcher B, VandenDriessche T, Ivics Z, Izsvák Z. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 626] [Cited by in RCA: 756] [Article Influence: 44.5] [Reference Citation Analysis (0)] |
| 51. | Doherty JE, Huye LE, Yusa K, Zhou L, Craig NL, Wilson MH. Hyperactive piggyBac gene transfer in human cells and in vivo. Hum Gene Ther. 2012;23:311-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 86] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 52. | Salewski RP, Buttigieg J, Mitchell RA, van der Kooy D, Nagy A, Fehlings MG. The generation of definitive neural stem cells from PiggyBac transposon-induced pluripotent stem cells can be enhanced by induction of the NOTCH signaling pathway. Stem Cells Dev. 2013;22:383-396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 53. | Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, Kaminski JM. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci USA. 2006;103:15008-15013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 274] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 54. | Ivics Z, Izsvák Z. Sleeping Beauty Transposition. Microbiol Spectr. 2015;3:MDNA3-0042-2014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 55. | Yusa K. piggyBac Transposon. Microbiol Spectr. 2015;3:MDNA3-0028-2014. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 70] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 56. | Wilson MH, Coates CJ, George AL. PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 384] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 57. | Belay E, Dastidar S, VandenDriessche T, Chuah MK. Transposon-mediated gene transfer into adult and induced pluripotent stem cells. Curr Gene Ther. 2011;11:406-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 58. | Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 686] [Cited by in RCA: 792] [Article Influence: 37.7] [Reference Citation Analysis (0)] |
| 59. | Collier LS, Largaespada DA. Transposons for cancer gene discovery: Sleeping Beauty and beyond. Genome Biol. 2007;8 Suppl 1:S15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 60. | Cadiñanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 240] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 61. | Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci USA. 2011;108:1531-1536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 448] [Cited by in RCA: 590] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 62. | Voigt F, Wiedemann L, Zuliani C, Querques I, Sebe A, Mátés L, Izsvák Z, Ivics Z, Barabas O. Sleeping Beauty transposase structure allows rational design of hyperactive variants for genetic engineering. Nat Commun. 2016;7:11126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 63. | Jursch T, Miskey C, Izsvák Z, Ivics Z. Regulation of DNA transposition by CpG methylation and chromatin structure in human cells. Mob DNA. 2013;4:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 64. | Claeys Bouuaert C, Chalmers R. Hsmar1 transposition is sensitive to the topology of the transposon donor and the target. PLoS One. 2013;8:e53690. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 65. | Vigdal TJ, Kaufman CD, Izsvák Z, Voytas DF, Ivics Z. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J Mol Biol. 2002;323:441-452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 208] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 66. | Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085-2094. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 245] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 67. | Meir YJ, Weirauch MT, Yang HS, Chung PC, Yu RK, Wu SC. Genome-wide target profiling of piggyBac and Tol2 in HEK 293: pros and cons for gene discovery and gene therapy. BMC Biotechnol. 2011;11:28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 68. | Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797-801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1896] [Cited by in RCA: 1721] [Article Influence: 101.2] [Reference Citation Analysis (0)] |
| 69. | Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, Longaker MT, Wu JC. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197-199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 575] [Cited by in RCA: 512] [Article Influence: 32.0] [Reference Citation Analysis (0)] |
| 70. | Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348-362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1024] [Cited by in RCA: 985] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 71. | Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618-630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2155] [Cited by in RCA: 1949] [Article Influence: 121.8] [Reference Citation Analysis (0)] |
