Published online Dec 26, 2018. doi: 10.4252/wjsc.v10.i12.183
Peer-review started: September 13, 2018
First decision: October 5, 2018
Revised: October 15, 2018
Accepted: November 15, 2018
Article in press: November 16, 2018
Published online: December 26, 2018
Processing time: 103 Days and 0.5 Hours
Cancer is a widespread worldwide chronic disease. In most cases, the high mortality rate from cancer correlates with a lack of clear symptoms, which results in late diagnosis for patients, and consequently, advanced tumor disease with poor probabilities for cure, since many patients will show chemo- and radio-resistance. Several mechanisms have been studied to explain chemo- and radio-resistance to anti-tumor therapies, including cell signaling pathways, anti-apoptotic mechanisms, stemness, metabolism, and cellular phenotypes. Interestingly, the presence of cancer stem cells (CSCs), which are a subset of cells within the tumors, has been related to therapy resistance. In this review, we focus on evaluating the presence of CSCs in different tumors such as breast cancer, gastric cancer, lung cancer, and hematological neoplasias, highlighting studies where CSCs were identified in patient samples. It is evident that there has been a great drive to identify the cell surface phenotypes of CSCs so that they can be used as a tool for anti-tumor therapy treatment design. We also review the potential effect of nanoparticles, drugs, natural compounds, aldehyde dehydrogenase inhibitors, cell signaling inhibitors, and antibodies to treat CSCs from specific tumors. Taken together, we present an overview of the role of CSCs in tumorigenesis and how research is advancing to target these highly tumorigenic cells to improve oncology patient outcomes.
Core tip: Tumor heterogeneity can explain the presence of cells that display high tumorigenic capacity along with chemo- and radio-resistance properties. These cells, identified as cancer stem cells (CSCs), are partially responsible for recurrence and tumor progression. Most tumors follow the CSC model, which indicates the existence of a subset of highly tumorigenic cells. This has been shown to be the case for several patients with several types of tumors. In this review, we focus on the phenotypes used for the study and identification of CSCs from human samples, as well as promising strategies to target CSCs.
- Citation: Toledo-Guzmán ME, Bigoni-Ordóñez GD, Ibáñez Hernández M, Ortiz-Sánchez E. Cancer stem cell impact on clinical oncology. World J Stem Cells 2018; 10(12): 183-195
- URL: https://www.wjgnet.com/1948-0210/full/v10/i12/183.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v10.i12.183
Cancer stem cells (CSCs) comprise a cell population within a tumor that, among other factors, is responsible for cancer initiation, propagation, metastasis and recurrence. It is known that solid tumors are composed of heterogeneous cell populations[1-3] with different phenotypic characteristics at different stages of development, with variable abilities to proliferate. However, only the CSC population is clonogenic in vitro and in vivo, suggesting that these cells are the only ones with the highest tumorigenic potential[4,5].
The existence of a subset of cancer cells that possesses an extensive proliferative capacity was reported in leukemia and multiple myeloma in the 1970s[6,7]. In both cancer types, only a cell population derived from a tumor was able to grow in clonogenic assays, where they formed spherical colonies, and induce tumors in mice that recapitulated the original tumor. At that time, the most reliable criterion for CSC identification was the capacity of these cells to produce colonies[6].
The first CSCs were isolated from acute myeloid leukemia (AML) by transplantation into severe combined immune-deficient (SCID) mice. They were identified as CD34+CD38- cells and named AML-initiating cells because of their ability to establish human leukemia in SCID mice. Since the identified CD34+CD38- cells were less differentiated than colony-forming cells, a hierarchy or heterogeneity in AML was proposed[1]. Later, in 1997, the model was reproduced in non-obese diabetic mice with severe combined immunodeficiency disease (NOD/SCID) mice, where CD34+CD38- CSCs were capable of differentiating into leukemic blasts in vivo, supporting the existence of a hierarchy in leukemia[8].
Some years later, enriched CSC populations were obtained from human brain tumors[9], using cells with a CD133+ phenotype that showed a higher capacity for proliferation, self-renewal, and differentiation. CD133+ cells were xenotransplanted into NOD/SCID mice and formed tumors that, when serially transplanted, recapitulated the original human tumor[10,11]. Since then, CSCs from various solid tumors have been reported[5].
In recent years, several research groups have focused on the identification and isolation of these cells. Besides leukemia and multiple myeloma, CSCs from solid tumors have been identified and isolated through the use of surface and functional markers[12-15], their growing capacity as spheroids in vitro[16,17], the evaluation of CSC clonogenic capacity[18,19] and their in vivo tumorigenic capacity in xenotransplant experiments[16,17,20,21].
Due to the reported participation of CSCs in chemo- and radio-resistance[22-24], an increasing interest in implementing strategies against CSCs in patients to improve their clinical outcome has grown in recent years because conventional therapies are effective in controlling tumor growth at the beginning, but over time, relapse is a main problem due to remaining CSCs[22,25,26].
A CSC is defined as a cell within a tumor that is able to produce an identical cell with the same properties to give rise heterogeneous differentiated progeny, and has the ability to modulate differentiation and self-renewal (homeostatic control). These CSCs possess the ability to propagate themselves, as well as recapitulate a tumor[2,3,27].
A major characteristic of CSCs relies on their ability to regulate stemness pathways such as Wnt/β-catenin, Sonic hedgehog (Shh), transforming growth factor beta (TGF-β), etc[28]. These pathways are dysregulated in CSCs, and targeting them has been proposed as a strategy to increase the effectiveness of cancer therapies.
The CSC model postulates that solid tumors and leukemia are hierarchically organized, with CSCs at the apex of this hierarchy, driving tumor growth, relapse, metastasis and drug resistance[5,29]. Cell heterogeneity is responsible for varying cell morphology, different proliferative index, genetic changes and therapeutic response[30]. For a successful therapy, all CSCs should be specifically eliminated to avoid relapse.
Typically, CSCs are defined as a small or a rare cell population[2,31] that forms tumors after being xenotransplanted into immunodeficient mice. However, recent reports have suggested that the percentage of CSCs within a tumor can vary from 0.02% to 25% depending on the tumor type, where higher CSC proportions are found in undifferentiated tumors[31-34]. Typically, higher CSC frequencies have been found in mouse models, leukemias and lymphomas, while lower frequencies are frequently found in solid tumors[35]. Based on this information, it has been suggested that not all cancers follow the CSC model[27]. Instead, a dynamic or plastic CSC model has been proposed, where CSCs and non-CSCs could alternate between two phenotypic states[36]. In this dynamic model, both cell types show varying levels of tumor-forming capacity, drug response and the ability to give rise to differentiated cells[29,35]. CSCs and non-CSCs can still be easily distinguished through surface and functional markers, but mainly by their self-renewal capacity.
It is very important to note that the CSC model is widely reported in several cancer types (Figure 1), although there are a few publications about cancers that do not follow a CSC model or a dynamic CSC model, specifically in lymphoma mice models[37] and melanoma[32], where the tumors are homogeneous. In 2007, Strasser and his group inoculated 10 to 105 pre-B/B lymphoma cells into recipient mice. All of the animals developed lymphoma within 35 d, regardless of the number of inoculated cells, differing only in tumor growth rate[37].
Although CSCs are able to self-renew and differentiate, they do not necessarily originate from the malignant transformation of stem cells[33]. The cell of origin refers only to the cell type that received the first genetic or epigenetic hit, which confers the ability for self-renewal or tumor growth[35]. Examples of these cells are: normal stem cells, restricted progenitor cells and more differentiated cells. All of them could have acquired or maintained self-renewal capacity, and some of them can even undergo epithelial to mesenchymal transition (EMT), giving rise to metastatic CSCs[36].
In conclusion, the variable phenotype of the CSC population in patients and tumor types proposed in the CSC dynamic model constitutes the main challenge for the possible use of anti-CSC therapy.
