Published online Nov 26, 2020. doi: 10.4252/wjsc.v12.i11.1354
Peer-review started: March 30, 2020
First decision: July 5, 2020
Revised: July 7, 2020
Accepted: September 8, 2020
Article in press: September 8, 2020
Published online: November 26, 2020
Processing time: 243 Days and 5.6 Hours
As the third most abundant element, aluminum is widespread in the environment. Previous studies have shown that aluminum has a neurotoxic effect and its exposure can impair neuronal development and cognitive function.
To study the effects of aluminum on epigenetic modification in neural stem cells and neurons.
Neural stem cells were isolated from the forebrain of adult mice. Neurons were isolated from the hippocampi tissues of embryonic day 16-18 mice. AlCl3 at 100 and 200 μmol/L was applied to stem cells and neurons.
Aluminum altered the differentiation of adult neural stem cells and caused apoptosis of newborn neurons while having no significant effects on the proliferation of neural stem cells. Aluminum application also significantly inhibited the dendritic development of hippocampal neurons. Mechanistically, aluminum exposure significantly affected the levels of DNA 5-hydroxy-methylcytosine, 5-methylcytosine, and N6-methyladenine in stem cells and neurons.
Our findings indicate that aluminum may regulate neuronal development by modulating DNA modifications.
Core Tip: Although the neurotoxic effects of aluminum have been known, it is still unclear regarding the effects of aluminum on epigenetic modifications in the context of neuronal development. Our present study revealed that aluminum exerted the neurotoxic effects including promoting cell death by regulating DNA modifications. These results highlight the crosstalk between the environment and epigenetics, and subsequent phenotypes. Our findings emphasize the importance of protecting the environment and improving food safety.
- Citation: Cheng XJ, Guan FL, Li Q, Dai G, Li HF, Li XK. AlCl3 exposure regulates neuronal development by modulating DNA modification. World J Stem Cells 2020; 12(11): 1354-1365
- URL: https://www.wjgnet.com/1948-0210/full/v12/i11/1354.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v12.i11.1354
Epigenetic modifications mainly include histone posttranslational modifications, DNA and RNA methylation and demethylation, and non-coding RNAs. Previous studies have indicated that epigenetic pathways play a critical function in diverse biological processes[1,2]. DNA methylation, mainly on the fifth carbon of cytosine [5-methylcytosine (5mC)] in mammalian, is established by DNA methyltransferases (DNMTs), including DNMT1, DNMT3A, and DNMT3B. In embryonic and postnatal neuronal development, the deficiency of DNMTs affects embryonic viability, cell survival, synaptic development, and learning and memory; however, neuronal activity could influence DNA methylation, suggesting that DNA methylation is important for normal neuronal function[3-5].
Recent studies have shown that 5mC can be further converted to 5-hydroxymethylcytosine (5hmC) by ten-eleven translocation (TET) proteins including TET1, TET2, and TET3[6,7]. 5hmC is significantly enriched in the brain relative to many other tissues and cell types, is acquired during postnatal neurodevelopment and aging, and displays spatial and temporal dynamics. Recent studies have shown consistently that Tet1 regulates neuronal activity, the formation and extinction of memory, and neurogenesis[8-13]. Recently, DNA N6-methyladenine (6mA) modification has been uncovered, which regulates gene expression and is involved in neuronal outcomes induced by environmental stress[14,15].
Aluminum is a neurotoxin and is associated with neuronal inflammation, memory impairment, and neurological disorders through different mechanisms[16]. AlCl3 exposure (50-100 mg/kg in vivo) significantly exacerbates amyloid beta (Aβ) deposition, plaque formation, and tau phosphorylation; causes cognitive dysfunction and mitochondria oxidative; and therefore induces Alzheimer’s disease-like phenotypes in rats[17-19]. Aluminum exposure (25 mg/kg in vivo or 0.5 mmol/L in vitro) also stimulates the expression of pro-inflammatory cytokines including TNF-α and IL-6, induces the production of reactive oxygen species (ROS), and then causes neuroinflammation and DNA damage[19-21]. However, it remains largely unknown whether aluminum has a neurotoxic effect by altering epigenetic states.
In the present study, we found that aluminum (AlCl3) skewed the differentiation of adult neural stem cells (aNSCs) toward glial cells and induced apoptosis of newborn neurons. Furthermore, aluminum inhibited the morphological development of neurons generated upon aNSC differentiation and hippocampal neurons. Finally, we found that AlCl3 exposure differentially altered the level of DNA methylation and hydroxymethylation in aNSCs and neurons by regulating the expression of DNA methyltransferases and dioxygenases. In summary, our results suggest that AlCl3 exerts a neurotoxic effect by modulating DNA modifications.
Mice were housed in a standard condition of the Animal Center of Zhejiang University on a 12 h light/dark cycle with free access to food and water. All animal experiments were performed according to the protocols approved by the Institutional Animal Care and Use Committee of Zhejiang University.