| 72. | Bire S, Gosset D, Jégot G, Midoux P, Pichon C, Rouleux-Bonnin F. Exogenous mRNA delivery and bioavailability in gene transfer mediated by piggyBac transposition. BMC Biotechnol. 2013;13:75. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 73. | Grabundzija I, Irgang M, Mátés L, Belay E, Matrai J, Gogol-Döring A, Kawakami K, Chen W, Ruiz P, Chuah MK, VandenDriessche T, Izsvák Z, Ivics Z. Comparative analysis of transposable element vector systems in human cells. Mol Ther. 2010;18:1200-1209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 182] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 74. | Izsvák Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J Mol Biol. 2000;302:93-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 253] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 75. | Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts A, Sivasubbu S, Wang X, Hackett PB, Largaespada DA, McIvor RS, Ekker SC. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2:e169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 273] [Cited by in RCA: 265] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 76. | Claeys Bouuaert C, Lipkow K, Andrews SS, Liu D, Chalmers R. The autoregulation of a eukaryotic DNA transposon. Elife. 2013;2:e00668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 77. | Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 822] [Cited by in RCA: 953] [Article Influence: 86.6] [Reference Citation Analysis (0)] |
| 78. | Ma H, Tu LC, Naseri A, Huisman M, Zhang S, Grunwald D, Pederson T. CRISPR-Cas9 nuclear dynamics and target recognition in living cells. J Cell Biol. 2016;214:529-537. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 153] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 79. | Kosicki M, Tomberg K, Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 2018;36:765-771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1145] [Cited by in RCA: 1274] [Article Influence: 159.3] [Reference Citation Analysis (0)] |
| 80. | Ivics Z, Katzer A, Stüwe EE, Fiedler D, Knespel S, Izsvák Z. Targeted Sleeping Beauty transposition in human cells. Mol Ther. 2007;15:1137-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 102] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 81. | Feng X, Bednarz AL, Colloms SD. Precise targeted integration by a chimaeric transposase zinc-finger fusion protein. Nucleic Acids Res. 2010;38:1204-1216. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 26] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 82. | Luo W, Galvan DL, Woodard LE, Dorset D, Levy S, Wilson MH. Comparative analysis of chimeric ZFP-, TALE- and Cas9-piggyBac transposases for integration into a single locus in human cells. Nucleic Acids Res. 2017;45:8411-8422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 83. | Owens JB, Mauro D, Stoytchev I, Bhakta MS, Kim MS, Segal DJ, Moisyadi S. Transcription activator like effector (TALE)-directed piggyBac transposition in human cells. Nucleic Acids Res. 2013;41:9197-9207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 84. | Mitra R, Fain-Thornton J, Craig NL. piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J. 2008;27:1097-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 169] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 85. | Liang Q, Kong J, Stalker J, Bradley A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis. 2009;47:404-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 118] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 86. | Bhatt S, Chalmers R. Targeted DNA transposition in vitro using a dCas9-transposase fusion protein. Nucleic Acids Res. 2019;47:8126-8135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 87. | Chen SP, Wang HH. An Engineered Cas-Transposon System for Programmable and Site-Directed DNA Transpositions. CRISPR J. 2019;2:376-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 88. | Hew BE, Sato R, Mauro D, Stoytchev I, Owens JB. RNA-guided piggyBac transposition in human cells. Synth Biol (Oxf). 2019;4:ysz018. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 89. | Strecker J, Ladha A, Gardner Z, Schmid-Burgk JL, Makarova KS, Koonin EV, Zhang F. RNA-guided DNA insertion with CRISPR-associated transposases. Science. 2019;365:48-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 413] [Cited by in RCA: 488] [Article Influence: 69.7] [Reference Citation Analysis (0)] |
