Published online Mar 26, 2019. doi: 10.4252/wjsc.v11.i3.196
Peer-review started: November 15, 2018
First decision: December 10, 2018
Revised: December 19, 2018
Accepted: January 26, 2019
Article in press: January 26, 2019
Published online: March 26, 2019
Processing time: 132 Days and 21.1 Hours
Intervertebral disc (IVD) degeneration is a condition characterized by a reduction in the water and extracellular matrix content of the nucleus pulposus (NP) and is considered as one of the dominating contributing factors to low back pain. Recent evidence suggests that stromal cell-derived factor 1α (SDF-1α) and its receptor C-X-C chemokine receptor type 4 (CXCR4) direct the migration of stem cells associated with injury repair in different musculoskeletal tissues.
To investigate the effects of SDF-1α on recruitment and chondrogenic differentiation of nucleus pulposus-derived stem cells (NPSCs).
We performed real-time RT-PCR and enzyme-linked immunosorbent assay to examine the expression of SDF-1α in nucleus pulposus cells after treatment with pro-inflammatory cytokines in vitro. An animal model of IVD degeneration was established using annular fibrosus puncture in rat coccygeal discs. Tissue samples were collected from normal control and degeneration groups. Differences in the expression of SDF-1α between the normal and degenerative IVDs were analyzed by immunohistochemistry. The migration capacity of NPSCs induced by SDF-1α was evaluated using wound healing and transwell migration assays. To determine the effect of SDF-1α on chondrogenic differentiation of NPSCs, we conducted cell micromass culture and examined the expression levels of Sox-9, aggrecan, and collagen II. Moreover, the roles of SDF-1/CXCR4 axis in the migration and chondrogenesis differentiation of NPSCs were analyzed by immunofluorescence, immunoblotting, and real-time RT-PCR.
SDF-1α was significantly upregulated in the native IVD cells cultured in vitro with pro-inflammatory cytokines, such as interleukin-1β and tumor necrosis factor-α, mimicking the degenerative settings. Immunohistochemical staining showed that the level of SDF-1α was also significantly higher in the degenerative group than in the normal group. SDF-1α enhanced the migration capacity of NPSCs in a dose-dependent manner. In addition, SDF-1α induced chondrogenic differentiation of NPSCs, as evidenced by the increased expression of chondrogenic markers using histological and immunoblotting analyses. Real-time RT-PCR, immunoblotting, and immunofluorescence showed that SDF-1α not only increased CXCR4 expression but also stimulated translocation of CXCR4 from the cytoplasm to membrane, accompanied by cytoskeletal rearrangement. Furthermore, blocking CXCR4 with AMD3100 effectively suppressed the SDF-1α-induced migration and differentiation capacities of NPSCs.
These findings demonstrate that SDF-1α has the potential to enhance recruitment and chondrogenic differentiation of NPSCs via SDF-1/CXCR4 chemotaxis signals that contribute to IVD regeneration.
Core tip: The chemokine stromal cell-derived factor-1α (SDF-1α) is upregulated in the pro-inflammatory microenvironment or degenerative intervertebral disc (IVD). The present study demonstrates for the first time that SDF-1α effectively promotes migration and chondrogenic differentiation of nucleus pulposus-derived stem cells (NPSCs), which serve as endogenous progenitor/stem cells residing in the IVD. In addition, SDF-1/CXCR4 chemotaxis signals may play a crucial role in the process of recruitment and chondrogenic differentiation of NPSCs that are involved in spontaneous IVD regeneration in the early stage of IVD degeneration.
- Citation: Ying JW, Wen TY, Pei SS, Su LH, Ruan DK. Stromal cell-derived factor-1α promotes recruitment and differentiation of nucleus pulposus-derived stem cells. World J Stem Cells 2019; 11(3): 196-211
- URL: https://www.wjgnet.com/1948-0210/full/v11/i3/196.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v11.i3.196
Intervertebral disc (IVD) degeneration is characterized by a gradual loss of cellular number and function, and a related breakdown of extracellular matrix[1,2]. This pathogenic process is one of the most common causes of low back pain (LBP)[3,4], resulting in an economic burden and reducing the quality of life of the patients[5]. Recently, transplantation of mesenchymal stem cells (MSCs) has emerged as one of the most promising therapeutic strategies for IVD degeneration[6,7]. Evidence from several studies has indicated that the regenerative effects of transplanted MSCs depend on the potential for differentiation toward nucleus pulposus (NP)-like cells, and the immunomodulatory and immunosuppressive properties via paracrine secretions[8,9]. In fact, the harsh IVD microenvironment negatively influences the survival and function of the transplanted MSCs, impairing their repair potential[10,11]. Moreover, the limited cell source, cell leakage, and osteophyte formation represent major obstacles to clinical applications of MSCs for IVD regeneration[12-14].
The release of cytokines and chemokines in response to cell or tissue damage has been shown to be involved in regulation of mobilization, trafficking, and homing of stem/progenitor cells, with the potential to facilitate tissue repair[15]. Stromal cell-derived factor-1α (SDF-1α, also known as C-X-C motif chemokine 12, CXCL12) is a potent chemoattractant cytokine, with a key role in the recruitment, proliferation, and differentiation of stem/progenitor cells through binding to its G-protein coupled transmembrane receptor, C-X-C chemokine receptor type 4 (CXCR4)[16,17]. It also has been reported that SDF-1α and its receptor CXCR4 are upregulated in the process of IVD degeneration[18,19]. Increased levels of SDF-1α in certain pathological situations can attract endogenous MSCs into the injured site, contributing to tissue repair in situ[20]. The hyaluronan-based delivery of SDF-1α has been proved to significantly boost the recruitment of MSCs into the degenerative IVD in organ culture ex vivo[21]. Sakai et al[22] provided evidence of elevated recruitment of labeled MSCs into the degenerative IVD in vivo using a mouse loop-disc degeneration model. It has also been shown that SDF-1α is capable of inducing osteogenic or chondrogenic differentiation of MSCs[23-26]. Therefore, in addition to migration, SDF-1/CXCR4 signaling might increase differentiation of endogenous progenitor/stem cells.
Although cell homing from an outer pool of progenitor cells may potentiate new therapeutic approaches, recruitment of circulating MSCs to the central IVD for regeneration purposes appears challenging owing to its avascular nature. Accumulating evidence indicates that progenitor/stem cells, which are present in different regions of the healthy and degenerative IVD and have the capacity for multilineage differentiation, have regenerative potential for tissue regeneration[27-29]. Therefore, activation and mobilization of these endogenous progenitor cell populations within the IVD represent an attractive target for future regenerative strategies for IVD degeneration in situ[11].
In the present study, we investigated whether upregulation of SDF-1α in the degenerative IVD could potentiate endogenous regeneration by regulating recruitment and differentiation of NP-derived stem cells (NPSCs) residing in the IVD stem niches. Thus, our study sheds light on the SDF-1/CXCR4 axis by which NPSCs are recruited into the injury sites of the degenerative IVD, enhancing the efficiency of stem cell-based therapies for IVD regeneration in situ.
All experiments and surgical procedures were performed in accordance with the Southern Medical University guidelines and approved by the Laboratory Animal Ethics Committee of PLA Navy General Hospital. Surgical procedures were performed on ten adult male Sprague-Dawley (SD) rats (3 mo old, weighing 200-250 g). All rats were housed in a stable animal care room with suitable temperature and humidity and a 12/12 h light/dark cycle, and had free access to food and water throughout the experiment.