The isolation and culture of adult neural stem cells were performed according to an established protocol[9]. The aNSCs were cultured in DMEM/F-12 medium containing 20 ng/mL FGF-2 (Catalog No. 100-18B-B, PeproTech), 20 ng/mL EGF (Catalog No. 100-15, PeproTech), 2% B27 supplement (Catalog No. 12587-010, Thermo Fisher Scientific), 1% antibiotic-antimycotic (Catalog No. 15140-122, Thermo Fisher Scientific), and 2 mmol/L L-glutamine (Catalog No. 25030–149, Thermo Fisher Scientific) in a humidified incubator supplied with 5% CO2 at 37 °C.
AlCl3 was dissolved with nuclease free water to 50 mmol/L and applied to cells at a final concentration of 100 μmol/L or 200 μmol/L. The cells were collected at scheduled time-point for in vitro assay.
For in vitro proliferation assay, aNSCs were cultured on coverslips with medium supplied with 5 mmol/L BrdU for 8 h. For in vitro differentiation assay, aNSCs were cultured on coverslips with proliferation medium, and then transferred into differentiation medium containing 1 mmol/L retinoic acid (Catalog No. R-2625, Sigma) and 5 mmol/L forskolin (Catalog No. F-6886, Sigma) for 48 h.
Primary neurons were isolated from the hippocampus of E16-E18 mice and seeded in cell climbing slices (Corning, 354087) or plates that were coated with poly-D-lysine (5 μg/mL, Sigma, P0899-10). Approximately 1 × 105 cells per well were seeded for a slice, while 1.5 million cells per well were seeded for a 6-well-plate. After growing in the plating medium for 4 h, which consisted of MEM (Gibco,11095-080), 10% FBS (Gibco,10091-148), 1% L-Glu (Gibco, 5030-149), 1% sodium pyruvate (Gibco, 11360-070), and 0.45% D-glucose (Amresco, 0188), the medium was changed to a maintaining medium that consisted of neurobasal (Gibco, 21103-049), 0.25% L-Glu (Gibco, 25030-149), 0.125% GlutaMax (Thermo, 35050061), and 2% B27 (Gibco, 17504-044). The medium was renewed half of the liquid volume every 3 d.
To detect the function of proliferation and differentiation, cell samples were washed with PBS for 30 min followed by blocking with PBS containing 3% normal goat serum and 0.1% triton X-100 for 1 h at room temperature. Samples were incubated with primary antibodies overnight at 4 °C. For BrdU immunostaining, samples were treated with 1M HCl at 37 °C for 30 min before blocking. The following primary antibodies were used: GFAP (Catalog No. Z0334, DAKO), Tuj1 (Catalog No. G712A, Promega), Caspase 3 (Catalog No. AB3623, Millipore), and BrdU (Catalog No. ab6326, Abcam). The second day, after being washed with PBS for 30 min, sections were incubated with fluorophore-conjugated secondary antibodies for 1 h at room temperature. After final washes, samples were mounted onto glass slides and cover slipped with mounting medium. Images were captured using a Nikon invert microscope, and the numbers of BrdU+, Tuj1+, GFAP+, and Caspase3+Tuj1+ cells were quantified with image J software (NIH).
Total RNA was extracted with TRIzol reagent (Catalog No. 15596018, Thermo Fisher Scientific) following the manufacturer’s protocol. The concentration was determined using a NanoDrop 2000 spectrophotometer, and 500 ng of total RNA was subjected to reverse transcription. All real-time PCR reactions were performed in triplicate using power SYBR Green PCR master Mix (Catalog No. Q71502, Vazyme), and the results were analyzed using the ∆∆Ct method. The sequence of all the used primers can be found in Supplementary Table 1.
Cells were washed with PBS and resuspended in RIPA (Catalog No. ab156034, Abcam) containing 1 × protease inhibitor cocktail (Catalog No. 04693124001, Sigma). The samples were centrifuged at 4 °C for 20 min at 14000 rpm, and the supernatants were collected for further experiments. Protein concentrations of the samples were measured with a BioPhotometer, and 20 μg of each sample was used for electrophoresis after denaturation for 5 min at 95 °C. Samples were subjected to SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. The following primary antibodies were used: Anti-Tuj1 (Catalog No. G712A, Promega), anti-GFAP (Catalog No. 3670, Cell Signaling), and anti-GAPDH (Catalog No. AM4300, Thermo Fisher Scientific). Secondary HRP conjugated antibodies were applied for 1 h at room temperature. The signal was detected with the Tanon 5200 Detection system, and the relative level of signal intensity was normalized to that of GAPDH.
DNA extraction and DNA dot-blot were performed as described previously[9]. The following primary antibodies were used: 5mC (Catalog No. 61255, Active Motif), 5hmC (Catalog No. 39769, Active Motif), and 6mA (Catalog No. 61496, Active Motif).
All data are expressed as the mean ± SE. GraphPad Prism (GraphPad Software Inc.) was used for statistical analyses. Unpaired student’s t-test was used to determine the differences between two groups with at least three replicates. P < 0.05 was considered statistically significant.