| 90. | Daniel R, Smith JA. Integration site selection by retroviral vectors: molecular mechanism and clinical consequences. Hum Gene Ther. 2008;19:557-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 91. | Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 695] [Cited by in RCA: 737] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 92. | Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, Beldjord K, Asnafi V, MacIntyre E, Dal Cortivo L, Radford I, Brousse N, Sigaux F, Moshous D, Hauer J, Borkhardt A, Belohradsky BH, Wintergerst U, Velez MC, Leiva L, Sorensen R, Wulffraat N, Blanche S, Bushman FD, Fischer A, Cavazzana-Calvo M. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132-3142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1295] [Cited by in RCA: 1430] [Article Influence: 79.4] [Reference Citation Analysis (0)] |
| 93. | Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, Brugman MH, Pike-Overzet K, Chatters SJ, de Ridder D, Gilmour KC, Adams S, Thornhill SI, Parsley KL, Staal FJ, Gale RE, Linch DC, Bayford J, Brown L, Quaye M, Kinnon C, Ancliff P, Webb DK, Schmidt M, von Kalle C, Gaspar HB, Thrasher AJ. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143-3150. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1055] [Cited by in RCA: 976] [Article Influence: 54.2] [Reference Citation Analysis (0)] |
| 94. | Wang G, Yang L, Grishin D, Rios X, Ye LY, Hu Y, Li K, Zhang D, Church GM, Pu WT. Efficient, footprint-free human iPSC genome editing by consolidation of Cas9/CRISPR and piggyBac technologies. Nat Protoc. 2017;12:88-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 101] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 95. | Querques I, Mades A, Zuliani C, Miskey C, Alb M, Grueso E, Machwirth M, Rausch T, Einsele H, Ivics Z, Hudecek M, Barabas O. A highly soluble Sleeping Beauty transposase improves control of gene insertion. Nat Biotechnol. 2019;37:1502-1512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 77] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 96. | Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, Siuzdak G, Schöler HR, Duan L, Ding S. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1309] [Cited by in RCA: 1238] [Article Influence: 72.8] [Reference Citation Analysis (0)] |
| 97. | Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472-476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1410] [Cited by in RCA: 1236] [Article Influence: 72.7] [Reference Citation Analysis (0)] |
| 98. | Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, Cui K, Qu X, Xiang T, Lu D, Chi X, Gao G, Ji W, Ding M, Deng H. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587-590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 334] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 99. | West FD, Terlouw SL, Kwon DJ, Mumaw JL, Dhara SK, Hasneen K, Dobrinsky JR, Stice SL. Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev. 2010;19:1211-1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 176] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 100. | Liu J, Balehosur D, Murray B, Kelly JM, Sumer H, Verma PJ. Generation and characterization of reprogrammed sheep induced pluripotent stem cells. Theriogenology. 2012;77:338-46.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 70] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 101. | Kawaguchi T, Tsukiyama T, Kimura K, Matsuyama S, Minami N, Yamada M, Imai H. Generation of Naïve Bovine Induced Pluripotent Stem Cells Using PiggyBac Transposition of Doxycycline-Inducible Transcription Factors. PLoS One. 2015;10:e0135403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 102. | Tanaka A, Woltjen K, Miyake K, Hotta A, Ikeya M, Yamamoto T, Nishino T, Shoji E, Sehara-Fujisawa A, Manabe Y, Fujii N, Hanaoka K, Era T, Yamashita S, Isobe K, Kimura E, Sakurai H. Efficient and reproducible myogenic differentiation from human iPS cells: prospects for modeling Miyoshi Myopathy in vitro. PLoS One. 2013;8:e61540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 135] [Cited by in RCA: 166] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 103. | Kim SI, Oceguera-Yanez F, Sakurai C, Nakagawa M, Yamanaka S, Woltjen K. Inducible Transgene Expression in Human iPS Cells Using Versatile All-in-One piggyBac Transposons. Methods Mol Biol. 2016;1357:111-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 81] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 104. | Shoji E, Woltjen K, Sakurai H. Directed Myogenic Differentiation of Human Induced Pluripotent Stem Cells. Methods Mol Biol. 2016;1353:89-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 105. | Inada E, Saitoh I, Watanabe S, Aoki R, Miura H, Ohtsuka M, Murakami T, Sawami T, Yamasaki Y, Sato M. PiggyBac transposon-mediated gene delivery efficiently generates stable transfectants derived from cultured primary human deciduous tooth dental pulp cells (HDDPCs) and HDDPC-derived iPS cells. Int J Oral Sci. 2015;7:144-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 106. | Kumar D, Talluri TR, Anand T, Kues WA. Induced pluripotent stem cells: Mechanisms, achievements and perspectives in farm animals. World J Stem Cells. 