The rats were euthanized by an intraperitoneal overdose injection of 10% chloral hydrate (Sigma Aldrich, St. Louis, MO, United States), and the gelatinous NP tissue was gently separated from surrounding tissues under a stereoscopic microscope in sterile conditions. After rinsing in phosphate-buffered saline (PBS) containing 200 units/mL penicillin and 200 µg/mL streptomycin (2% Pen/Strep; Solarbio, Beijing, China) three times, the NP tissue was minced into 1 mm3 pieces and then digested with 0.2% collagenase type II (Sigma Aldrich) for 4 h at 37 °C. The mixture was filtered through a 40 µm strainer and re-suspended in Low Glucose-Dulbecco's Modified Eagle Medium (LG-DMEM; Hyclone, Logan, Utah, United States) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, United States) to inactivate collagenase. After centrifugation at 800 g for 5 min, the supernatant was removed and the remaining cells were re-suspended in LG-DMEM with 10% FBS and 1% Pen/Strep. Single-cell suspension was then seeded into 25-cm2 flasks (Costar, Cambridge, MA, United States) and cultured at 37 °C and 5% CO2 in a humidified incubator (Thermo Fisher Scientific, Waltham, MA, United States).
To isolate NPSCs, a previously described method of differential adhesion was used[30]. Briefly, the culture medium with non-adherent cells and fragments was discarded, and the adherent cells were supplemented with the complete medium after 24 h. Thereafter, the culture medium was replaced every three days. When cell colony-forming units reached 80%–90% confluence, the cells were digested with 0.05%/0.02% trypsin-EDTA (Gibco), passaged at a ratio of 1:3, and cultured in the complete medium.
The extraction process of nucleus pulposus cells (NPCs) was similar to the method used for NPSCs isolation. The primary NPCs were cultured in LG-DMEM supplemented with 10% FBS and 1% Pen/Strep at 37 °C in a humidified atmosphere containing 5% CO2. The complete medium was changed every three days. The cells were passaged by 0.05%/0.02% trypsin-EDTA treatment at approximately 80% confluence. Cells at passages (P) 3-5 were used for subsequent experiments.
Specific surface markers: According to the basic criteria for the definition of MSCs proposed by the International Society for Cellular Therapy, the expression of specific surface markers like CD34, CD45, CD73, CD90, and CD105 was evaluated for NPSCs in this study[31]. About 2 × 105 cells were washed and then resuspended with 100 µL PBS to produce a single cell solution. Then, NPSCs were incubated with fluorescein isothiocyanate (FITC) or phycoerythrin (PE)-conjugated antibodies CD34, CD105 (Abcam, Cambridge, MA, United States), CD45, CD73, and CD90 (BD Pharmingen, San Diego, CA, United States) in the dark at room temperature for 30 min. FITC/PE-conjugated non-specific mouse IgG (Invitrogen, Carlsbad, CA, United States) was used as a negative control. Finally, labeled cells were rinsed three times with PBS and resuspended in 500 µL PBS, and surface marker expression was analyzed using flow cytometry (BD Biosciences, United States).
Multilineage differentiation: For osteogenic differentiation, P3 NPSCs at 80%–90% confluence were cultured in Osteogenic Differentiation Medium (Cyagen Biosciences, Guangzhou, China) containing 10% FBS, 1% penicillin-streptomycin, 0.01% dexamethasone, 1% β-glycerophosphate, and 0.2% ascorbate, which was changed every three days. After 3 wk, the calcified deposits in the extracellular matrix were assessed by staining with Alizarin Red (Cyagen Biosciences).
For adipogenic differentiation, when P3 NPSCs reached approximately 100% confluence, the medium was replaced by Adipogenic Differentiation Induction Medium (Cyagen Biosciences) containing 10% FBS, 1% penicillin-streptomycin, 1% glutamine, 0.2% insulin, 0.1% IBMX, 0.1% rosiglitazone, and 0.1% dexametha-sone. Three days later, the medium was changed to Adipogenic Differentiation Maintenance Medium (Cyagen Biosciences) containing 10% FBS, 1% penicillin-streptomycin, 1% glutamine, and 0.2% insulin. After 24 h, the medium was changed back to the induction medium. The cycle of induction/maintenance was repeated four times before the cells were cultured in the maintenance medium for additional seven days. To confirm triglyceride deposits, the induced cells were finally fixed with 4% paraformaldehyde and stained with Oil Red O (Cyagen Biosciences).
For chondrogenesis, cell micromass culture was performed as previously[32,33]. In brief, cells were suspended to a concentration of 1 × 106/mL in serum-free DMEM, and 200 µL aliquots (2 × 105 cells) were added to a 15 mL polypropylene culture tube (Costar), and then centrifuged at 800 g for 5 min. The supernatant was aspirated and replaced with Complete Chondrogenic Medium (Cyagen Biosciences) containing 0.01% dexamethasone, 0.3% ascorbate, 1% ITS+ Supplement, 0.1% sodium pyruvate, 0.1% proline, and 1% transforming growth factor (TGF)-β1. The cells formed free-floating aggregates within the first 24 h. The medium was changed every three days. After 4 wk, chondrogenic pellets were fixed with 4% paraformaldehyde and embedded in paraffin for Alcian blue (Cyagen Biosciences) staining to evaluate the synthesis of proteoglycans.
Colony-forming assay: To evaluate the colony-forming ability of NPSCs, single-cell suspension at a density of 1000 cells/10 cm2 was seeded into a 6-well plate (Costar). After seven days, cells were fixed with 4% paraformaldehyde and then stained with 0.1% crystal violet (Solarbio). Aggregates of more than 50 cells were identified as colonies.
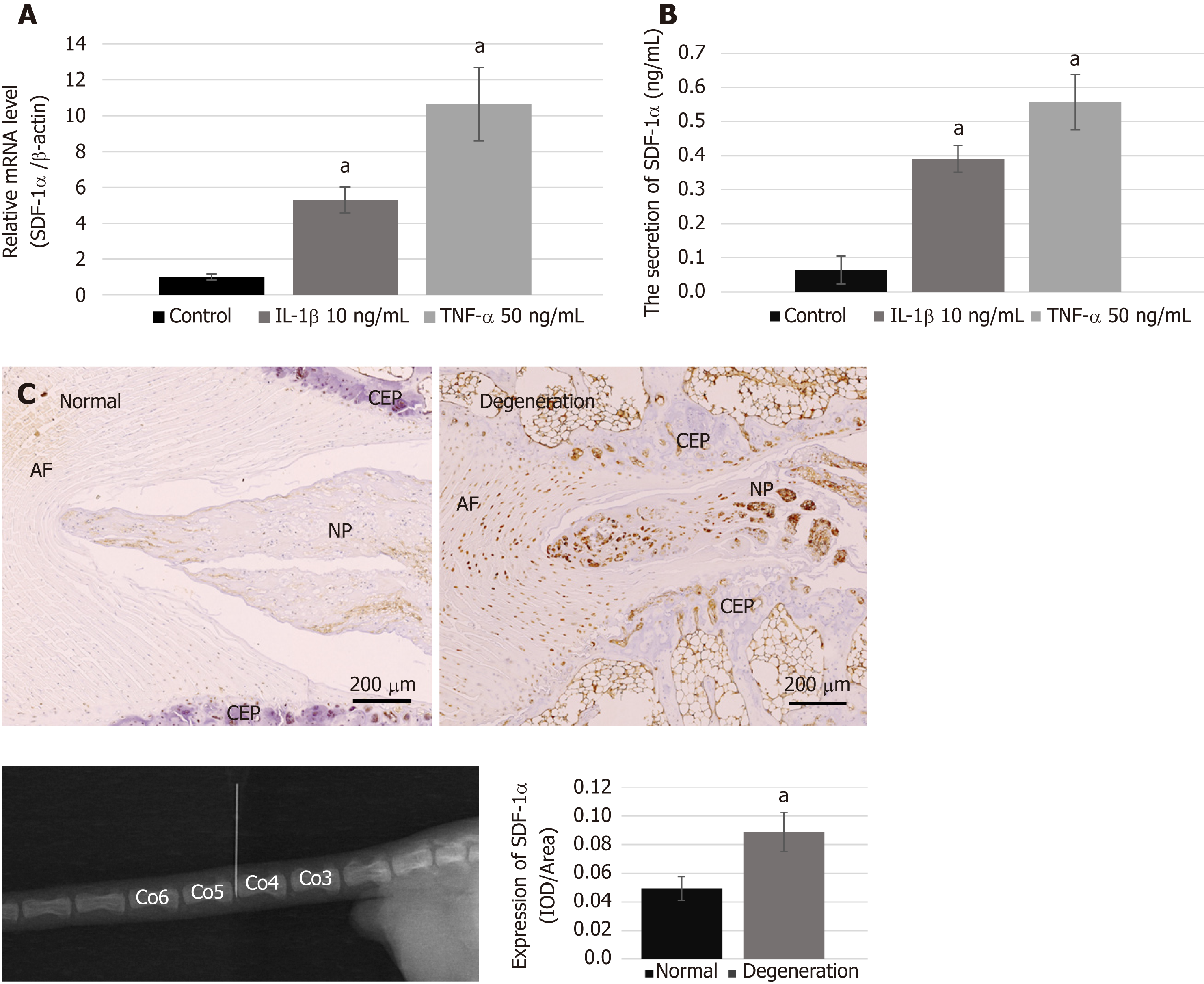
NPCs (1 × 105 cells/well) were plated in 6-well plates and cultured in complete medium for one day. To simulate the pro-inflammatory and poor nutrition microenvironment of the degenerative disc, the medium was then changed to serum-free medium containing LG-DMEM and 10 ng/mL interleukin (IL)-1β (PeproTech, Rocky Hill, NJ, United States) or 50 ng/mL tumor necrosis factor-α (TNF-α) (PeproTech). After incubation for 48 h, the protein level of SDF-1α secreted in the cell culture supernatant was measured with an enzyme-linked immunosorbent assay (ELISA) kit (USCN, Wuhan, China), according to the manufacturer’s instructions. In addition, the remaining cells were used for extracting total RNA using TRIzol reagent (Invitrogen, Carlsbad, CA, United States). The amount of synthesized mRNA of SDF-1α was then determined by real-time RT-PCR.