To determine the effects of AlCl3 on the proliferation of aNSCs, aNSCs were exposed to AlCl3 for 48 h, and BrdU was administered at 5 μmol/L for 8 h followed by immunofluorescence staining (Supplementary Figure 1A). The quantification results showed that the number of BrdU positive (BrdU+) cells did not show a significant difference between control and AlCl3 exposure aNSCs (Supplementary Figure 1B), suggesting that AlCl3 does not affect the proliferation of aNSCs.
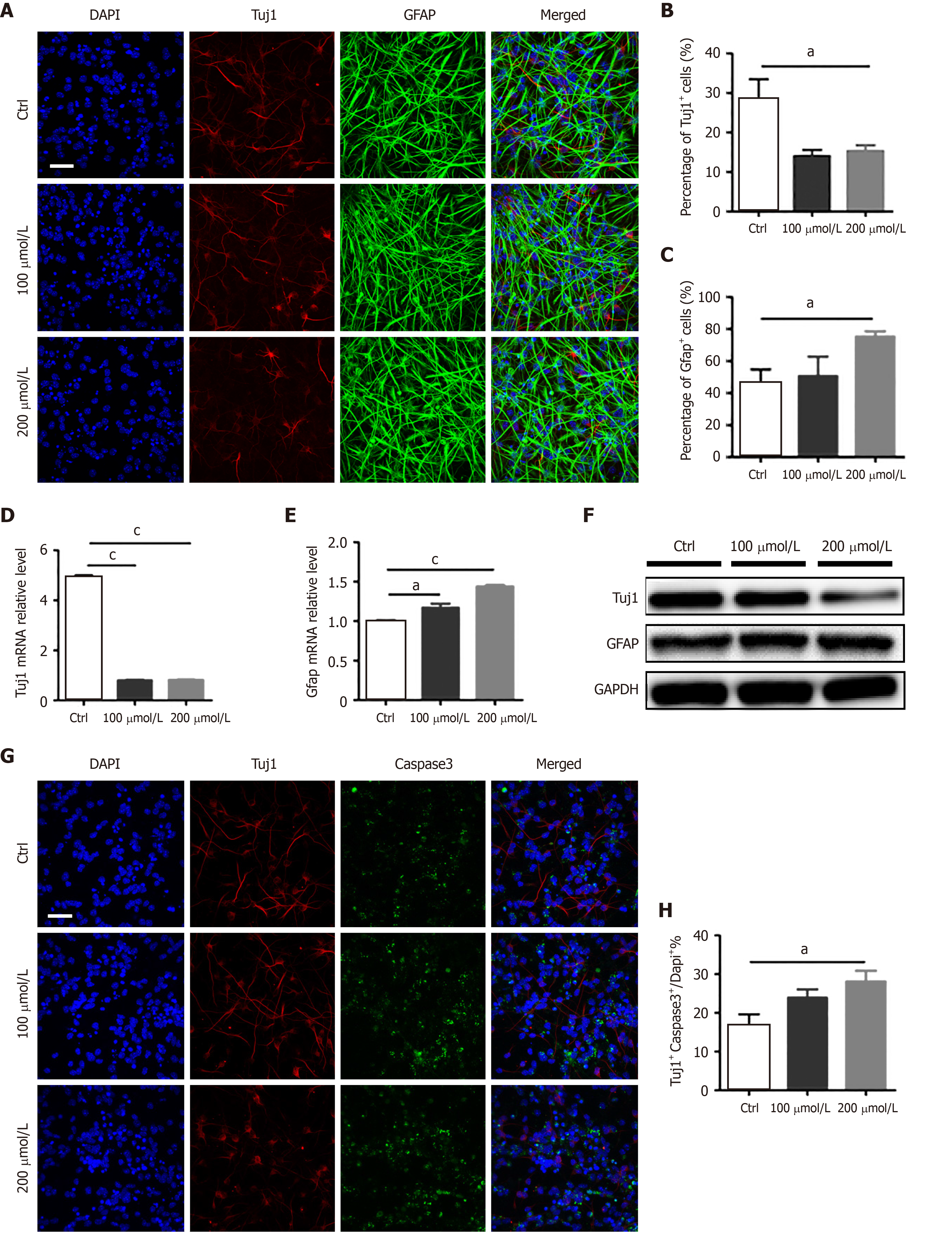
To examine the effects of AlCl3 on the differentiation of aNSCs, aNSCs were exposed to AlCl3 for 2 d and then underwent differentiation induction. Immunostaining for neuronal cell marker Tuj1 and astrocyte marker GFAP was performed (Figure 1A). The quantification results of immunofluorescence staining showed that the number of neuronal marker Tuj1 positive cells was significantly decreased (Figure 1B), but the number of glial cell marker GFAP positive cells increased after AlCl3 application (200 μmol/L) (Figure 1C). We also detected the expression levels of Tuj1 and GFAP by qRT-PCR and Western blot, and we found that the level of Tuj1 decreased, while the level of GFAP increased (Figure 1D-F). Taken together, these results suggest that AlCl3 regulates the differentiation of aNSCs.