The CSC population possesses several characteristics that can be useful for cancer therapy development, primarily focusing on the elimination of these cells.
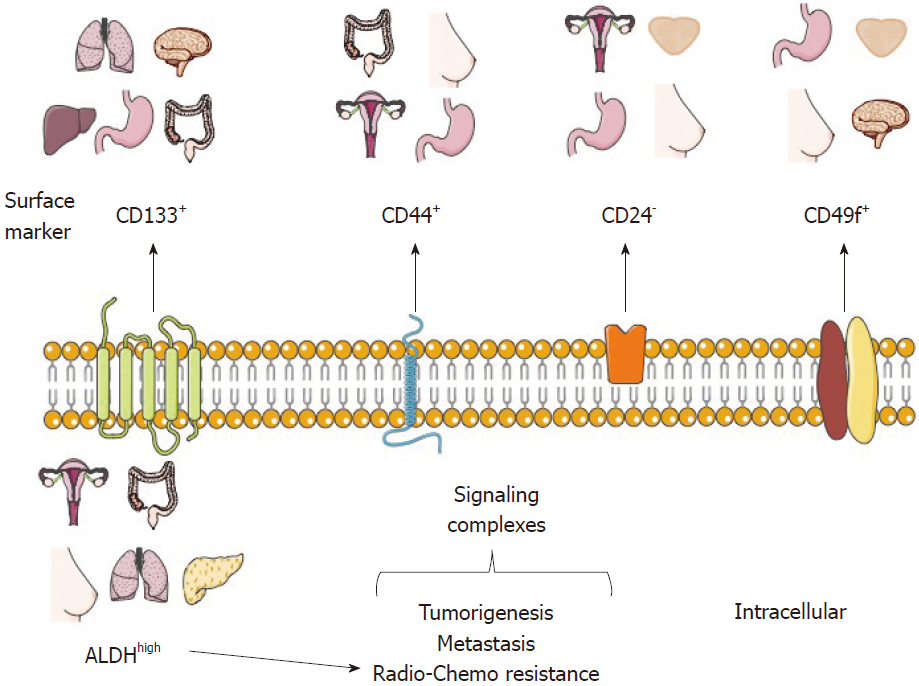
Usually, a distinctive profile of surface and functional markers characterizes the CSC population, and their identification and purification usually begins with the description of such markers[3,29]. Moreover, there is an increasing interest in identifying the role of each marker in CSCs, as well as targeting CSC-specific pathways, which could increase the radio- and chemo-sensitivity of CSCs.
To date, several CSC markers from distinct tumor types have been proposed and validated through different experimental models (Table 1 and Figure 1). Some of these markers are discussed below.
| Cancer type | Phenotype | Model | References |
| Prostate cancer | CD44+ | PCa cell line and tumor xenograft in mice | [58] |
| Breast cancer | CD44+ CD24-/low | Patient-derived tumor xenograft in mice | [5] |
| Cervical cancer | CD44+ CD24+ | SiHa cell line | [55] |
| Gastric cancer | CD44+ CD24+ | AGS cell line and patient tissue samples | [56] |
| Nasopharyngeal carcinoma | CD24- | NPC cell lnes, mice | [54] |
| Gastric adenocarcinoma | CD44+ CD133+ | Patient tissue samples | [51] |
| Oral squamous cell carcinoma | CD44+ ALDH1 | Metastatic lymph nodes | [153] |
| Breast cancer | CD44v | Clinical samples | [154] |
| Prostate cancer | CD133 | Primary prostate cancer cell lines | [155] |
| Endometrial cancer | CD133 | Human endometrial cell lines | [42] |
| Liver cancer | CD133 | Huh-7 cells and tumor xenograft in mice | [47] |
| Prostate cancer | CD133 | Primary human prostate cancer cell lines | [155] |
| Cervical cancer | CD49f | SiHa and HeLa cell lines | [156] |
| Non-small cell lung cancer | CD49f | Patient-derived sphere-forming assays | [157] |
| Gastric cancer | CD49f | Gastric tumor tissues and tumor xenograft in mice | [75] |
| Colon cancer | CD49f | HT29 and Caco2 cell lines, clinical samples | [77] |
| Cervical cancer | ALDH | SiHa and HeLa cell lines, mice model | [85] |
| Colon cancer | ALDH1A3 | HT29 cell line | [158] |
| Colon cancer | ALDH1A1 | HT29 cell line and tumor xenograft in mice | [159] |
| Breast cancer | ALDH | Breast cancer tumor tissues | [160] |
Nowadays, there are CSC markers that are widely used to identify several tumor types. Such markers have been reported in CSC-enrichment culture models from cell lines or primary cultures derived from patient samples and serial xenotransplantation of putative CSCs in mouse models, which must be able to recapitulate the original heterogeneous populations and be directly validated in human tumor samples. It is important to note that the use of a single marker to define a CSC population is not recommended. For this purpose, a phenotypic profile that combines various markers should be established, as well as carrying out self-renewal assays (Figure 1)[2,25].
CD133, also known as prominin-1, is a transmembrane cell surface glycoprotein traditionally used as a hematopoietic stem cell marker that is effective for detection of non-stem cells from various tumor and tissue samples. The Dirks laboratory used the CSC marker CD133 for brain CSC identification. The purified CD133+ population from primary human brain tumors samples showed higher proliferation and self-renewal capacity in neurosphere formation assays than CD133- cells[10]. Moreover, the inoculation of only a few CD133+ cells was sufficient to produce a tumor, which was then successfully transplanted[11]. In 2013, the Pelicci laboratory reported that CD133 was found in an interconvertible state in glioblastoma patient-derived neurospheres and that the use of short hairpin RNA (shRNA) against CD133 diminished their self-renewal and tumorigenicity potential[18]. Interestingly, some studies have proposed that CD133 could maintain CSC properties through the Wnt/β-catenin signaling pathway[38].
CD133 has also been tested in colorectal cancer cell lines and tumor tissue samples[39,40] through the use of various techniques, including flow cytometry and serial xenotransplantation in mice[41]. Additionally, CD133+ CSCs have been reported in many other solid cancer models, including endometrial cancer[42], lung cancer[43], small cell lung cancer[44], laryngeal cancer[45,46], liver cancer[47], colorectal cancer[48], and gastric cancer[49].
CD133 has been found in samples that represent higher stage tumors and are predictors of poor prognosis. For this reason, CD133 is considered a promising therapeutic target. This year, a phase I trial for testing the efficacy of CD133-directed CAR-T cells showed that CD133+ cells were successfully eliminated after CART-133 infusion[50].
CD44 is a multifunctional glycoprotein involved in cell adhesion, signaling, proliferation, migration, hematopoiesis, and lymphocyte activation[51]. It functions as a receptor for hyaluronan and other extracellular matrix components[52]. CD44 is widely used as a CSC marker, especially for tumors of epithelial origin, and it is used alone or in combination with CD24 for the identification of breast CSCs[5]. CD24 is a small surface protein that is found in many tumor types. However, reports from cancer cell lines show that there is a substantial variation in CD24 expression even among the same tumor types[53].
Though CD24- cells are commonly associated with CSC phenotypes, there are some cases in which CD24+ has been found to be a marker for cell populations with CSC features. For example, in nasopharyngeal carcinoma (NPC) cell lines[54] and in HPV-16 SiHa cervical cancer cells, isolated CD44+CD24+ cells were radioresistant and more tumorigenic than those negative for the same markers[55]. The same CD44+CD24+ phenotype was used to identify gastric CSCs[56].