In the present study, needle puncture in the tail disc was used to induce a disc degeneration model. The procedure was performed as previously described[34]. Briefly, rats were anesthetized intraperitoneally with 10% chloral hydrate (Sigma Aldrich) at a dose of 3.5 mL/kg under sterile conditions. Two consecutive rat tail discs (Co4/Co5 and Co5/Co6) were randomly assigned to the control and degeneration groups. The disc selected for degeneration was punctured with a sterile 22-gauge needle from the dorsal to the ventral side (depth 5 mm, rotated 360°, and held for 5 s). After 4 wk, normal and degeneration-induced tail discs were harvested for histological analysis.
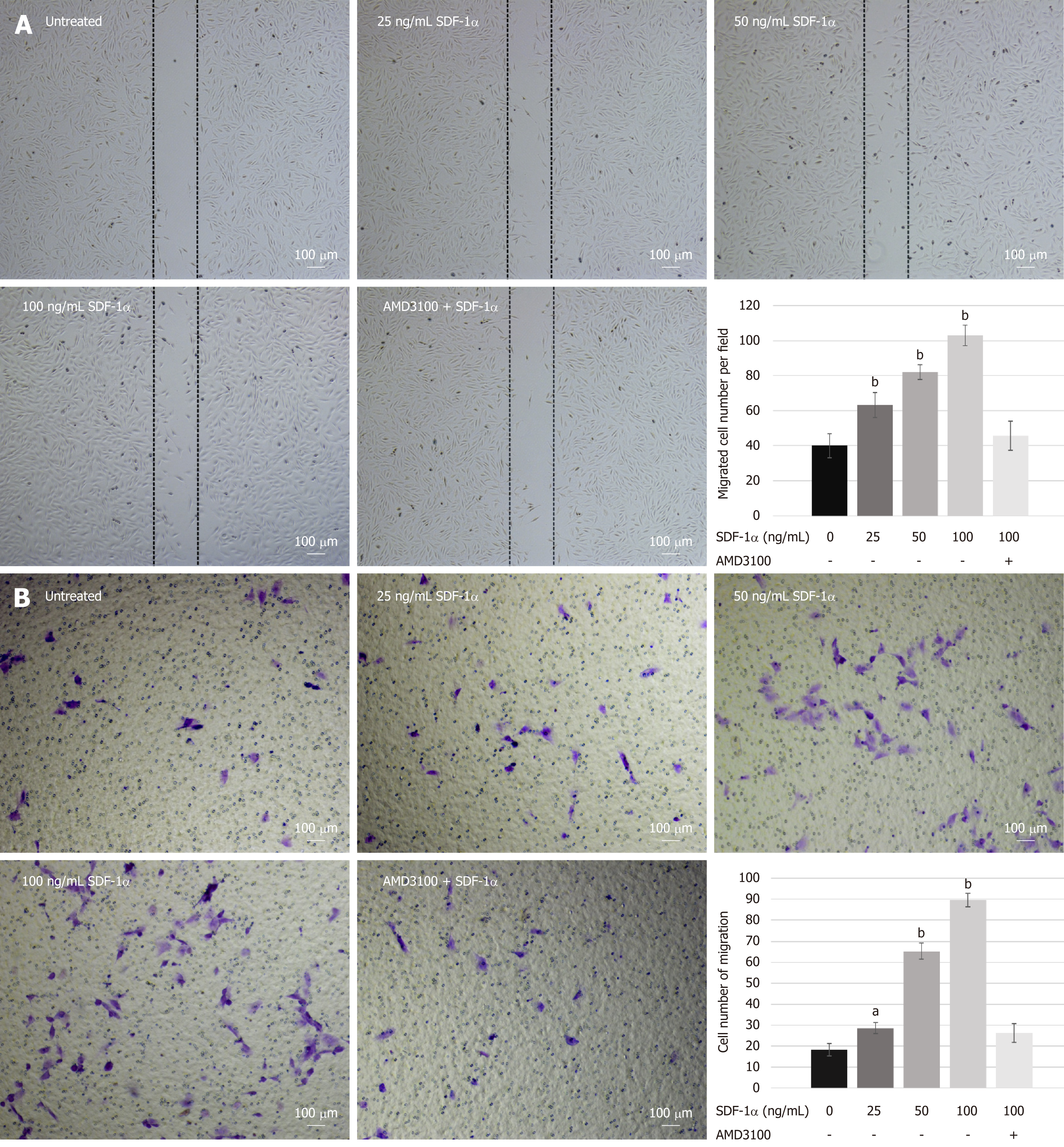
Wound healing assay: NPSCs were seeded into 6-well plates at a concentration of 1 × 105/mL and cultured until nearly 100% confluence. The straight wounds were created by scratching the monolayer with a 200 μL pipet tip. After washing with PBS twice, the cells were incubated with 0, 25, 50, and 100 ng/mL SDF-1α in serum-free medium for 12 h to allow migration back to the wound area. For the inhibition experiment, cells were pre-treated with 10 µg/mL AMD3100 for 2 h before incubation with 100 ng/mL SDF-1α. Cells from three random low-power fields of each group were counted to assess the average number of migrated cells.
Transwell migration assay: For transmigration assay, we used 24-well transwell chamber plates (pore size 8 μm; Costar). A total of 2 × 104 cells in 200 μL serum-free medium were added into the upper chamber, and 600 μL serum-free medium supplemented with different concentrations (0, 25, 50, and 100 ng/mL) of SDF-1α was placed into the lower chamber. For the inhibition experiment, cells were pre-incubated with 10 µg/mL AMD3100 for 2 h before being added into the upper chamber. After incubating for 12 h, the cells remained on the upside were gently removed with a cotton swab, and the cells adhered on the underside of the membrane were fixed and then stained with 0.1% crystal violet. The mean number of the migrated cells in three random high-power fields was counted under a light microscope (Olympus, Tokyo, Japan).