To determine whether AlCl3 affects the survival of newborn neurons, we performed immunofluorescence staining for Tuj1 and caspase 3. Representative images (Figure 1G) and quantification results (Figure 1H) show that AlCl3 at a dosage of 200 μmol/L significantly increased the number of Caspase3 positive cells, suggesting that AlCl3 exposure induces apoptosis of newborn neurons.
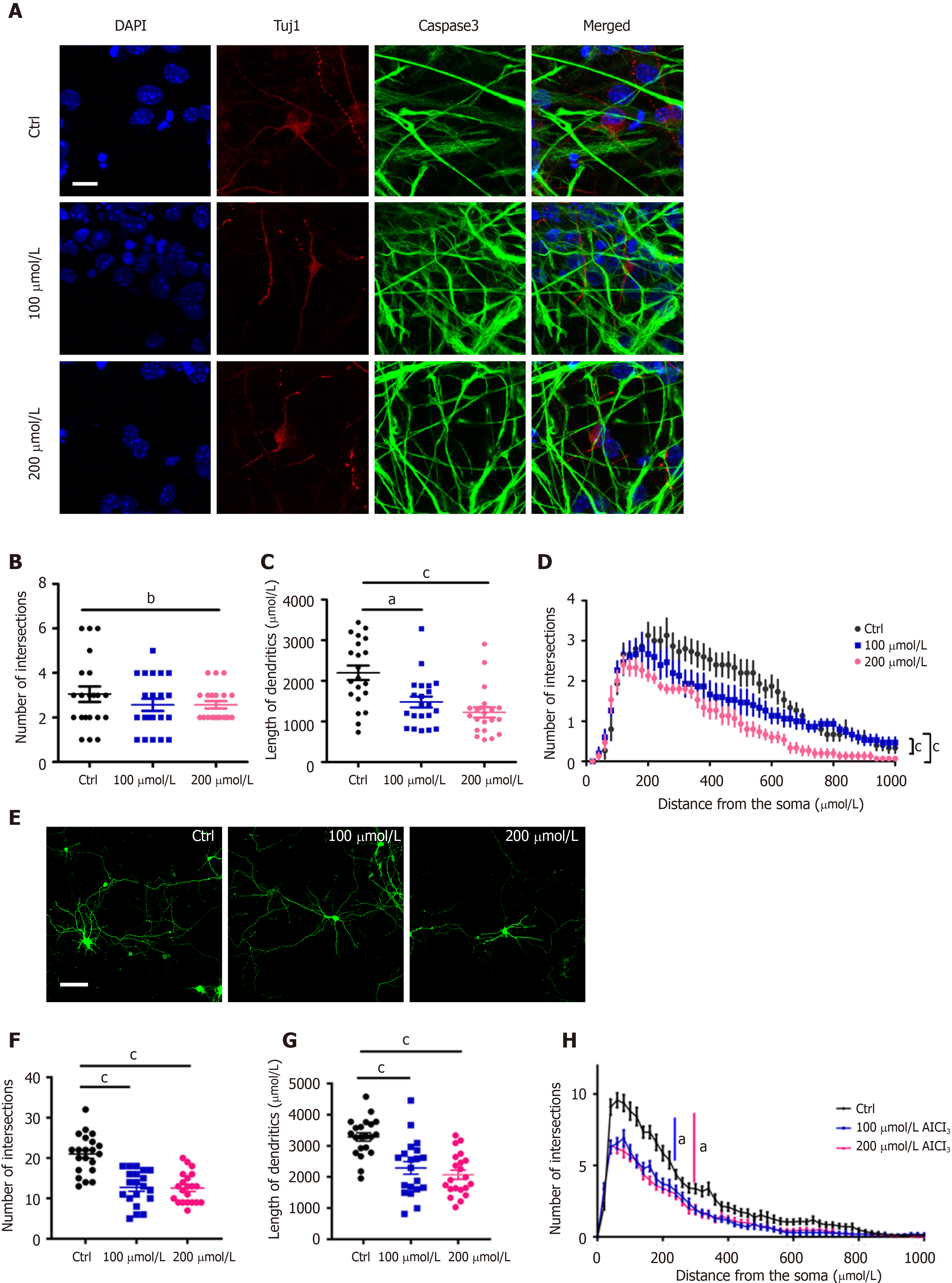
Next, we aimed to investigate whether AlCl3 regulates neuronal development. We first analyzed the effects of AlCl3 on the development of newborn neurons generated upon the differentiation of aNSCs. Immunostaining (Figure 2A) and Sholl analysis showed that AlCl3 at a dosage of 200 μmol/L significantly decreased dendritic length and the number of intersections (Figure 2B-D).