A known classic publication demonstrated that only a small population isolated from breast tumors, defined as CD44+CD24-/low, has the capacity to sustain tumor growth in NOD/SCID mice and generate heterogeneous cell populations as the original breast tumor[5]. Later, in human prostate cancer samples, CSCs characterized through immunofluorescence with the CD44+/β2β1hi/CD133+ phenotype were identified and characterized[57]. The next year, CD44+ prostate cancer cell populations were obtained[58]. Also, CD44 and CD133 expression was evaluated in gastric adenocarcinoma tumors by immunohistochemistry, and it was found that both markers could be correlated with clinical and pathological parameters[51].
Although CD44 is widely reported as a CSC marker, it is very important to note that it is a ubiquitously expressed molecule derived from a gene with 18 exons. When all variable exons are spliced out, the standard form (CD44s) is expressed, and when alternative splicing occurs, variant forms (CD44v) are expressed[59]. In spite of this, there are only a few reports in which CD44 isoforms are considered when evaluating CSCs. In 2005, Mackenzie and his group demonstrated the existence of two CSC populations, both expressing CD44high (and CD44+), derived from head and neck cutaneous squamous cell carcinoma. One was associated with EMT properties and the other one possessed an epithelial phenotype[60]. They demonstrated that the CD44high cells that undergo EMT preferably expressed the CD44s isoform; while the epithelial CD44high cells expressed the CD44v isoform. Using RNAseq, another group later confirmed these results. The CD44v6 isoform was identified as the predominant isoform in a prostate cancer epithelial cell line[61].
A very important contribution from the Mackenzie laboratory is that they demonstrated that the use of enzymes (for example, trypsin or collagenase) for cell extraction from tissues caused destruction of cell surface CD44v isoforms, leaving only the CD44s isoform[62]. Moreover, CD44-specific antibodies are not able to distinguish between all isoforms. Specifically, in breast cancer, CD44v was found to be associated with better prognosis while CD44s was related to poor prognosis[63]. As a consequence, CD44 is the most frequently found CSC marker[64,65]. Other examples are found in colorectal cancer, in which CD44 was found together with CD133[66,67], head and neck squamous cell carcinoma[68,69], ovarian CSCs[70], and gastric cancer using the specific isoform CD44v8-10[71].
CD49f or integrin α6, is a transmembrane glycoprotein that functions as the receptor for the extracellular matrix protein laminin[72,73]. CD49f is widely distributed in stem cells and in the brain[73]; because of its role in tumor cell proliferation, survival, self-renewal and tumor growth, it has been proposed that it could be used as a CSC marker[73].
In sphere colony forming cell culture using prostate cancer cells, CD49f was shown to be a better marker than CD133 and CD44[74]. In gastric cancer, CD49high cells displayed CSC characteristics, including resistance to doxorubicin, 5-fluorouracil and doxifluridine[75].This has also been reported in breast[76] and colon cancer[77].
Besides the examples mentioned above, there are other surface markers that have been proposed as CSC markers, such as CXCR4 and LGR5, among others.
Another strategy for CSC identification and purification is the use of functional or intracellular markers (Figure 1), which are considered to be more stable than surface markers. The principal functional CSC marker is aldehyde dehydrogenase or ALDH, part of an enzyme superfamily encoded by 19 genes that metabolize endogenous and exogenous aldehydes. It is present in practically all organisms, and its levels and isozymes vary depending on tissue and organ[78].
For ALDH identification, specific ALDH antibodies are available; nonetheless, we suggest that the most appropriate way for ALDH identification is the measurement of its activity using the commercial ALDH fluorescent substrate ALDEFLUOR® kit assay by Stem Cells Technologies, Inc. (Vancouver, BC, Canada). Cells that display high ALDH activity, (named ALDHhigh or ALDH+ or ALDHbr), can be identified and isolated using flow cytometry[79]. Several works have shown that high ALDH activity is often associated with CSCs derived from solid tumor types[80]. These cells are generally characterized by a higher proliferation potential, colony-forming capacity, self-renewal, in vivo tumorigenic capacity, metastasis, and drug resistance. For instance, ALDHhigh CSCs have been identified in colon cancer[81,82], lung cancer[83], cervical cancer[14,84,85], breast cancer[86], pancreatic cancer[87,88], and melanoma[89,90], to mention some examples.
As for surface markers, ALDH is often reported in combination with other cell markers to increase the accuracy of CSC validation. In some cases, high ALDH activity is found together with high expression of markers like CD133. Some cases have been identified in ovarian cancer[91,92], invasive ductal breast carcinoma tumors[93], and lung cancer[94]. The combination ALDH+/CD44+ has been evaluated in various tumors such as breast cancer[95] and lung cancer[96].
Several cancers acquire drug resistance during or after treatment, which is the case for cancers that possess cells that are more resistant than the rest of the tumor. Generally, resistant cells have proteins that remove drugs from cells[97]. One of the most studied mechanisms of drug resistance in CSCs is their ability to actively expel therapeutic drugs via transport proteins. Such proteins are a family known as ATP-binding cassette transporters. These proteins use ATP-dependent drug efflux pumps for drug elimination, mostly into the extracellular space, and they have been found to be overexpressed in CSCs using side population assays[41,98-100].
Additionally, high ALDH activity is directly related to a higher resistance to several drugs, for example, cyclophosphamide, temozolomide, irinotecan, paclitaxel, and doxorubicin[101-103]. Resistance conferred by ALDH has been observed in numerous cell lines and patient samples[97,104]. A well known case is the resistance to cyclophosphamide, where ALDH irreversibly oxidizes aldophosphamide, an active metabolite of cyclophosphamide, into an inert compound[105]. In breast cancer, the inhibition of ALDH activity in ALDHhigh CD44+ cells leads to a reduction in chemoresistance to doxorubicin and paclitaxel[106]. This information suggests that the inhibition of ALDH activity leads to cell sensitization to chemotherapeutics[99].
Besides higher resistance to conventional cancer treatments, evidence shows that highly metastatic tumors correlate with a higher percentage of CSCs[28].
Most publications about the identification of CSCs have been performed in cell lines. However, in this section, we will discuss the cases in which CSCs were identified in patient samples.
CD133 was analyzed in a meta-analysis of 32 studies of non-small cell lung cancer, and a higher CD133 expression was associated with poor tumor differentiation and lymph node metastasis[107].
Gastric CSCs have been identified in tumor tissues and peripheral blood using the CD44+CD54+ phenotype[108]. Nevertheless, in another study, CD133+/CD44+ cells sorted from 44 patients who underwent gastrostomy failed to produce tumors in mice and did not show any CSC properties[109].
The presence of ALDH has been analyzed in normal mammary and breast cancer tissues[110]. The activity of ALDH1A3 is associated with metastasis in patient breast cancer samples by microarray analysis[86]. In another analysis of formalin-fixed paraffin-embedded tissue samples from primary stage IV breast cancer, ALDH and CD44/CD24 expression was correlated with response to endocrine therapy and clinical outcome but was not statistically significant[111].
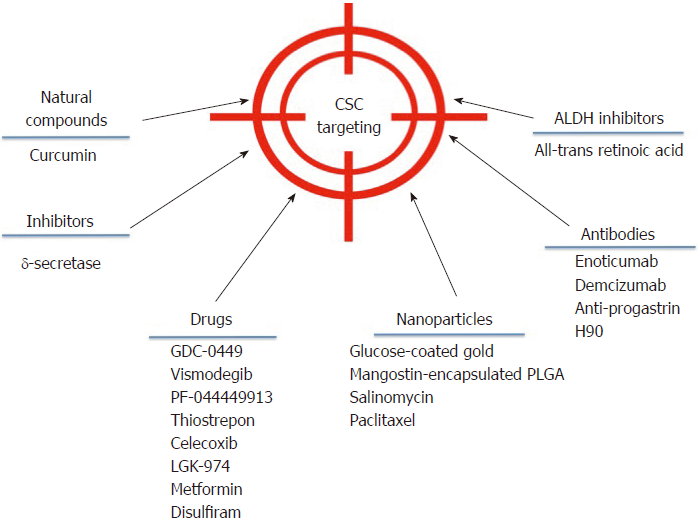
Despite the broad variety of CSC publications in the last years, the discovery of effective therapies has remained elusive. However, some advances have been made in the field that could be getting us closer to direct CSC elimination. A brief outline of some of these strategies is showed in Figure 2.