NPSCs were exposed to 0, 50, and 100 ng/mL SDF-1α for 72 h with or without pre-treatment with 10 µg/mL AMD3100 for 2 h. Total protein was extracted using PIPA lysis buffer (Applygen, Beijing, China) containing PMSF and then analyzed by immunoblotting. Total RNA extracted using TRIzol reagent was analyzed by real-time RT-PCR.
For aggregate chondrogenesis model, NPSCs were centrifuged into cell micromass and induced with chondrogenic medium as described above. To determine whether SDF-1α enhanced TGF-1β-induced chondrogenesis of NPSCs, SDF-1α was added into the Complete Chondrogenic Medium (Cyagen Biosciences). CXCR4 antagonist AMD3100 (10 µg/mL) was used for the inhibition assay. After 28 d, aggregates were collected for histological analysis, as described below.
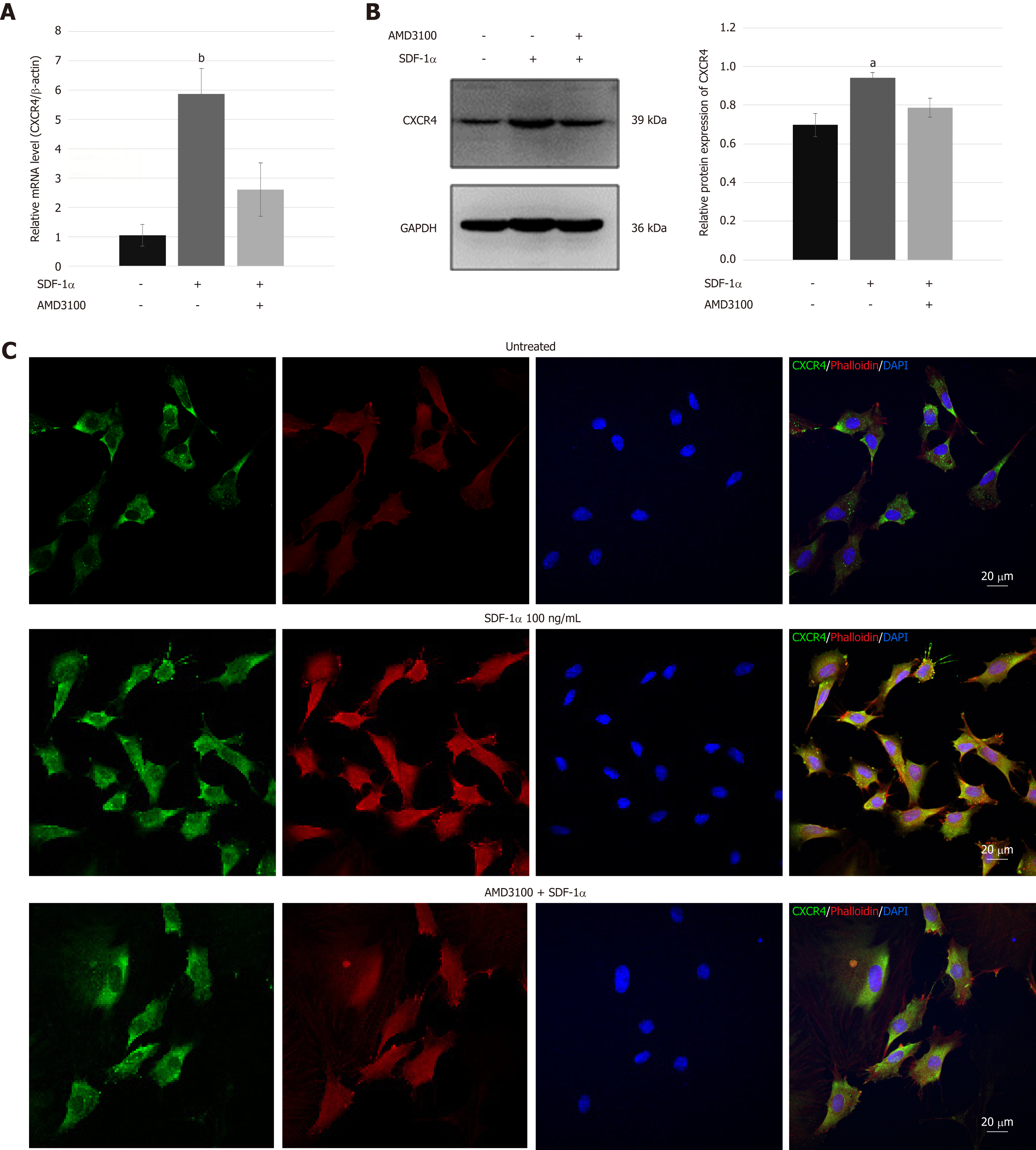
NPSCs were seeded on slides and treated with or without 100 ng/mL SDF-1α for 24 h. For inhibition, the cells were pre-treated with 10 µg/mL AMD3100 for 2 h before treatment with 100 ng/mL SDF-1α. Then, cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton-X-100 in PBS, and blocked with 10% goat serum. Subsequently, the cells were incubated with primary antibody rabbit-anti-rat CXCR4 (1:50 dilution; Abcam) at 4 °C overnight. After PBS washing, cells were incubated with FITC-conjugated goat-anti-rabbit secondary antibody (Abcam) and red fluorescence-conjugate phalloidin working solution using a CytoPainter F-actin staining kit (Abcam) at room temperature for 60 min before counterstaining with DAPI (Beyotime, Shanghai, China). Cells that were incubated only with secondary antibody served as the negative control. Images were obtained using a confocal laser scanning microscope (LSM780, Zeiss, Germany).
The specimens were fixed in 10% formaldehyde for 1 wk, decalcified in 0.05% EDTA for 4 wk, and then embedded in paraffin. Serial sagittal sections of discs (5 μm thickness) were prepared, deparaffinized in dimethylbenzene, and rehydrated, before being incubated with rabbit primary antibodies for SDF-1α (1:100 dilution; Cell Signaling Technology, MA, United States), collagen II (dilution 1:200; Abcam), and aggrecan (dilution 1:400; Bioss, Beijing, China) overnight at 4 °C. The next day, all sections were washed with PBS and incubated with secondary antibody horseradish peroxidase conjugated goat anti-rabbit IgG (1:200 dilution; Abcam) at 37 °C for 20 min, followed by color development with diaminobenzidine (DAB, Beyotime). The sections were then counterstained with hematoxylin (Beyotime), mounted on coverslips, and examined using a light microscope (Nikon, Japan). Integrated optical density, as the evaluation parameter for immunohistochemistry, was measured using Image-Pro Plus 6.0 software (Media Cybernetics, Silver Spring, MD, United States).