Next, we isolated neurons from the hippocampal tissues of embryonic mice and examined the effects of AlCl3 on the development of primary neurons. Immunofluorescence (Figure 2E) and Sholl analysis showed that AlCl3 exposure at both 100 μmol/L and 200 μmol/L dosages significantly decreased the intersection number and dendritic length of hippocampal neurons (Figure 2F-H). Collectively, these results indicate that AlCl3 exposure inhibits neuronal development.
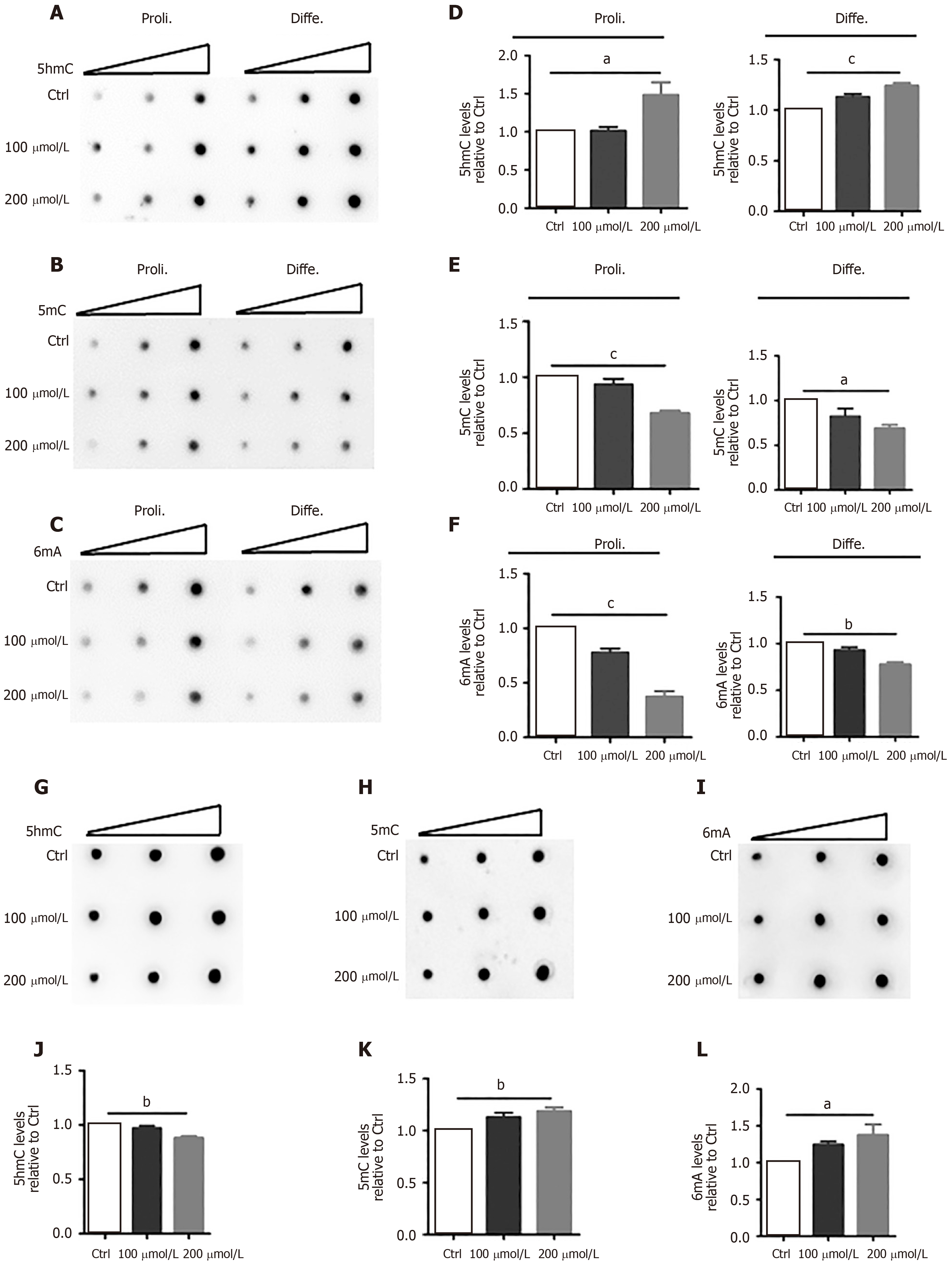
Previous studies have shown the important function of DNA modifications in neurogenesis and neuronal development[22,23]. To dissect the molecular mechanisms by which AlCl3 regulates neuronal development, we first analyzed the effects of AlCl3 on the DNA modifications of aNSCs and neurons. DNA dot-blots and quantification results showed that AlCl3 exposure increased the global level of 5-hmC in proliferating and differentiated aNSCs, but AlCl3 exposure decreased the global levels of 5-mC and 6mA (Figure 3A-F).
Next, we analyzed the effects of AlCl3 on the DNA modifications in neurons. DNA dot-blot and quantification results showed that AlCl3 exposure increased the global levels of 5-mC and 6mA but decreased the global level of 5-hmC in hippocampal neurons (Figure 3G-L). Taken together, these results indicate that AlCl3 alters the epigenetic state in aNSCs and neurons.