Targeting deregulated pathways in CSCs aims at developing effective strategies against CSCs. In adult pancreas, the Hedgehog (Hh) signaling pathway is dormant, but it is upregulated in pancreatic ductal adenocarcinoma, specifically in CD44+/CD24+/ESA+CSCs. In a phase I study, 68 patients were treated with GDC-0449 or Vismodegib, a Hh pathway antagonist[112], alone or in combination with gemcitabine. GDC-0449 inhibited Hh signaling, but there was no correlation with survival or other parameters[113]. Other drugs that show promising results in inhibiting this pathway are PF-044449913[114] and thiostrepon, which attenuates CD44+/CD24- triple-negative breast CSCs[115].
In addition, γ–secretase inhibitors target the Notch pathway and possess a stronger anti-neoplastic activity when combined with chemotherapeutic agents[116]. Nevertheless, adverse effects have been reported, as patients developed cutaneous rash in phase I clinical trials[117,118].
Several drugs that aim to inhibit the Wnt/β-catenin signaling pathway are being developed. One such drug is Celecoxib, a non-steroidal anti-inflammatory drug that inhibits β-catenin signaling by cyclo-oxygenase (commonly known as COX)-dependent and COX-independent mechanisms[116]. This drug downregulates CD133 expression in colon cancer cells by inhibiting Wnt signaling[119] and intestinal cancer growth[120]. The Wnt inhibitor LGK-974 inhibits porcupine, an O-acyltransferase required for Wnt secretion. In liver cancer cells, LGK-974 blocks secretion of the Wnt3A protein, and as a consequence, cells become more sensitive to radiation[121]. A recent study showed that LGK-974 downregulates ALDH1A3 and reduces chemoresistance in glioblastoma cells[122].
Curcumin is an antioxidant derived from turmeric whose anti-cancer effect is well documented. Referring specifically to CSCs, curcumin has shown the potential to regulate the CSC self-renewal pathways, as well as specific microRNAs[123]. In CD133+ lung CSCs, curcumin suppresses the activation of Wnt/β-catenin and Shh pathways, as well as other CSC traits[124]. It has been demonstrated that in bladder cancer, curcumin suppress the Shh pathway[125] and in laryngeal carcinoma treatment, curcumin enhances the effectiveness of cisplatin, reducing CD133+ cells in vitro[46]. Additionally, a combination of curcumin and FOLFOX chemotherapy inhibits colorectal CSCs in ex vivo models[126].
An interesting strategy is to target CSCs using nanoparticles to reduce side effects on surrounding normal cells. In 2015, construction of glucose-coated gold nanoparticles (Glu-GNPs) that used glucose to facilitate GNP entry into leukemic stem cells overexpressing CD44 (TH1-P) was reported. Leukemic cells were cultured for one hour in the absence of glucose for better Glu-GNP uptake, and then X-ray irradiation tests were performed. Results showed that Glu-GNPs enhanced cell death compared to either irradiation or GNPs alone[127]. Formulated mangostin-encapsulated poly(lactic-co-glycolic acid) nanoparticles (Mang-NPs) successfully downregulated the known stemness genes c-Myc, Nanog and Oct4, two CSC markers, CD24 and CD133, and the Shh pathway[128]. Salinomycin and paclitaxel nanoparticles are also being used to eliminate breast cancer cells including CD44 breast CSCs[129].
Interestingly, CSCs have a strict dependence on mitochondrial biogenesis. Five classes of FDA-approved antibiotics that inhibit mitochondrial biogenesis were used on eight different cancer cell lines, and the results suggested that the observed therapeutic effects were infection-independent[130]. Clinical trials using doxycycline showed positive results in cancer patients[131]. Another drug that has been shown to specifically eliminate CSCs is metformin, and its effects are enhanced when it is used in combination with doxorubicin[132]. Moreover, it has been observed that metformin reduces metastasis by targeting both EMT and CSCs[133]. In the ovarian cancer cell line SKOV3, low doses of metformin diminished CD44+CD117+ CSCs in xenograft tissue and enhanced the effect of cisplatin[134]. In esophageal cancer, metformin reduced the number of ALDH+ cells, tumor growth in vivo[135], and in pancreatic cancer, it increased radiation sensitivity[136].
Using antibodies is another strategy to block CSC signaling pathways and reduce tumor activity in different models. For instance, the anti-DLL4 (Enoticumab) antibody that targets the dominant Notch ligand DLL4 has shown anti-tumor activity, especially in VEGF-resistant tumors in human phase I studies[137]. Furthermore, another anti-DLL4 antibody (Demcizumab) is effective in decreasing tumor size but produces hypertension[138]. In colon cancer patients, increased progastrin levels in the blood have been observed, which is a tumor-promoting peptide that participates in colon CSC self-renewal and is also a direct target gene of β–catenin/Tcf4. Based on this information, specific anti-progastrin antibodies have been developed and tested in colon cancer cell lines and in mice. The antibodies, alone or in combination with chemotherapy, decreased self-renewal, migration and invasion. Moreover, they mitigated Wnt-driven intestinal neoplasia and induced tumor cell differentiation in vivo[139]. H90 is a mouse IgG1 mAb against human CD44 that directly targets CSCs to induce differentiation and proliferation in AML xenograft mouse models[140]. Additionally, anti-CD44s-specific antibodies are effective in eliminating pancreatic stem cells[141]. For more extensive information about antibodies against CSCs, we recommend reference[142].
ALDH is an important CSC marker that is overexpressed in several cancers. Specific ALDH inhibitors are effective in modulating cell growth, apoptosis and differentiation. Additionally, increased chemo- and radio-sensitivity is usually observed. All-trans retinoic acid (commonly known as ATRA) is a first generation systemic retinoid that promotes cell differentiation[143,144] and has been used in clinical trials[145]. ATRA has also been tested in breast cancer cells[106,146,147] and in gastric cancer, where it inhibited tumor growth[148], and in head and neck cancer, where it suppressed Wnt/β-catenin signaling[149]. In a phase I/II trial, advanced breast cancer patients did not show a significant improvement when treated with ATRA and tamoxifen compared with tamoxifen alone[150].
Disulfiram is a drug used for treating alcoholism, and it shows anti-cancer activity in vitro and in vivo, further potentiating the chemotherapeutic response. Its effectiveness has been demonstrated on paclitaxel-resistant triple-negative breast cancer cells[151], in non-small cell lung cancer cells[152], and glioblastoma.
CSCs are potential cancer therapy targets due to their tumorigenic capabilities, such as chemo- and radio-resistance, phenomena involved in tumor relapse in patients. Several efforts have been made to continue to identify the CSCs in several tumors to better understand the mechanisms related to tumor resistance in oncologic patients. It is known that de-regulated cell signaling pathways are partially responsible for maintaining CSC stemness. Consequently, Wnt, Notch and Hh signaling pathways have been studied to develop an efficient anti-CSC therapy. However, innovative anti-cancer treatments need to be developed to improve the lifespan and quality of life of cancer patients. Finally, we suggest that there cannot be a generalized CSC phenotype to design and promote new drugs, antibodies, nanoparticles, and cellular treatments to treat oncological patients. Taken together, we suggest the full characterization of phenotypes and capabilities of CSCs in patients, a cellular component responsible for tumor progression, tumor relapse and metastasis.