Total mRNA (1 µg) was used to synthesize cDNA using the GoScriptTM Reverse Transcription System (Promega, Madison, WI, United States) following the manufacturer’s instructions. Subsequently, a PCR assay was performed using a LightCycler® 480 real-time PCR system with SYBR Green Master I (Roche, Indianapolis, IN, United States). A 20 µL reaction volume was used. The reactions were run on a Peltier Thermal Cycler (Bio-Rad, California, United States) under the following conditions: 95 °C for 5 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 20 s. Custom-specific primers (Sangon Biotech, Shanghai, China) are described in Table 1. β-actin was employed as the internal reference gene. The relative gene expression was calculated using the 2–ΔΔCt method and was normalized to the gene expression of the control.
| Gene name | Forward sequence (5’-3’) | Reverse sequence (5’-3’) |
| SDF-1α | TTCTATTGAGGACTAGCACGTC | CTGTCCTAAGGAAAGGTAGGTG |
| CXCR4 | GCTCCAGAATGTGTGGTAAATC | CACCAAGCAAGTTTACCATTGA |
| Aggrecan | TATGATGTCTACTGCTACGTGG | GTAGAGGTAGACAGTTCTCACG |
| Collagen II | CTGGAGTCAAGGGTGATCG | CTCTCCTCTGTCTCCTTGTTTG |
| Sox-9 | GACAACTTTACCAGTTTCGGTC | GAGGGAAAACAGAGAACGAAAC |
| β-actin | GGAGATTACTGCCCTGGCTCCTA | GGAGATTACTGCCCTGGCTCCTA |
Protein samples were separated by 8% or 10% (wt/vol) sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) electrophoresis and transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, United States). After blocking with 5% non-fat milk in Tris-buffered saline with Tween 20 (TBS-T) for 2 h at 37 °C, the membranes were incubated with primary antibodies for CXCR4 (1:100 dilution), Sox-9 (1:1000 dilution; Abcam), aggrecan (1:1000 dilution), collagen II (1:1000 dilution), and GAPDH (1:10000 dilution; Abcam) overnight at 4 °C. The membranes were then incubated with horseradish peroxidase-conjugated secondary antibody (Abcam) for 1 h at room temperature. An electrochemiluminescence reagent was added to the surfaces of membranes, and immunolabeling was visualized using an ImageQuant LAS4000 (GE Healthcare, United States). The relative protein concentrations were measured using Image-Pro Plus 6.0 software. The expression level of GAPDH was used as an internal control.
All experiments were performed at least three times. The data are presented as the mean ± SD. All statistical analyses were performed using SPSS 22.0 software (Chicago, Illinois, United States). One-way analysis of variance (ANOVA) was conducted to analyze any significant differences among different groups, followed by the Student-Newman-Keul (homogeneity of variance) or Tamhane’s test (heterogeneity of variance) for post hoc multiple comparisons. P-values less than 0.05 were considered to indicate statistical significance.
Primary cultured cells adhered to the culture flask within 24 h, presenting a typical fibroblast-like morphology and vortex-like pattern (Figure 1A). The isolated P3 NPSCs showed colony-like growth after seven days of culture (Figure 1B). Multilineage differentiation assays revealed that the isolated cells had successfully differentiated into osteogenic, adipogenic, and chondrogenic lineages after specific differentiation induction (Figure 1C). In addition, we used flow cytometric analysis to examine a series of specific cell surface markers commonly used to identify MSCs. The cells were positive for widely used MSC markers CD73 and CD90, positive for CD105, which is expressed in certain populations of MSCs to serve as an auxiliary receptor for the TGF-β receptor complex, and negative for the hematopoietic markers CD45 and CD34 (Figure 1D).
We performed real-time RT-PCR and ELISA to examine the mRNA synthesis and protein secretion of the chemokine SDF-1α in NPCs after treatment with pro-inflammatory cytokines, and immunohistochemical analysis to compare the protein expression levels of SDF-1α between the degenerative group and the normal group. The data showed that the levels of mRNA and protein secretion of SDF-1α in NPCs were significantly increased in vitro after treatment with IL-1β and TNF-α (Figure 2A and B). In the animal experiment, immunohistochemical staining showed that the expression of SDF-1α in the degenerative IVD was also significantly higher than that in the control group (Figure 2C).
To investigate the effects of SDF-1α on NPSCs migration in vitro, we performed wound healing and transwell migration assays. SDF-1α significantly promoted NPSCs migration in a dose-dependent manner, whereas pretreatment with AMD3100 significantly suppressed the migration capacity that had been enhanced by SDF-1α (Figure 3).
Real-time RT-PCR and immunoblotting demonstrated that SDF-1α significantly increased the expression of CXCR4 that had been inhibited by AMD3100 (Figure 4A and B). As shown in Figure 4C, immunofluorescence assays showed that CXCR4 was co-expressed in the cell membrane and cytoplasm of NPSCs and translocated to the cell surface by the stimulation with SDF-1α. In addition, cytoskeleton organization associated with cell migration was analyzed by immunostaining of fibrous actin. We found that SDF-1α enhanced F-actin cytoskeletal remodeling and lamellipodia formation in NPSCs. However, AMD3100 was able to inhibit the expression and translocation of CXCR4, and cytoskeletal rearrangements of NPSCs.
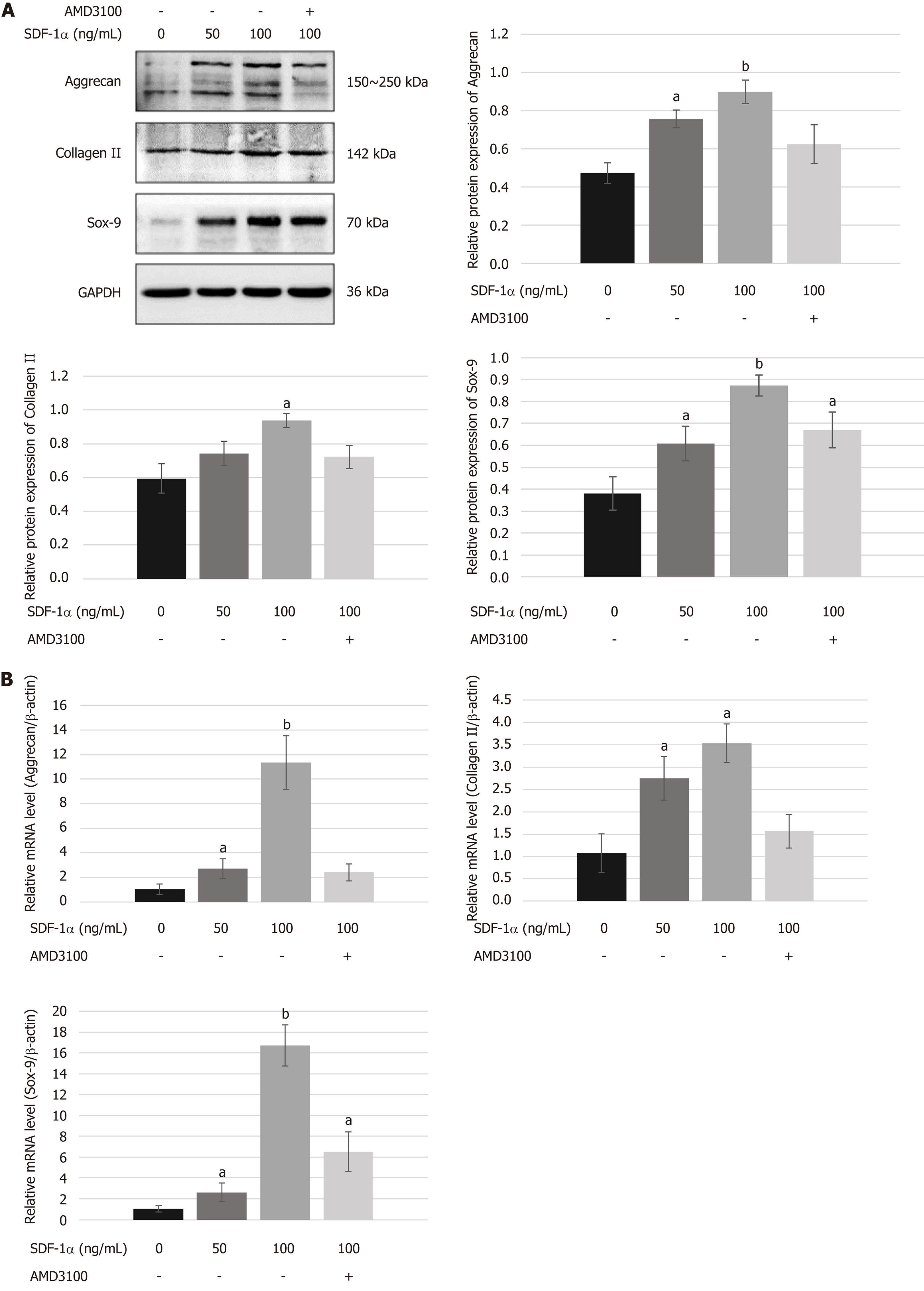
To determine the role of the SDF-1/CXCR4 axis in chondrogenic differentiation of NPSCs, we examined the mRNA and protein expression levels of chondrogenic markers in vitro. As shown in Figure 5, real-time RT-PCR and Western blot analyses revealed that treating NPSCs with SDF-1α significantly increased the mRNA and protein levels of aggrecan, collagen II, and Sox-9. However, pretreatment with AMD3100 effectively blocked the SDF-1α-induced expression of aggrecan and collagen II.