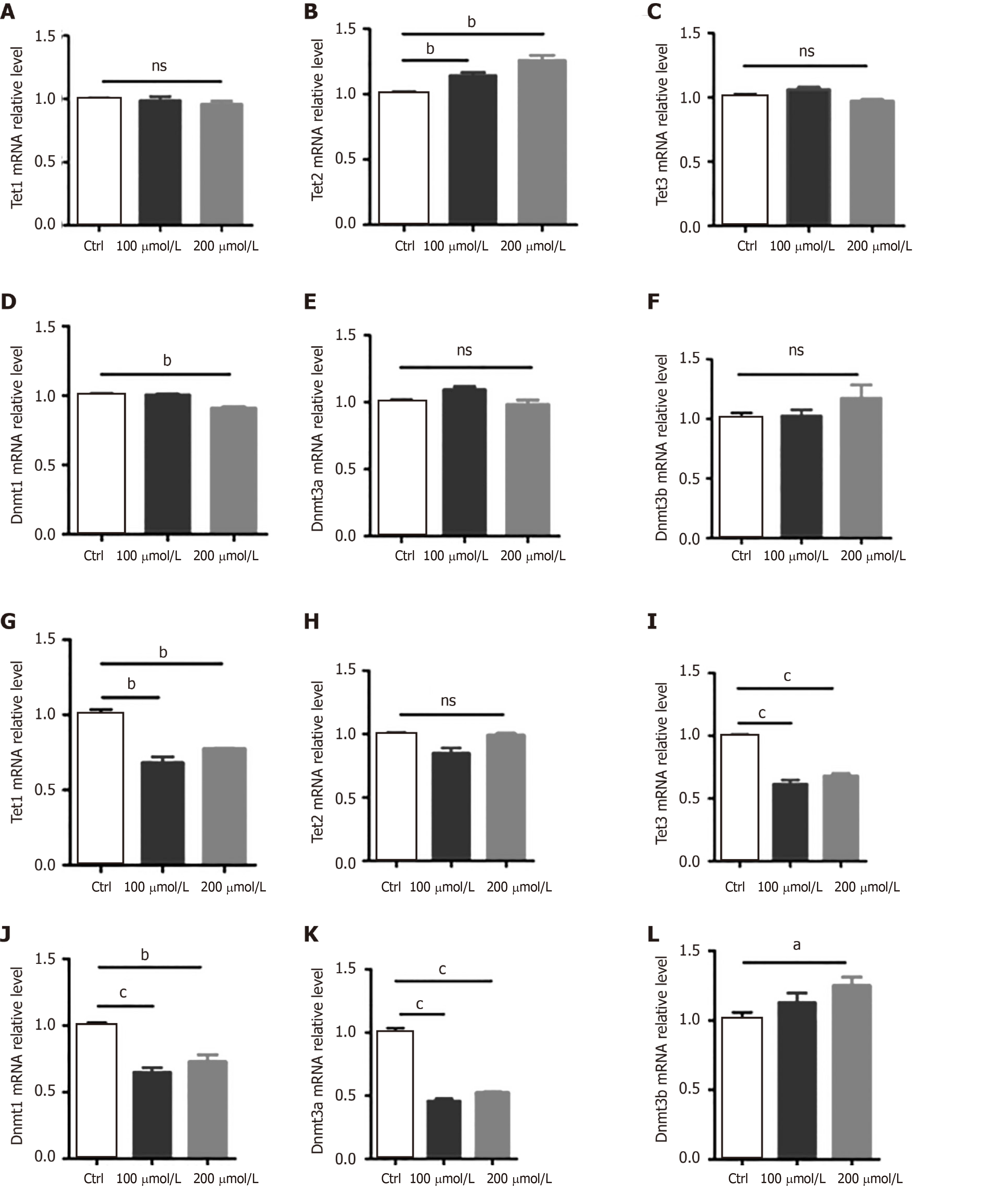
Next, we examined the expression of genes related to DNA modifications. We found that AlCl3 exposure significantly increased the mRNA level of Tet2 but did not affect the mRNA levels of Tet1 and Tet3 in aNSCs (Figure 4A-C). Meanwhile, AlCl3 exposure significantly decreased the mRNA level of DNA methyltransferases Dnmt1 (Figure 4D) but did not affect the levels of Dnmt3a and Dnmt3b (Figure 4E and F).
We then aimed to determine the effects of AlCl3 on the expression of Tets and DNMTs in neurons. We found that AlCl3 exposure decreased the mRNA levels of Tet1 and Tet3 while not affecting the level of Tet2 (Figure 4G-I) in neurons. Furthermore, AlCl3 exposure led to a decrease in hte mRNA levels of Dnmt1 and Dnmt3a but induced an increase in Dnmt3b (Figure 4J-L). Taken together, these results suggest that AlCl3 differentially regulates the expression of genes relating to DNA methylation and demethylation in aNSCs and neurons.