The authors thank Marco Antonio Meraz Rodríguez for his constructive suggestions and Dra. Elizabeth Langley McCarron for her editing and we thank Intituto Nacional de Cancerología, Instituto Politécnico Nacional and Posgrado de Biomedicina y Biotecnología Molecular.
| 1. | Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3316] [Cited by in RCA: 3436] [Article Influence: 107.4] [Reference Citation Analysis (0)] |
| 2. | Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339-9344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2176] [Cited by in RCA: 2214] [Article Influence: 110.7] [Reference Citation Analysis (0)] |
| 3. | Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 927] [Cited by in RCA: 928] [Article Influence: 48.8] [Reference Citation Analysis (0)] |
| 4. | Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105-111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6844] [Cited by in RCA: 6980] [Article Influence: 279.2] [Reference Citation Analysis (0)] |
| 5. | Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983-3988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7830] [Cited by in RCA: 7800] [Article Influence: 339.1] [Reference Citation Analysis (0)] |
| 6. | Park CH, Bergsagel DE, McCulloch EA. Mouse myeloma tumor stem cells: a primary cell culture assay. J Natl Cancer Inst. 1971;46:411-422. [PubMed] |
| 7. | Hamburger AW, Salmon SE. Primary bioassay of human tumor stem cells. Science. 1977;197:461-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1318] [Cited by in RCA: 1293] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 8. | Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4851] [Cited by in RCA: 4928] [Article Influence: 169.9] [Reference Citation Analysis (1)] |
| 9. | Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178-15183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1432] [Cited by in RCA: 1400] [Article Influence: 60.9] [Reference Citation Analysis (1)] |
| 10. | Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821-5828. [PubMed] |
| 11. | Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5422] [Cited by in RCA: 5627] [Article Influence: 255.8] [Reference Citation Analysis (0)] |
| 12. | Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555-567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3173] [Cited by in RCA: 3106] [Article Influence: 163.5] [Reference Citation Analysis (0)] |
| 13. | Greve B, Kelsch R, Spaniol K, Eich HT, Götte M. Flow cytometry in cancer stem cell analysis and separation. Cytometry A. 2012;81:284-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (1)] |
| 14. | Rao QX, Yao TT, Zhang BZ, Lin RC, Chen ZL, Zhou H, Wang LJ, Lu HW, Chen Q, Di N. Expression and functional role of ALDH1 in cervical carcinoma cells. Asian Pac J Cancer Prev. 2012;13:1325-1331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 15. | Shackleton M. Normal stem cells and cancer stem cells: similar and different. Semin Cancer Biol. 2010;20:85-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 111] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 16. | Yu SC, Ping YF, Yi L, Zhou ZH, Chen JH, Yao XH, Gao L, Wang JM, Bian XW. Isolation and characterization of cancer stem cells from a human glioblastoma cell line U87. Cancer Lett. 2008;265:124-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 178] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 17. | Qiang L, Yang Y, Ma YJ, Chen FH, Zhang LB, Liu W, Qi Q, Lu N, Tao L, Wang XT. Isolation and characterization of cancer stem like cells in human glioblastoma cell lines. Cancer Lett. 2009;279:13-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 159] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 18. | Brescia P, Ortensi B, Fornasari L, Levi D, Broggi G, Pelicci G. CD133 is essential for glioblastoma stem cell maintenance. Stem Cells. 2013;31:857-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 187] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 19. | Wang L, Guo H, Lin C, Yang L, Wang X. Enrichment and characterization of cancer stemlike cells from a cervical cancer cell line. Mol Med Rep. 2014;9:2117-2123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 20. | Fillmore CM, Kuperwasser C. Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res. 2008;10:R25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 755] [Cited by in RCA: 810] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 21. | Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, Pratesi G, Fabbri A, Andriani F, Tinelli S. Highly tumorigenic lung cancer CD133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci USA. 2009;106:16281-16286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 562] [Cited by in RCA: 648] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 22. | Li X, Lewis MT, Huang J, Gutierrez C, Osborne CK, Wu MF, Hilsenbeck SG, Pavlick A, Zhang X, Chamness GC. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100:672-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1319] [Cited by in RCA: 1379] [Article Influence: 76.6] [Reference Citation Analysis (0)] |
| 23. | Liao J, Qian F, Tchabo N, Mhawech-Fauceglia P, Beck A, Qian Z, Wang X, Huss WJ, Lele SB, Morrison CD. Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism. PLoS One. 2014;9:e84941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 277] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 24. | Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4467] [Cited by in RCA: 4850] [Article Influence: 242.5] [Reference Citation Analysis (0)] |
| 25. | Clarke MF. A self-renewal assay for cancer stem cells. Cancer Chemother Pharmacol. 2005;56 Suppl 1:64-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 32] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Eyler CE, Rich JN. Survival of the fittest: cancer stem cells in therapeutic resistance and angiogenesis. J Clin Oncol. 2008;26:2839-2845. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 579] [Cited by in RCA: 553] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 27. | Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat Med. 2009;15:1010-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 718] [Cited by in RCA: 718] [Article Influence: 42.2] [Reference Citation Analysis (0)] |
| 28. | Ajani JA, Song S, Hochster HS, Steinberg IB. Cancer stem cells: the promise and the potential. Semin Oncol. 2015;42 Suppl 1:S3-S17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 240] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 29. | Vlashi E, Pajonk F. Cancer stem cells, cancer cell plasticity and radiation therapy. Semin Cancer Biol. 2015;31:28-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 239] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 30. | Visvader JE. Cells of origin in cancer. Nature. 2011;469:314-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1008] [Cited by in RCA: 1103] [Article Influence: 73.5] [Reference Citation Analysis (0)] |
| 31. | Ishizawa K, Rasheed ZA, Karisch R, Wang Q, Kowalski J, Susky E, Pereira K, Karamboulas C, Moghal N, Rajeshkumar NV. Tumor-initiating cells are rare in many human tumors. Cell Stem Cell. 2010;7:279-282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 183] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 32. | Quintana E, Shackleton M, Foster HR, Fullen DR, Sabel MS, Johnson TM, Morrison SJ. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 2010;18:510-523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 497] [Cited by in RCA: 452] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 33. | Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717-728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 916] [Cited by in RCA: 1005] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 34. | Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, Metzeler KH, Poeppl A, Ling V, Beyene J. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086-1093. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 712] [Cited by in RCA: 846] [Article Influence: 56.4] [Reference Citation Analysis (0)] |
| 35. | Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2552] [Cited by in RCA: 2650] [Article Influence: 147.2] [Reference Citation Analysis (0)] |
| 36. | Khan IN, Al-Karim S, Bora RS, Chaudhary AG, Saini KS. Cancer stem cells: a challenging paradigm for designing targeted drug therapies. Drug Discov Today. 2015;20:1205-1216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 37. | Kelly PN, Dakic A, Adams JM, Nutt SL, Strasser A. Tumor growth need not be driven by rare cancer stem cells. Science. 2007;317:337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 612] [Cited by in RCA: 581] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 38. | Alvarado-Ortiz E, Sarabia-Sanchez MA, Garcia-Carranca A. Molecular mechanisms underlying the functions of cellular markers associated with the phenotype of Cancer Stem Cells. Curr Stem Cell Res Ther. 2018;. [PubMed] |