Using cell micromass culture, we examined the effects of SDF-1α on TGF-β1-induced chondrogenic differentiation of NPSCs. Cells within these aggregates were surrounded by a proteoglycan-rich extracellular matrix, as demonstrated by Alcian Blue staining. The weight and proteoglycan content of the aggregates increased significantly in the presence of SDF-1α, but this effect was counteracted by AMD3100 (Figure 6A and B). Immunohistochemical analysis further confirmed that the synthesis of collagen II and aggrecan at the protein level was increased after treatment with SDF-1α but blocked by AMD3100 (Figure 6C). This suggests that SDF-1α synergistically promotes TGF-β1-induced chondrogenic differentiation of NPSCs through the SDF-1/CXCR4 axis.
Stem cell therapy has been recognized as a promising strategy for disc regeneration[35-37]. However, the potential for long-term survival and biological function of the transplanted cells in the harsh microenvironment remains unknown[10,38]. To overcome the problems associated with stem cell transplantation, mobilization of endogenous progenitor/stem cells to the injury site has been considered as an alternative approach. Many studies have demonstrated that progenitor cells residing in the stem niches of the IVD migrate toward the annular fibrosus and the inner parts of the IVD that may contribute to disc regeneration in situ[27-29,39]. In the natural healing process, the affected cells and tissues release a variety of cytokines/chemokines to activate the dormant progenitor cells and then trigger their migration from their niches to the injury sites[40]. However, little is currently known about the possibility of endogenous tissue repair of the degenerative IVD by targeting the local microenvironment, or the mechanism by which this might occur. The present study sought to investigate the role of chemokine SDF-1α in regulating the migration and differentiation capacities of endogenous stem cells within the IVD for regeneration.
Recent studies have shown that the degenerative IVD releases several cytokines and chemokines for effective recruitment of endogenous stem cells during the repair process[41]. SDF-1α is considered to be a potent chemokine with a role in trafficking endogenous progenitor cells to repair injury[42]. It has been documented that upregulation of SDF-1α and of its receptor CXCR4 are closely related to IVD degeneration[19]. In the present study, we also demonstrated high expression and release of SDF-1α in vitro in native cells affected by the pro-inflammatory mediators IL-1β and TNF-α. Moreover, upregulation of SDF-1α in the degenerative IVD induced by needle puncture was confirmed by immunohistochemistry.
In vitro migration assays demonstrated that SDF-1α promoted migration capacity of NPSCs in a dose-dependent manner. Although neutralization of CXCR4, using a pharmaceutical inhibitor AMD3100, decreased the SDF-1α-induced enhancement of migration of NPSCs, it could not completely abolish the migration capacity. This suggests that the SDF-1/CXCR4 axis is essential for endogenous recruitment of NPSCs, although there may be other factors involved in their specific migration. Furthermore, in the immunofluorescence study, we found that SDF-1α promoted the formation of F-actin and lamellipodia in NPSCs, which was suppressed by the CXCR4 antagonist AMD3100. F-actin cytoskeletal networks are widely accepted to be key regulators of cellular shape and force generation in cell migration, and are involved in lamellipodia formation, cell adhesion, and cellular shape changes[43]. Herein, we hypothesized that the formation of F-actin and lamellipodia was compatible with the SDF-1α-induced migration of NPSCs, suggesting that SDF-1α rearranges F-actin cytoskeletal networks in NPSCs to increase their migratory capability.
The collected evidence demonstrated that bone marrow-derived mesenchymal stem cells (BMSCs) expressing CXCR4 showed powerful chemotaxis in response to SDF-1α, and that SDF-1α can promote the migration of the BMSCs by increasing the expression of CXCR4[44]. The present study, for the first time, showed the expression of SDF-1α receptor CXCR4 in NPSCs. The immunoblotting and real-time RT-PCR results demonstrated that the expression of CXCR4 was upregulated by SDF-1α and abolished by pretreatment with the antagonist AMD3100. The majority of CXCR4 is localized in endosomal compartments of MSCs; it cycles continuously to the cell surface and re-enters the cytoplasm via endocytosis[45]. Immunofluorescence revealed that CXCR4 was mainly expressed in the cytoplasm of NPSCs, and SDF-1α not only increased the CXCR4 expression level but also induced translocation of intracellular CXCR4 to the cell surface. This phenomenon also appeared in the regulation of the SDF-1/CXCR4 axis in the chemoattraction of MSCs. These results suggest that SDF-1α affects cellular production and surface expression of CXCR4 in NPSCs, contributing to their migration capacity[46].
The role of the SDF-1/CXCR4 axis in chondrogenic differentiation of NPSCs is poorly understood. A recent report suggested a novel role of SDF-1α in regulating bone morphogenetic protein (BMP)-2-induced osteogenic or TGF-β1-induced chondrogenic differentiation of MSCs[23-26]. Wang et al[47] demonstrated that SDF-1α can recruit MSCs and promote their chondrogenic differentiation for the repair of cartilage defects. On the contrary, Kim et al[33] found that the SDF-1/CXCR4 axis had little effect on early chondrogenic differentiation of MSCs. In the present study, we found that SDF-1α upregulated the expression of its transmembrane receptor CXCR4 in NPSCs, which modulated the chondrogenic differentiation of NPSCs in the presence of TGF-β1. After micromass culture of NPSCs for chondrogenesis in vitro, histological analysis showed that SDF-1α could not only increase the weight and size of chondrogenic pellets but also promote NPSCs to synthesize aggrecan and collagen II in a dose-dependent manner. However, the effect of SDF-1α-stimulated chondrogenic differentiation of NPSCs was neutralized by the CXCR4 antagonist AMD3100, which demonstrated that blocking of the receptor CXCR4 inhibits the differentiation of NPSCs into the chondrocyte lineages in response to TGF-β1 stimulation, evidenced by the reduced expression of chondrogenic markers such as aggrecan, collagen II, and Sox-9. Therefore, we propose that SDF-1/CXCR4 axis synergy with TGF-β1 signaling activates intracellular R-Smads and Erk1/2, which translocate to the nucleus for regulating the transcription of chondrogenic genes like Sox-9[25]. These findings may suggest that the SDF-1/CXCR4 axis plays an important role in mediating NPSC differentiation toward an NP-like phenotype in cooperation with TGF-β1 for facilitating IVD regeneration.
In summary, the current research demonstrated a novel role of SDF-1α, which pathogenically accumulates in the damaged or degenerative IVD, in effectively promoting NPSC chondrogenic differentiation and migration toward injury sites via the SDF-1/CXCR4 axis. This provides a new therapeutic target and strategy to improve endogenous repair or regeneration of the degenerative IVD in situ. Future work will focus on understanding the possible downstream signaling pathways of the SDF-1/CXCR4 axis and their role in regulating the biological behaviors of NPSCs.