2015;7:315-328. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 107. | Hannoun Z, Steichen C, Dianat N, Weber A, Dubart-Kupperschmitt A. The potential of induced pluripotent stem cell derived hepatocytes. J Hepatol. 2016;65:182-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 78] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 108. | Bruyneel AA, McKeithan WL, Feyen DA, Mercola M. Will iPSC-cardiomyocytes revolutionize the discovery of drugs for heart disease? Curr Opin Pharmacol. 2018;42:55-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 109. | Rowe RG, Daley GQ. Induced pluripotent stem cells in disease modelling and drug discovery. Nat Rev Genet. 2019;20:377-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 462] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 110. | Moradi S, Mahdizadeh H, Šarić T, Kim J, Harati J, Shahsavarani H, Greber B, Moore JB 4th. Research and therapy with induced pluripotent stem cells (iPSCs): social, legal, and ethical considerations. Stem Cell Res Ther. 2019;10:341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 224] [Cited by in RCA: 179] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 111. | Woltjen K, Hämäläinen R, Kibschull M, Mileikovsky M, Nagy A. Transgene-free production of pluripotent stem cells using piggyBac transposons. Methods Mol Biol. 2011;767:87-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 112. | Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 544] [Article Influence: 38.9] [Reference Citation Analysis (0)] |
| 113. | Hwang NS, Varghese S, Elisseeff J. Controlled differentiation of stem cells. Adv Drug Deliv Rev. 2008;60:199-214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 272] [Cited by in RCA: 230] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 114. | Katayama M, Hirayama T, Horie K, Kiyono T, Donai K, Takeda S, Nishimori K, Fukuda T. Induced Pluripotent Stem Cells With Six Reprogramming Factors From Prairie Vole, Which Is an Animal Model for Social Behaviors. Cell Transplant. 2016;25:783-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 115. | Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods. 2009;6:363-369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 513] [Cited by in RCA: 467] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 116. | Igawa K, Kokubu C, Yusa K, Horie K, Yoshimura Y, Yamauchi K, Suemori H, Yokozeki H, Toyoda M, Kiyokawa N, Okita H, Miyagawa Y, Akutsu H, Umezawa A, Katayama I, Takeda J. Removal of reprogramming transgenes improves the tissue reconstitution potential of keratinocytes generated from human induced pluripotent stem cells. Stem Cells Transl Med. 2014;3:992-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 117. | Itoh M, Kiuru M, Cairo MS, Christiano AM. Generation of keratinocytes from normal and recessive dystrophic epidermolysis bullosa-induced pluripotent stem cells. Proc Natl Acad Sci USA. 2011;108:8797-8802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 216] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 118. | Anand T, Talluri TR, Kumar D, Garrels W, Mukherjee A, Debowski K, Behr R, Kues WA. Differentiation of Induced Pluripotent Stem Cells to Lentoid Bodies Expressing a Lens Cell-Specific Fluorescent Reporter. PLoS One. 2016;11:e0157570. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 119. | Damjanov I, Clark RK, Andrews PW. Cytoskeleton of human embryonal carcinoma cells. Cell Differ. 1984;15:133-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 120. | Rossant J, McBurney MW. The developmental potential of a euploid male teratocarcinoma cell line after blastocyst injection. J Embryol Exp Morphol. 1982;70:99-112. [PubMed] |
| 121. | Kebriaei P, Huls H, Jena B, Munsell M, Jackson R, Lee DA, Hackett PB, Rondon G, Shpall E, Champlin RE, Cooper LJ. Infusing CD19-directed T cells to augment disease control in patients undergoing autologous hematopoietic stem-cell transplantation for advanced B-lymphoid malignancies. Hum Gene Ther. 2012;23:444-450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 122. | Kebriaei P, Singh H, Huls MH, Figliola MJ, Bassett R, Olivares S, Jena B, Dawson MJ, Kumaresan PR, Su S, Maiti S, Dai J, Moriarity B, Forget MA, Senyukov V, Orozco A, Liu T, McCarty J, Jackson RN, Moyes JS, Rondon G, Qazilbash M, Ciurea S, Alousi A, Nieto Y, Rezvani K, Marin D, Popat U, Hosing C, Shpall EJ, Kantarjian H, Keating M, Wierda W, Do KA, Largaespada DA, Lee DA, Hackett PB, Champlin RE, Cooper LJ. Phase I trials using Sleeping Beauty to generate CD19-specific CAR T cells. J Clin Invest. 2016;126:3363-3376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 398] [Article Influence: 39.8] [Reference Citation Analysis (0)] |