| 39. | Ren F, Sheng WQ, Du X. CD133: a cancer stem cells marker, is used in colorectal cancers. World J Gastroenterol. 2013;19:2603-2611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 106] [Cited by in RCA: 135] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 40. | Lin L, Fuchs J, Li C, Olson V, Bekaii-Saab T, Lin J. STAT3 signaling pathway is necessary for cell survival and tumorsphere forming capacity in ALDH+/CD133+ stem cell-like human colon cancer cells. Biochem Biophys Res Commun. 2011;416:246-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 94] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 41. | Catalano V, Di Franco S, Iovino F, Dieli F, Stassi G, Todaro M. CD133 as a target for colon cancer. Expert Opin Ther Targets. 2012;16:259-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 42. | Nakamura M, Zhang X, Mizumoto Y, Maida Y, Bono Y, Takakura M, Kyo S. Molecular characterization of CD133+ cancer stem-like cells in endometrial cancer. Int J Oncol. 2014;44:669-677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 43. | Wang S, Xu ZY, Wang LF, Su W. CD133+ cancer stem cells in lung cancer. Front Biosci (Landmark Ed). 2013;18:447-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 44. | Sarvi S, Mackinnon AC, Avlonitis N, Bradley M, Rintoul RC, Rassl DM, Wang W, Forbes SJ, Gregory CD, Sethi T. CD133+ cancer stem-like cells in small cell lung cancer are highly tumorigenic and chemoresistant but sensitive to a novel neuropeptide antagonist. Cancer Res. 2014;74:1554-1565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 165] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 45. | Wu CP, Du HD, Gong HL, Li DW, Tao L, Tian J, Zhou L. Hypoxia promotes stem-like properties of laryngeal cancer cell lines by increasing the CD133+ stem cell fraction. Int J Oncol. 2014;44:1652-1660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 46. | Zhang H, Yu T, Wen L, Wang H, Fei D, Jin C. Curcumin enhances the effectiveness of cisplatin by suppressing CD133+ cancer stem cells in laryngeal carcinoma treatment. Exp Ther Med. 2013;6:1317-1321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 47. | Piao LS, Hur W, Kim TK, Hong SW, Kim SW, Choi JE, Sung PS, Song MJ, Lee BC, Hwang D. CD133+ liver cancer stem cells modulate radioresistance in human hepatocellular carcinoma. Cancer Lett. 2012;315:129-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 159] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 48. | Abbasian M, Mousavi E, Arab-Bafrani Z, Sahebkar A. The most reliable surface marker for the identification of colorectal cancer stem-like cells: A systematic review and meta-analysis. J Cell Physiol. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 49. | Lee HH, Seo KJ, An CH, Kim JS, Jeon HM. CD133 expression is correlated with chemoresistance and early recurrence of gastric cancer. J Surg Oncol. 2012;106:999-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 50. | Wang Y, Chen M, Wu Z, Tong C, Dai H, Guo Y, Liu Y, Huang J, Lv H, Luo C. CD133-directed CAR T cells for advanced metastasis malignancies: A phase I trial. Oncoimmunology. 2018;7:e1440169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 266] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 51. | Nosrati A, Naghshvar F, Khanari S. Cancer Stem Cell Markers CD44, CD133 in Primary Gastric Adenocarcinoma. Int J Mol Cell Med. 2014;3:279-286. [PubMed] |
| 52. | Yan Y, Zuo X, Wei D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Transl Med. 2015;4:1033-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 484] [Cited by in RCA: 506] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 53. | Jaggupilli A, Elkord E. Significance of CD44 and CD24 as cancer stem cell markers: an enduring ambiguity. Clin Dev Immunol. 2012;2012:708036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 282] [Cited by in RCA: 374] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 54. | Yang CH, Wang HL, Lin YS, Kumar KP, Lin HC, Chang CJ, Lu CC, Huang TT, Martel J, Ojcius DM. Identification of CD24 as a cancer stem cell marker in human nasopharyngeal carcinoma. PLoS One. 2014;9:e99412. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 50] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 55. | Liu H, Wang YJ, Bian L, Fang ZH, Zhang QY, Cheng JX. CD44+/CD24+ cervical cancer cells resist radiotherapy and exhibit properties of cancer stem cells. Eur Rev Med Pharmacol Sci. 2016;20:1745-1754. [PubMed] |
| 56. | Zhang C, Li C, He F, Cai Y, Yang H. Identification of CD44+CD24+ gastric cancer stem cells. J Cancer Res Clin Oncol. 2011;137:1679-1686. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 169] [Cited by in RCA: 161] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 57. | Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946-10951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1998] [Cited by in RCA: 2045] [Article Influence: 102.3] [Reference Citation Analysis (0)] |
| 58. | Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696-1708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 727] [Cited by in RCA: 731] [Article Influence: 36.6] [Reference Citation Analysis (10)] |
| 59. | Prochazka L, Tesarik R, Turanek J. Regulation of alternative splicing of CD44 in cancer. Cell Signal. 2014;26:2234-2239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 146] [Article Influence: 12.2] [Reference Citation Analysis (1)] |
| 60. | Biddle A, Liang X, Gammon L, Fazil B, Harper LJ, Emich H, Costea DE, Mackenzie IC. Cancer stem cells in squamous cell carcinoma switch between two distinct phenotypes that are preferentially migratory or proliferative. Cancer Res. 2011;71:5317-5326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 276] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 61. | Hernandez JR, Kim JJ, Verdone JE, Liu X, Torga G, Pienta KJ, Mooney SM. Alternative CD44 splicing identifies epithelial prostate cancer cells from the mesenchymal counterparts. Med Oncol. 2015;32:159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 62. | Biddle A, Gammon L, Fazil B, Mackenzie IC. CD44 staining of cancer stem-like cells is influenced by down-regulation of CD44 variant isoforms and up-regulation of the standard CD44 isoform in the population of cells that have undergone epithelial-to-mesenchymal transition. PLoS One. 2013;8:e57314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 63. | Inoue K, Fry EA. Aberrant Splicing of Estrogen Receptor, HER2, and CD44 Genes in Breast Cancer. Genet Epigenet. 2015;7:19-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 56] [Reference Citation Analysis (0)] |
| 64. | Zöller M. CD44: can a cancer-initiating cell profit from an abundantly expressed molecule? Nat Rev Cancer. 2011;11:254-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 785] [Cited by in RCA: 887] [Article Influence: 59.1] [Reference Citation Analysis (0)] |
| 65. | Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 733] [Cited by in RCA: 751] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 66. | Zhang L, Zheng W, Wang Y, Wang Y, Huang H. Human bone marrow mesenchymal stem cells support the derivation and propagation of human induced pluripotent stem cells in culture. Cell Reprogram. 2013;15:216-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 67. | Jing F, Kim HJ, Kim CH, Kim YJ, Lee JH, Kim HR. Colon cancer stem cell markers CD44 and CD133 in patients with colorectal cancer and synchronous hepatic metastases. Int J Oncol. 2015;46:1582-1588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 87] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 68. | Joshua B, Kaplan MJ, Doweck I, Pai R, Weissman IL, Prince ME, Ailles LE. Frequency of cells expressing CD44, a head and neck cancer stem cell marker: correlation with tumor aggressiveness. Head Neck. 2012;34:42-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 121] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 69. | Faber A, Barth C, Hörmann K, Kassner S, Schultz JD, Sommer U, Stern-Straeter J, Thorn C, Goessler UR. CD44 as a stem cell marker in head and neck squamous cell carcinoma. Oncol Rep. 2011;26:321-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 22] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 70. | Meng E, Long B, Sullivan P, McClellan S, Finan MA, Reed E, Shevde L, Rocconi RP. CD44+/CD24- ovarian cancer cells demonstrate cancer stem cell properties and correlate to survival. Clin Exp Metastasis. 