Intervertebral disc (IVD) degeneration, the primary cause of low back pain, is an irreversible disease with no currently effective treatment. Nucleus pulposus-derived stem cells (NPSCs) with the capacity of multilineage differentiation have the regenerative potential to repair the degenerative IVD. Stromal cell-derived factor 1α (SDF-1α) released in the injury tissues displays a remarkable ability to attract stem cells for repairing tissues.
Mobilization and differentiation of NPSCs within the IVD represent an attractive target for future regenerative strategies for IVD degeneration in situ.
In the present study, the aim was to detect SDF-1α in the degenerative IVD and to determine its roles in the migration and differentiation of NPSCs.
Enzyme-linked immunosorbent assay (ELISA), RT-qPCR, and immunohistochemistry were performed to demonstrate the levels of SDF-1α generated in the condition of IVD degeneration. The capacity of SDF-1α to recruit NPSCs was evaluated using wound healing and transwell migration assays. Western blot, RT-qPCR, and immunofluorescence were employed to determine the expression and location of C-X-C chemokine receptor type 4 (CXCR4) in response to SDF-1α. Cell micromass culture was used to assess the effect of SDF-1α on chondrogenic differentiation of NPSCs. Western blot, RT-qPCR, and histological analysis were conducted to examine the expression of chondrogenic markers such as aggrecan, collagen II, and Sox-9. CXCR4 antagonist AMD3100 was used to inhibit the effects of the SDF-1/CXCR4 axis on the migration and chondrogenic differentiation capacities of NPSCs.
SDF-1α was highly expressed in the inflammatory microenvironment of the degenerative IVD. SDF-1α promoted migration and chondrogenic differentiation of NPSCs in a dose-dependent manner. SDF-1α not only increased CXCR4 expression but also stimulated translocation of CXCR4 from the cytoplasm to membrane, accompanied by cytoskeletal rearrangement. Inhibition of the SDF-1/CXCR4 axis using CXCR4 antagonist AMD3100 effectively abolished the SDF-1α-induced migration and differentiation capacities of NPSCs.
The present study demonstrated that the chemokine SDF-1α was pathogenically produced and secreted in the degenerative IVD, which plays a crucial role in promoting NPSC chondrogenic differentiation and migration toward injury sites via the SDF-1/CXCR4 axis.
In summary, SDF-1α is highly released in the damaged or degenerative IVD and its receptor CXCR4 is expressed in the cytomembrane and cytoplasm of NPSCs. SDF-1α has the potential to enhance recruitment and chondrogenic differentiation of NPSCs via SDF-1/CXCR4 chemotaxis signals, which provides a novel insight into IVD regeneration mechanism and a promising therapeutic strategy to improve endogenous repair or regeneration of the degenerative IVD in situ. Future work will focus on understanding the possible downstream signaling pathways of the SDF-1/CXCR4 axis and their role in regulating the biological behaviors of NPSCs.
We thank Li-Jun Zhou and Da-Jin Zhang in the Research Laboratory Center, Navy General Hospital for providing technical support
| 1. | Buckwalter JA. Aging and degeneration of the human intervertebral disc. Spine (Phila Pa 1976). 1995;20:1307-1314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 922] [Cited by in RCA: 866] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 2. | Roughley PJ. Biology of intervertebral disc aging and degeneration: involvement of the extracellular matrix. Spine (Phila Pa 1976). 2004;29:2691-2699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 528] [Cited by in RCA: 562] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 3. | Deyo RA, Weinstein JN. Low back pain. N Engl J Med. 2001;344:363-370. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1365] [Cited by in RCA: 1253] [Article Influence: 50.1] [Reference Citation Analysis (0)] |
| 4. | Balagué F, Mannion AF, Pellisé F, Cedraschi C. Non-specific low back pain. Lancet. 2012;379:482-491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1022] [Cited by in RCA: 1169] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 5. | Katz JN. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. J Bone Joint Surg Am. 2006;88 Suppl 2:21-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 493] [Cited by in RCA: 785] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 6. | Risbud MV, Albert TJ, Guttapalli A, Vresilovic EJ, Hillibrand AS, Vaccaro AR, Shapiro IM. Differentiation of mesenchymal stem cells towards a nucleus pulposus-like phenotype in vitro: implications for cell-based transplantation therapy. Spine (Phila Pa 1976). 2004;29:2627-2632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 238] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 7. | Yim RL, Lee JT, Bow CH, Meij B, Leung V, Cheung KM, Vavken P, Samartzis D. A systematic review of the safety and efficacy of mesenchymal stem cells for disc degeneration: insights and future directions for regenerative therapeutics. Stem Cells Dev. 2014;23:2553-2567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 8. | Murphy MB, Moncivais K, Caplan AI. Mesenchymal stem cells: environmentally responsive therapeutics for regenerative medicine. Exp Mol Med. 2013;45:e54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 786] [Cited by in RCA: 910] [Article Influence: 70.0] [Reference Citation Analysis (0)] |
| 9. | Yang H, Cao C, Wu C, Yuan C, Gu Q, Shi Q, Zou J. TGF-βl Suppresses Inflammation in Cell Therapy for Intervertebral Disc Degeneration. Sci Rep. 2015;5:13254. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 10. | Liang C, Li H, Tao Y, Zhou X, Li F, Chen G, Chen Q. Responses of human adipose-derived mesenchymal stem cells to chemical microenvironment of the intervertebral disc. J Transl Med. 2012;10:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 11. | Sakai D, Grad S. Advancing the cellular and molecular therapy for intervertebral disc disease. Adv Drug Deliv Rev. 2015;84:159-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 242] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 12. | Chik TK, Ma XY, Choy TH, Li YY, Diao HJ, Teng WK, Han SJ, Cheung KM, Chan BP. Photochemically crosslinked collagen annulus plug: a potential solution solving the leakage problem of cell-based therapies for disc degeneration. Acta Biomater. 2013;9:8128-8139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 13. | Vadalà G, Sowa G, Hubert M, Gilbertson LG, Denaro V, Kang JD. Mesenchymal stem cells injection in degenerated intervertebral disc: cell leakage may induce osteophyte formation. J Tissue Eng Regen Med. 2012;6:348-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 234] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 14. | Omlor GW, Bertram H, Kleinschmidt K, Fischer J, Brohm K, Guehring T, Anton M, Richter W. Methods to monitor distribution and metabolic activity of mesenchymal stem cells following in vivo injection into nucleotomized porcine intervertebral discs. Eur Spine J. 2010;19:601-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 40] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Liu ZJ, Zhuge Y, Velazquez OC. Trafficking and differentiation of mesenchymal stem cells. J Cell Biochem. 2009;106:984-991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 249] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 16. | Liekens S, Schols D, Hatse S. CXCL12-CXCR4 axis in angiogenesis, metastasis and stem cell mobilization. Curr Pharm Des. 2010;16:3903-3920. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 176] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 17. | Lapidot T, Kollet O. The essential roles of the chemokine SDF-1 and its receptor CXCR4 in human stem cell homing and repopulation of transplanted immune-deficient NOD/SCID and NOD/SCID/B2m(null) mice. Leukemia. 2002;16:1992-2003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 340] [Article Influence: 14.2] [Reference Citation Analysis (1)] |