Previous studies have shown that DNA modifications play an important role in neuronal development and function and that dysregulation of DNA modifications is involved in neurological disorders[24-26]. Machineries of regulating DNA modifications have been identified. In the present study, we found that aluminum inhibits the differentiation of aNSCs and the development of neurons. Furthermore, aluminum can induce apoptosis of newborn neurons derived upon the differentiation of aNSCs. Mechanistically, aluminum affects the global level of 5mC, 5hmC, and 6mA in aNSCs and neurons by regulating the expression of DNA modification associated genes including Tets and DNMTs. Taken together, our results reveal a novel mechanism for regulating adult neurogenesis.
In adult mammalian brain, two regions, the subventricular zone in the lateral ventricle and subgranular zone in the dentate gyrus of hippocampus, maintain the neurogenic capacity[27]. Adult neurogenesis is driven by aNSCs and regulated by multiple mechanisms including environmental stimuli, genetics, and epigenetics including DNA modifications[22,27,28]. Our results show that aluminum can affect the differentiation of aNSCs and induce apoptosis of newborn neurons. Therefore, our study reveals the roles of aluminum in regulating neuronal development and associated mechanisms.
DNA modifications are regulated by diverse factors, such as environmental stimuli and food nutrients. Nutrient Vitamin C can serve as a cofactor for Tet and improves the reprogramming and neuronal differentiation by enhancing the expression level of Tets and therefore increasing the global level of 5hmC[29,30]. As one type of environmental pollution, the excessive intake of aluminum could induce inflammatory responses and oxidative stress, and then cause toxic effects on the neural, immune, and reproductive systems. Aluminum exposure also increases apoptosis and impairs learning and memory in adult rats[31]. These findings indicate the crosstalk between environmental signal and epigenetic modifications.
In summary, our findings show the neurotoxic effect of aluminum on neuronal development. One limitation of the present study is that the data were collected in vitro. A further study should be performed to examine the effects of aluminum on neuronal development and DNA modifications in vivo.
With the industrial development of society, environmental pollution is becoming a serious challenge for humans. Previous studies have revealed the crosstalk between environment and epigenetics and consequent phenotypes.
Aluminum pollution is a common issue and its exposure induces neurotoxic effects and impairs neuronal development and cognitive function.
To study the effects of aluminum on epigenetics in the context of neuronal development.
Neural stem cells were isolated from the brain of adult mice. Hippocampal neurons were isolated from the brain of embryonic mouse pups. The levels of DNA modifications were detected by dot-blot. The levels of DNA modification related genes were examined by qRT-PCR.
Our present findings uncovered the roles of aluminum in inhibiting neuronal development and promoting cell death. Our results also showed that aluminum exposure can display significant effects on DNA modifications.
Our study indicated that aluminum exposure regulates neuronal development by modulating DNA modifications.
Future studies should be performed to examine whether DNA modification could be a target for the treatment of neurological disorders induced by aberrant neuronal development.
We thank Dr. Dorazio RM for editing the manuscript. We thank Li YZ (Li Yanze) for narrating the core tip.
| 1. | Li X, Zhao X. Epigenetic regulation of mammalian stem cells. Stem Cells Dev. 2008;17:1043-1052. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 56] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 2. | Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5074] [Cited by in RCA: 4953] [Article Influence: 206.4] [Reference Citation Analysis (0)] |
| 3. | LaPlant Q, Vialou V, Covington HE 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iñiguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolaños CA, Kabbaj M, Xiao G, Neve RL, Hurd YL, Oosting RS, Fan G, Morrison JH, Nestler EJ. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137-1143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 524] [Cited by in RCA: 477] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 4. | Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423-430. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 861] [Cited by in RCA: 790] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 5. | Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y, Song H. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345-1351. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 494] [Cited by in RCA: 516] [Article Influence: 34.4] [Reference Citation Analysis (10)] |
| 6. | Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929-930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2321] [Cited by in RCA: 2074] [Article Influence: 122.0] [Reference Citation Analysis (0)] |
| 7. | Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930-935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4860] [Cited by in RCA: 4478] [Article Influence: 263.4] [Reference Citation Analysis (0)] |