2012;29:939-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 71. | Lau WM, Teng E, Chong HS, Lopez KA, Tay AY, Salto-Tellez M, Shabbir A, So JB, Chan SL. CD44v8-10 is a cancer-specific marker for gastric cancer stem cells. Cancer Res. 2014;74:2630-2641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 177] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 72. | Watt FM. Role of integrins in regulating epidermal adhesion, growth and differentiation. EMBO J. 2002;21:3919-3926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 508] [Cited by in RCA: 496] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 73. | Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, Macswords J, Wu Q, Vasanji A, McLendon RE, Hjelmeland AB. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421-432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 546] [Cited by in RCA: 503] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 74. | Yamamoto H, Masters JR, Dasgupta P, Chandra A, Popert R, Freeman A, Ahmed A. CD49f is an efficient marker of monolayer- and spheroid colony-forming cells of the benign and malignant human prostate. PLoS One. 2012;7:e46979. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 75. | Fukamachi H, Seol HS, Shimada S, Funasaka C, Baba K, Kim JH, Park YS, Kim MJ, Kato K, Inokuchi M. CD49f(high) cells retain sphere-forming and tumor-initiating activities in human gastric tumors. PLoS One. 2013;8:e72438. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 76. | Ye F, Zhong X, Qiu Y, Yang L, Wei B, Zhang Z, Bu H. CD49f Can Act as a Biomarker for Local or Distant Recurrence in Breast Cancer. J Breast Cancer. 2017;20:142-149. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 77. | Haraguchi N, Ishii H, Mimori K, Ohta K, Uemura M, Nishimura J, Hata T, Takemasa I, Mizushima T, Yamamoto H. CD49f-positive cell population efficiently enriches colon cancer-initiating cells. Int J Oncol. 2013;43:425-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 78. | Sládek NE. Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact. J Biochem Mol Toxicol. 2003;17:7-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 202] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 79. | Storms RW, Trujillo AP, Springer JB, Shah L, Colvin OM, Ludeman SM, Smith C. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci USA. 1999;96:9118-9123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 407] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 80. | Toledo-Guzmán ME, Ibañez Hernández M, Gomez-Gallegos AA, Ortiz-Sánchez E. ALDH as a Stem Cell marker in solid tumors. Curr Stem Cell Res Ther. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 81. | Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382-3389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 831] [Cited by in RCA: 834] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 82. | Shenoy A, Butterworth E, Huang EH. ALDH as a marker for enriching tumorigenic human colonic stem cells. Methods Mol Biol. 2012;916:373-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 83. | Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, Wang H, Liu Z, Su Y, Stass SA. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 598] [Cited by in RCA: 626] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 84. | Liu SY, Zheng PS. High aldehyde dehydrogenase activity identifies cancer stem cells in human cervical cancer. Oncotarget. 2013;4:2462-2475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 108] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 85. | Ortiz-Sánchez E, Santiago-López L, Cruz-Domínguez VB, Toledo-Guzmán ME, Hernández-Cueto D, Muñiz-Hernández S, Garrido E, Cantú De León D, García-Carrancá A. Characterization of cervical cancer stem cell-like cells: phenotyping, stemness, and human papilloma virus co-receptor expression. Oncotarget. 2016;7:31943-31954. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 63] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 86. | Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform ALDH1A3 and its expression is predictive of metastasis. Stem Cells. 2011;29:32-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 366] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 87. | Kim SK, Kim H, Lee DH, Kim TS, Kim T, Chung C, Koh GY, Kim H, Lim DS. Reversing the intractable nature of pancreatic cancer by selectively targeting ALDH-high, therapy-resistant cancer cells. PLoS One. 2013;8:e78130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 88. | Hoshino Y, Nishida J, Katsuno Y, Koinuma D, Aoki T, Kokudo N, Miyazono K, Ehata S. Smad4 Decreases the Population of Pancreatic Cancer-Initiating Cells through Transcriptional Repression of ALDH1A1. Am J Pathol. 2015;185:1457-1470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 89. | Luo Y, Nguyen N, Fujita M. Isolation of human melanoma stem cells using ALDH as a marker. Curr Protoc Stem Cell Biol. 2013;26:Unit 3.8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 90. | Luo Y, Dallaglio K, Chen Y, Robinson WA, Robinson SE, McCarter MD, Wang J, Gonzalez R, Thompson DC, Norris DA. ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells. 2012;30:2100-2113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 233] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 91. | Kryczek I, Liu S, Roh M, Vatan L, Szeliga W, Wei S, Banerjee M, Mao Y, Kotarski J, Wicha MS. Expression of aldehyde dehydrogenase and CD133 defines ovarian cancer stem cells. Int J Cancer. 2012;130:29-39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 214] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 92. | Silva IA, Bai S, McLean K, Yang K, Griffith K, Thomas D, Ginestier C, Johnston C, Kueck A, Reynolds RK. Aldehyde dehydrogenase in combination with CD133 defines angiogenic ovarian cancer stem cells that portend poor patient survival. Cancer Res. 2011;71:3991-4001. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 420] [Cited by in RCA: 415] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 93. | Mansour SF, Atwa MM. Clinicopathological Significance of CD133 and ALDH1 Cancer Stem Cell Marker Expression in Invasive Ductal Breast Carcinoma. Asian Pac J Cancer Prev. 2015;16:7491-7496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 94. | Roudi R, Korourian A, Shariftabrizi A, Madjd Z. Differential Expression of Cancer Stem Cell Markers ALDH1 and CD133 in Various Lung Cancer Subtypes. Cancer Invest. 2015;33:294-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 95. | Qiu Y, Pu T, Guo P, Wei B, Zhang Z, Zhang H, Zhong X, Zheng H, Chen L, Bu H. ALDH(+)/CD44(+) cells in breast cancer are associated with worse prognosis and poor clinical outcome. Exp Mol Pathol. 2016;100:145-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 18] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 96. | Liu J, Xiao Z, Wong SK, Tin VP, Ho KY, Wang J, Sham MH, Wong MP. Lung cancer tumorigenicity and drug resistance are maintained through ALDH(hi)CD44(hi) tumor initiating cells. Oncotarget. 2013;4:1698-1711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 82] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 97. | Januchowski R, Wojtowicz K, Zabel M. The role of aldehyde dehydrogenase (ALDH) in cancer drug resistance. Biomed Pharmacother. 2013;67:669-680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 160] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 98. | Leonard GD, Fojo T, Bates SE. The role of ABC transporters in clinical practice. Oncologist. 2003;8:411-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 550] [Cited by in RCA: 589] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 99. | Abdullah LN, Chow EK. Mechanisms of chemoresistance in cancer stem cells. Clin Transl Med. 2013;2:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 477] [Cited by in RCA: 570] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 100. | Eyre R, Harvey I, Stemke-Hale K, Lennard TW, Tyson-Capper A, Meeson AP. Reversing paclitaxel resistance in ovarian cancer cells via inhibition of the ABCB1 expressing side population. Tumour Biol. 2014;35:9879-9892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 101. | Zhao J. Cancer stem cells and chemoresistance: The smartest survives the raid. Pharmacol Ther. 2016;160:145-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 350] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 102. | Pearce DJ, Taussig D, Simpson C, Allen K, Rohatiner AZ, Lister TA, Bonnet D. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells. 2005;23:752-760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 213] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 103. | Tanei T, Morimoto K, Shimazu K, Kim SJ, Tanji Y, Taguchi T, Tamaki Y, Noguchi S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential Paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin Cancer Res. 2009;15:4234-4241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 406] [Cited by in RCA: 441] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 104. | Sreerama L, Sladek NE. Cellular levels of class 1 and class 3 aldehyde dehydrogenases and certain other drug-metabolizing enzymes in human breast malignancies. Clin Cancer Res. 1997;3:1901-1914. [PubMed] |