| 18. | Jia CQ, Zhao JG, Zhang SF, Qi F. Stromal cell-derived factor-1 and vascular endothelial growth factor may play an important role in the process of neovascularization of herniated intervertebral discs. J Int Med Res. 2009;37:136-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 19. | Zhang H, Zhang L, Chen L, Li W, Li F, Chen Q. Stromal cell-derived factor-1 and its receptor CXCR4 are upregulated expression in degenerated intervertebral discs. Int J Med Sci. 2014;11:240-245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 20. | Marquez-Curtis LA, Janowska-Wieczorek A. Enhancing the migration ability of mesenchymal stromal cells by targeting the SDF-1/CXCR4 axis. Biomed Res Int. 2013;2013:561098. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 165] [Cited by in RCA: 221] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 21. | Pereira CL, Gonçalves RM, Peroglio M, Pattappa G, D'Este M, Eglin D, Barbosa MA, Alini M, Grad S. The effect of hyaluronan-based delivery of stromal cell-derived factor-1 on the recruitment of MSCs in degenerating intervertebral discs. Biomaterials. 2014;35:8144-8153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 22. | Sakai D, Nishimura K, Tanaka M, Nakajima D, Grad S, Alini M, Kawada H, Ando K, Mochida J. Migration of bone marrow-derived cells for endogenous repair in a new tail-looping disc degeneration model in the mouse: a pilot study. Spine J. 2015;15:1356-1365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 58] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 23. | Kortesidis A, Zannettino A, Isenmann S, Shi S, Lapidot T, Gronthos S. Stromal-derived factor-1 promotes the growth, survival, and development of human bone marrow stromal stem cells. Blood. 2005;105:3793-3801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 283] [Cited by in RCA: 289] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 24. | Zhu W, Boachie-Adjei O, Rawlins BA, Frenkel B, Boskey AL, Ivashkiv LB, Blobel CP. A novel regulatory role for stromal-derived factor-1 signaling in bone morphogenic protein-2 osteogenic differentiation of mesenchymal C2C12 cells. J Biol Chem. 2007;282:18676-18685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 84] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 25. | Guang LG, Boskey AL, Zhu W. Regulatory role of stromal cell-derived factor-1 in bone morphogenetic protein-2-induced chondrogenic differentiation in vitro. Int J Biochem Cell Biol. 2012;44:1825-1833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | Chim H, Miller E, Gliniak C, Alsberg E. Stromal-cell-derived factor (SDF) 1-alpha in combination with BMP-2 and TGF-β1 induces site-directed cell homing and osteogenic and chondrogenic differentiation for tissue engineering without the requirement for cell seeding. Cell Tissue Res. 2012;350:89-94. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 27. | Sakai D, Nakamura Y, Nakai T, Mishima T, Kato S, Grad S, Alini M, Risbud MV, Chan D, Cheah KS, Yamamura K, Masuda K, Okano H, Ando K, Mochida J. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat Commun. 2012;3:1264. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 263] [Cited by in RCA: 363] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 28. | Risbud MV, Guttapalli A, Tsai TT, Lee JY, Danielson KG, Vaccaro AR, Albert TJ, Gazit Z, Gazit D, Shapiro IM. Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine (Phila Pa 1976). 2007;32:2537-2544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 234] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 29. | Henriksson H, Thornemo M, Karlsson C, Hägg O, Junevik K, Lindahl A, Brisby H. Identification of cell proliferation zones, progenitor cells and a potential stem cell niche in the intervertebral disc region: a study in four species. Spine (Phila Pa 1976). 2009;34:2278-2287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 158] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 30. | Tao YQ, Liang CZ, Li H, Zhang YJ, Li FC, Chen G, Chen QX. Potential of co-culture of nucleus pulposus mesenchymal stem cells and nucleus pulposus cells in hyperosmotic microenvironment for intervertebral disc regeneration. Cell Biol Int. 2013;37:826-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 31. | Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop Dj, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11055] [Cited by in RCA: 13034] [Article Influence: 686.0] [Reference Citation Analysis (12)] |
| 32. | Wehling N, Palmer GD, Pilapil C, Liu F, Wells JW, Müller PE, Evans CH, Porter RM. Interleukin-1beta and tumor necrosis factor alpha inhibit chondrogenesis by human mesenchymal stem cells through NF-kappaB-dependent pathways. Arthritis Rheum. 2009;60:801-812. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 211] [Cited by in RCA: 203] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 33. | Kim GW, Han MS, Park HR, Lee EJ, Jung YK, Usmani SE, Ulici V, Han SW, Beier F. CXC chemokine ligand 12a enhances chondrocyte proliferation and maturation during endochondral bone formation. Osteoarthritis Cartilage. 2015;23:966-974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 34. | Han B, Zhu K, Li FC, Xiao YX, Feng J, Shi ZL, Lin M, Wang J, Chen QX. A simple disc degeneration model induced by percutaneous needle puncture in the rat tail. Spine (Phila Pa 1976). 2008;33:1925-1934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 353] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 35. | Hohaus C, Ganey TM, Minkus Y, Meisel HJ. Cell transplantation in lumbar spine disc degeneration disease. Eur Spine J. 2008;17 Suppl 4:492-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 134] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 36. | Iwashina T, Mochida J, Sakai D, Yamamoto Y, Miyazaki T, Ando K, Hotta T. Feasibility of using a human nucleus pulposus cell line as a cell source in cell transplantation therapy for intervertebral disc degeneration. Spine (Phila Pa 1976). 2006;31:1177-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 56] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 37. | Kregar Velikonja N, Urban J, Fröhlich M, Neidlinger-Wilke C, Kletsas D, Potocar U, Turner S, Roberts S. Cell sources for nucleus pulposus regeneration. Eur Spine J. 2014;23 Suppl 3:S364-S374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 38. | Wuertz K, Godburn K, Neidlinger-Wilke C, Urban J, Iatridis JC. Behavior of mesenchymal stem cells in the chemical microenvironment of the intervertebral disc. Spine (Phila Pa 1976). 2008;33:1843-1849. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 128] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 39. | Henriksson HB, Svala E, Skioldebrand E, Lindahl A, Brisby H. Support of concept that migrating progenitor cells from stem cell niches contribute to normal regeneration of the adult mammal intervertebral disc: a descriptive study in the New Zealand white rabbit. Spine (Phila Pa 1976). 2012;37:722-732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 40. | Vanden Berg-Foels WS. In situ tissue regeneration: chemoattractants for endogenous stem cell recruitment. Tissue Eng Part B Rev. 2014;20:28-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 115] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 41. | Li Z, Peroglio M, Alini M, Grad S. Potential and limitations of intervertebral disc endogenous repair. Curr Stem Cell Res Ther. 2015;10:329-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 42. | Jaerve A, Schira J, Müller HW. Concise review: the potential of stromal cell-derived factor 1 and its receptors to promote stem cell functions in spinal cord repair. Stem Cells Transl Med. 2012;1:732-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 43. | Stricker J, Falzone T, Gardel ML. Mechanics of the F-actin cytoskeleton. J Biomech. 2010;43:9-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 336] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 44. | Li X, Gao Z, Wang J. Single percutaneous injection of stromal cell-derived factor-1 induces bone repair in mouse closed tibial fracture model. Orthopedics. 2011;34:450. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 45. | Pelekanos RA, Ting MJ, Sardesai VS, Ryan JM, Lim YC, Chan JK, Fisk NM. Intracellular trafficking and endocytosis of CXCR4 in fetal mesenchymal stem/stromal cells. BMC Cell Biol. 2014;15:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 46. | Iannone M, Ventre M, Pagano G, Giannoni P, Quarto R, Netti PA. Defining an optimal stromal derived factor-1 presentation for effective recruitment of mesenchymal stem cells in 3D. Biotechnol Bioeng. 2014;111:2303-2316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 47. | Wang Y, Sun X, Lv J, Zeng L, Wei X, Wei L. Stromal Cell-Derived Factor-1 Accelerates Cartilage Defect Repairing by Recruiting Bone Marrow Mesenchymal Stem Cells and Promoting Chondrogenic Differentiation. Tissue Eng Part A. 2017;23:1160-1168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Grawish ME, Saeki K, Tanabe S S-Editor: Ji FF L-Editor: Wang TQ E-Editor: Song H