| 8. | Zhang RR, Cui QY, Murai K, Lim YC, Smith ZD, Jin S, Ye P, Rosa L, Lee YK, Wu HP, Liu W, Xu ZM, Yang L, Ding YQ, Tang F, Meissner A, Ding C, Shi Y, Xu GL. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell Stem Cell. 2013;13:237-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 291] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 9. | Li X, Yao B, Chen L, Kang Y, Li Y, Cheng Y, Li L, Lin L, Wang Z, Wang M, Pan F, Dai Q, Zhang W, Wu H, Shu Q, Qin Z, He C, Xu M, Jin P. Ten-eleven translocation 2 interacts with forkhead box O3 and regulates adult neurogenesis. Nat Commun. 2017;8:15903. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 77] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 10. | Hahn MA, Qiu R, Wu X, Li AX, Zhang H, Wang J, Jui J, Jin SG, Jiang Y, Pfeifer GP, Lu Q. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep. 2013;3:291-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 364] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 11. | Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming GL, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086-1093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 323] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 12. | Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai LH. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109-1122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 385] [Cited by in RCA: 350] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 13. | Zhu X, Girardo D, Govek EE, John K, Mellén M, Tamayo P, Mesirov JP, Hatten ME. Role of Tet1/3 Genes and Chromatin Remodeling Genes in Cerebellar Circuit Formation. Neuron. 2016;89:100-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 65] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 14. | Yao B, Li Y, Wang Z, Chen L, Poidevin M, Zhang C, Lin L, Wang F, Bao H, Jiao B, Lim J, Cheng Y, Huang L, Phillips BL, Xu T, Duan R, Moberg KH, Wu H, Jin P. Active N6-Methyladenine Demethylation by DMAD Regulates Gene Expression by Coordinating with Polycomb Protein in Neurons. Mol Cell 2018; 71: 848-857. e6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 15. | Yao B, Cheng Y, Wang Z, Li Y, Chen L, Huang L, Zhang W, Chen D, Wu H, Tang B, Jin P. DNA N6-methyladenine is dynamically regulated in the mouse brain following environmental stress. Nat Commun. 2017;8:1122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 171] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 16. | Liaquat L, Sadir S, Batool Z, Tabassum S, Shahzad S, Afzal A, Haider S. Acute aluminum chloride toxicity revisited: Study on DNA damage and histopathological, biochemical and neurochemical alterations in rat brain. Life Sci. 2019;217:202-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 75] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 17. | Prakash A, Shur B, Kumar A. Naringin protects memory impairment and mitochondrial oxidative damage against aluminum-induced neurotoxicity in rats. Int J Neurosci. 2013;123:636-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 56] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 18. | Haider S, Liaquat L, Ahmad S, Batool Z, Siddiqui RA, Tabassum S, Shahzad S, Rafiq S, Naz N. Naringenin protects AlCl3/D-galactose induced neurotoxicity in rat model of AD via attenuation of acetylcholinesterase levels and inhibition of oxidative stress. PLoS One. 2020;15:e0227631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 19. | Praticò D, Uryu K, Sung S, Tang S, Trojanowski JQ, Lee VM. Aluminum modulates brain amyloidosis through oxidative stress in APP transgenic mice. FASEB J. 2002;16:1138-1140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 181] [Cited by in RCA: 188] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 20. | Zaky A, Mohammad B, Moftah M, Kandeel KM, Bassiouny AR. Apurinic/apyrimidinic endonuclease 1 is a key modulator of aluminum-induced neuroinflammation. BMC Neurosci. 2013;14:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 21. | Zaky A, Bassiouny A, Farghaly M, El-Sabaa BM. A Combination of Resveratrol and Curcumin is Effective Against Aluminum Chloride-Induced Neuroinflammation in Rats. J Alzheimers Dis. 2017;60:S221-S235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 42] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 22. | Yao B, Christian KM, He C, Jin P, Ming GL, Song H. Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci. 2016;17:537-549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 308] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 23. | Sun W, Guan M, Li X. 5-hydroxymethylcytosine-mediated DNA demethylation in stem cells and development. Stem Cells Dev. 2014;23:923-930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 24. | Sun W, Zang L, Shu Q, Li X. From development to diseases: the role of 5hmC in brain. Genomics. 2014;104:347-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 74] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 25. | Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33 Suppl:245-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4367] [Cited by in RCA: 4320] [Article Influence: 187.8] [Reference Citation Analysis (0)] |
| 26. | Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol. 2012;74:41-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 164] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 27. | Christian KM, Song H, Ming GL. Functions and dysfunctions of adult hippocampal neurogenesis. Annu Rev Neurosci. 2014;37:243-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 310] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 28. | Hsieh J, Zhao X. Genetics and Epigenetics in Adult Neurogenesis. Cold Spring Harb Perspect Biol. 2016;8:a018911. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 29. | Chen J, Guo L, Zhang L, Wu H, Yang J, Liu H, Wang X, Hu X, Gu T, Zhou Z, Liu J, Liu J, Wu H, Mao SQ, Mo K, Li Y, Lai K, Qi J, Yao H, Pan G, Xu GL, Pei D. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat Genet. 2013;45:1504-1509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 247] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 30. | He XB, Kim M, Kim SY, Yi SH, Rhee YH, Kim T, Lee EH, Park CH, Dixit S, Harrison FE, Lee SH. Vitamin C facilitates dopamine neuron differentiation in fetal midbrain through TET1- and JMJD3-dependent epigenetic control manner. Stem Cells. 2015;33:1320-1332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 31. | Qin X, Li L, Nie X, Niu Q. Effects of Chronic Aluminum Lactate Exposure on Neuronal Apoptosis and Hippocampal Synaptic Plasticity in Rats. Biol Trace Elem Res. 2020;197:571-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Litofsky NS S-Editor: Yan JP L-Editor: Wang TQ P-Editor: Xing YX