| 123. | Sayed N, Liu C, Wu JC. Translation of Human-Induced Pluripotent Stem Cells: From Clinical Trial in a Dish to Precision Medicine. J Am Coll Cardiol. 2016;67:2161-2176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 210] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 124. | Shi Y, Inoue H, Wu JC, Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov. 2017;16:115-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 960] [Cited by in RCA: 1080] [Article Influence: 120.0] [Reference Citation Analysis (0)] |
| 125. | Filareto A, Parker S, Darabi R, Borges L, Iacovino M, Schaaf T, Mayerhofer T, Chamberlain JS, Ervasti JM, McIvor RS, Kyba M, Perlingeiro RC. An ex vivo gene therapy approach to treat muscular dystrophy using inducible pluripotent stem cells. Nat Commun. 2013;4:1549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 126. | Belay E, Mátrai J, Acosta-Sanchez A, Ma L, Quattrocelli M, Mátés L, Sancho-Bru P, Geraerts M, Yan B, Vermeesch J, Rincón MY, Samara-Kuko E, Ivics Z, Verfaillie C, Sampaolesi M, Izsvák Z, Vandendriessche T, Chuah MK. Novel hyperactive transposons for genetic modification of induced pluripotent and adult stem cells: a nonviral paradigm for coaxed differentiation. Stem Cells. 2010;28:1760-1771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 127. | Sun N, Zhao H. Seamless correction of the sickle cell disease mutation of the HBB gene in human induced pluripotent stem cells using TALENs. Biotechnol Bioeng. 2014;111:1048-1053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 128. | Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, Kan YW. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24:1526-1533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 353] [Cited by in RCA: 313] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 129. | Xu JY, Ye ZL, Jiang DQ, He JC, Ding YM, Li LF, Lv SQ, Wang Y, Jin HJ, Qian QJ. Mesothelin-targeting chimeric antigen receptor-modified T cells by piggyBac transposon system suppress the growth of bile duct carcinoma. Tumour Biol. 2017;39:1010428317695949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 130. | Sebe A, Ivics Z. Reprogramming of Human Fibroblasts to Induced Pluripotent Stem Cells with Sleeping Beauty Transposon-Based Stable Gene Delivery. Methods Mol Biol. 2016;1400:419-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 131. | Schumann GG, Fuchs NV, Tristán-Ramos P, Sebe A, Ivics Z, Heras SR. The impact of transposable element activity on therapeutically relevant human stem cells. Mob DNA. 2019;10:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 132. | Yusa K. Seamless genome editing in human pluripotent stem cells using custom endonuclease-based gene targeting and the piggyBac transposon. Nat Protoc. 2013;8:2061-2078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 133. | Gün G, Kues WA. Current progress of genetically engineered pig models for biomedical research. Biores Open Access. 2014;3:255-264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (1)] |
| 134. | Gogol-Döring A, Ammar I, Gupta S, Bunse M, Miskey C, Chen W, Uckert W, Schulz TF, Izsvák Z, Ivics Z. Genome-wide Profiling Reveals Remarkable Parallels Between Insertion Site Selection Properties of the MLV Retrovirus and the piggyBac Transposon in Primary Human CD4(+) T Cells. Mol Ther. 2016;24:592-606. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 126] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 135. | Ivics Z, Izsvák Z. The expanding universe of transposon technologies for gene and cell engineering. Mob DNA. 2010;1:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 136. | Li MA, Pettitt SJ, Eckert S, Ning Z, Rice S, Cadiñanos J, Yusa K, Conte N, Bradley A. The piggyBac transposon displays local and distant reintegration preferences and can cause mutations at noncanonical integration sites. Mol Cell Biol. 2013;33:1317-1330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 72] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 137. | Huang X, Guo H, Tammana S, Jung YC, Mellgren E, Bassi P, Cao Q, Tu ZJ, Kim YC, Ekker SC, Wu X, Wang SM, Zhou X. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and piggyBac transposons in human primary T cells. Mol Ther. 2010;18:1803-1813. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 144] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 138. | Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, Wang X, Bradley A, Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:9290-9295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 310] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 139. | Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, Chang JC, Bao G, Muench MO, Yu J, Levy JA, Kan YW. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc Natl Acad Sci USA. 2014;111:9591-9596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 245] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 140. | Geurts AM, Collier LS, Geurts JL, Oseth LL, Bell ML, Mu D, Lucito R, Godbout SA, Green LE, Lowe SW, Hirsch BA, Leinwand LA, Largaespada DA. Gene mutations and genomic rearrangements in the mouse as a result of transposon mobilization from chromosomal concatemers. PLoS Genet. 2006;2:e156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 81] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 141. | Galla M, Schambach A, Falk CS, Maetzig T, Kuehle J, Lange K, Zychlinski D, Heinz N, Brugman MH, Göhring G, Izsvák Z, Ivics Z, Baum C. Avoiding cytotoxicity of transposases by dose-controlled mRNA delivery. Nucleic Acids Res. 2011;39:7147-7160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 142. | Lin T, Wu S. Reprogramming with Small Molecules instead of Exogenous Transcription Factors. Stem Cells Int. 2015;2015:794632. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 143. | Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, Ge J, Xu J, Zhang Q, Zhao Y, Deng H. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 938] [Cited by in RCA: 1055] [Article Influence: 81.2] [Reference Citation Analysis (0)] |
| 144. | Tsutsui H, Valamehr B, Hindoyan A, Qiao R, Ding X, Guo S, Witte ON, Liu X, Ho CM, Wu H. An optimized small molecule inhibitor cocktail supports long-term maintenance of human embryonic stem cells. Nat Commun. 2011;2:167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 133] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 145. | Kawamata M, Ochiya T. Generation of genetically modified rats from embryonic stem cells. Proc Natl Acad Sci USA. 2010;107:14223-14228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 88] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 146. | Jones PA, Ohtani H, Chakravarthy A, De Carvalho DD. Epigenetic therapy in immune-oncology. Nat Rev Cancer. 2019;19:151-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 401] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 147. | Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795-797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1320] [Cited by in RCA: 1215] [Article Influence: 67.5] [Reference Citation Analysis (0)] |
| 148. | Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, Huangfu D, Akutsu H, Liu DR, Rubin LL, Eggan K. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 632] [Cited by in RCA: 630] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 149. | Li W, Tian E, Chen ZX, Sun G, Ye P, Yang S, Lu D, Xie J, Ho TV, Tsark WM, Wang C, Horne DA, Riggs AD, Yip ML, Shi Y. Identification of Oct4-activating compounds that enhance reprogramming efficiency. Proc Natl Acad Sci USA. 2012;109:20853-20858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 60] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 150. | Rodríguez-Martínez JA, Peterson-Kaufman KJ, Ansari AZ. Small-molecule regulators that mimic transcription factors. Biochim Biophys Acta. 2010;1799:768-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 151. | Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749-1751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1010] [Cited by in RCA: 995] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 152. | Singh H, Figliola MJ, Dawson MJ, Huls H, Olivares S, Switzer K, Mi T, Maiti S, Kebriaei P, Lee DA, Champlin RE, Cooper LJ. Reprogramming CD19-specific T cells with IL-21 signaling can improve adoptive immunotherapy of B-lineage malignancies. Cancer Res. 2011;71:3516-3527. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 170] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 153. | Peng PD, Cohen CJ, Yang S, Hsu C, Jones S, Zhao Y, Zheng Z, Rosenberg SA, Morgan RA. Efficient nonviral Sleeping Beauty transposon-based TCR gene transfer to peripheral blood lymphocytes confers antigen-specific antitumor reactivity. Gene Ther. 2009;16:1042-1049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 80] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 154. | Tipanee J, Chai YC, VandenDriessche T, Chuah MK. Preclinical and clinical advances in transposon-based gene therapy. Biosci Rep. 2017;37. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Liu XG S-Editor: Ma YJ L-Editor: Webster JR E-Editor: Xing YX