| 105. | Vasiliou V, Pappa A, Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev. 2004;36:279-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 234] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 106. | Croker AK, Allan AL. Inhibition of aldehyde dehydrogenase (ALDH) activity reduces chemotherapy and radiation resistance of stem-like ALDHhiCD44+ human breast cancer cells. Breast Cancer Res Treat. 2012;133:75-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 238] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 107. | Chen E, Zeng Z, Bai B, Zhu J, Song Z. The prognostic value of CSCs biomarker CD133 in NSCLC: a meta-analysis. Oncotarget. 2016;7:56526-56539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 108. | Chen T, Yang K, Yu J, Meng W, Yuan D, Bi F, Liu F, Liu J, Dai B, Chen X. Identification and expansion of cancer stem cells in tumor tissues and peripheral blood derived from gastric adenocarcinoma patients. Cell Res. 2012;22:248-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 151] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 109. | Rocco A, Liguori E, Pirozzi G, Tirino V, Compare D, Franco R, Tatangelo F, Palaia R, D’Armiento FP, Pollastrone G. CD133 and CD44 cell surface markers do not identify cancer stem cells in primary human gastric tumors. J Cell Physiol. 2012;227:2686-2693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 110. | Singer CF, Zabkova P, Rappaport C, Muhr D, Pfeiler G, Gschwantler-Kaulich D, Fink-Retter A, Staudigl C, Walter I, Hudelist G. Presence of intratumoral stem cells in breast cancer patients with or without BRCA germline mutations. Curr Cancer Drug Targets. 2012;12:44-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 111. | Hashimoto K, Shimizu C, Tsuda H, Saji S, Osaki A, Shigekawa T, Aogi K. Immunohistochemical detection of breast cancer stem cells in hormone receptor-positive breast cancer and their role in response to endocrine therapy and clinical outcome. Oncology. 2012;82:168-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 112. | Singh BN, Fu J, Srivastava RK, Shankar S. Hedgehog signaling antagonist GDC-0449 (Vismodegib) inhibits pancreatic cancer stem cell characteristics: molecular mechanisms. PLoS One. 2011;6:e27306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 156] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 113. | Kim EJ, Sahai V, Abel EV, Griffith KA, Greenson JK, Takebe N, Khan GN, Blau JL, Craig R, Balis UG. Pilot clinical trial of hedgehog pathway inhibitor GDC-0449 (vismodegib) in combination with gemcitabine in patients with metastatic pancreatic adenocarcinoma. Clin Cancer Res. 2014;20:5937-5945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 245] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 114. | Sadarangani A, Pineda G, Lennon KM, Chun HJ, Shih A, Schairer AE, Court AC, Goff DJ, Prashad SL, Geron I. GLI2 inhibition abrogates human leukemia stem cell dormancy. J Transl Med. 2015;13:98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 115. | Yang N, Zhou TC, Lei XX, Wang C, Yan M, Wang ZF, Liu W, Wang J, Ming KH, Wang BC. Inhibition of Sonic Hedgehog Signaling Pathway by Thiazole Antibiotic Thiostrepton Attenuates the CD44+/CD24-Stem-Like Population and Sphere-Forming Capacity in Triple-Negative Breast Cancer. Cell Physiol Biochem. 2016;38:1157-1170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 116. | Takebe N, Miele L, Harris PJ, Jeong W, Bando H, Kahn M, Yang SX, Ivy SP. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat Rev Clin Oncol. 2015;12:445-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 916] [Cited by in RCA: 1040] [Article Influence: 94.5] [Reference Citation Analysis (2)] |
| 117. | Krop I, Demuth T, Guthrie T, Wen PY, Mason WP, Chinnaiyan P, Butowski N, Groves MD, Kesari S, Freedman SJ. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J Clin Oncol. 2012;30:2307-2313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 259] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 118. | Messersmith WA, Shapiro GI, Cleary JM, Jimeno A, Dasari A, Huang B, Shaik MN, Cesari R, Zheng X, Reynolds JM. A Phase I, dose-finding study in patients with advanced solid malignancies of the oral γ-secretase inhibitor PF-03084014. Clin Cancer Res. 2015;21:60-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 184] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 119. | Deng Y, Su Q, Mo J, Fu X, Zhang Y, Lin EH. Celecoxib downregulates CD133 expression through inhibition of the Wnt signaling pathway in colon cancer cells. Cancer Invest. 2013;31:97-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 120. | Egashira I, Takahashi-Yanaga F, Nishida R, Arioka M, Igawa K, Tomooka K, Nakatsu Y, Tsuzuki T, Nakabeppu Y, Kitazono T. Celecoxib and 2,5-dimethylcelecoxib inhibit intestinal cancer growth by suppressing the Wnt/β-catenin signaling pathway. Cancer Sci. 2017;108:108-115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 56] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 121. | Tian D, Shi Y, Chen D, Liu Q, Fan F. The Wnt inhibitor LGK-974 enhances radiosensitivity of HepG2 cells by modulating Nrf2 signaling. Int J Oncol. 2017;51:545-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 122. | Suwala AK, Koch K, Rios DH, Aretz P, Uhlmann C, Ogorek I, Felsberg J, Reifenberger G, Köhrer K, Deenen R. Inhibition of Wnt/beta-catenin signaling downregulates expression of aldehyde dehydrogenase isoform 3A1 (ALDH3A1) to reduce resistance against temozolomide in glioblastoma in vitro. Oncotarget. 2018;9:22703-22716. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 123. | Li Y, Zhang T. Targeting cancer stem cells by curcumin and clinical applications. Cancer Lett. 2014;346:197-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 124. | Zhu JY, Yang X, Chen Y, Jiang Y, Wang SJ, Li Y, Wang XQ, Meng Y, Zhu MM, Ma X. Curcumin Suppresses Lung Cancer Stem Cells via Inhibiting Wnt/β-catenin and Sonic Hedgehog Pathways. Phytother Res. 2017;31:680-688. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 130] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 125. | Wang D, Kong X, Li Y, Qian W, Ma J, Wang D, Yu D, Zhong C. Curcumin inhibits bladder cancer stem cells by suppressing Sonic Hedgehog pathway. Biochem Biophys Res Commun. 2017;493:521-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 49] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 126. | James MI, Iwuji C, Irving G, Karmokar A, Higgins JA, Griffin-Teal N, Thomas A, Greaves P, Cai H, Patel SR. Curcumin inhibits cancer stem cell phenotypes in ex vivo models of colorectal liver metastases, and is clinically safe and tolerable in combination with FOLFOX chemotherapy. Cancer Lett. 2015;364:135-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 140] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 127. | Hu C, Niestroj M, Yuan D, Chang S, Chen J. Treating cancer stem cells and cancer metastasis using glucose-coated gold nanoparticles. Int J Nanomedicine. 2015;10:2065-2077. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 128. | Verma RK, Yu W, Shrivastava A, Shankar S, Srivastava RK. α-Mangostin-encapsulated PLGA nanoparticles inhibit pancreatic carcinogenesis by targeting cancer stem cells in human, and transgenic (Kras(G12D), and Kras(G12D)/tp53R270H) mice. Sci Rep. 2016;6:32743. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 60] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 129. | Muntimadugu E, Kumar R, Saladi S, Rafeeqi TA, Khan W. CD44 targeted chemotherapy for co-eradication of breast cancer stem cells and cancer cells using polymeric nanoparticles of salinomycin and paclitaxel. Colloids Surf B Biointerfaces. 2016;143:532-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 146] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 130. | Lamb R, Ozsvari B, Lisanti CL, Tanowitz HB, Howell A, Martinez-Outschoorn UE, Sotgia F, Lisanti MP. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: treating cancer like an infectious disease. Oncotarget. 2015;6:4569-4584. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 298] [Cited by in RCA: 390] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 131. | Lamb R, Fiorillo M, Chadwick A, Ozsvari B, Reeves KJ, Smith DL, Clarke RB, Howell SJ, Cappello AR, Martinez-Outschoorn UE. Doxycycline down-regulates DNA-PK and radiosensitizes tumor initiating cells: Implications for more effective radiation therapy. Oncotarget. 2015;6:14005-14025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 132. | Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells, and acts together with chemotherapy to block tumor growth and prolong remission. Cancer Res. 2009;69:7507-7511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 803] [Cited by in RCA: 884] [Article Influence: 52.0] [Reference Citation Analysis (0)] |
| 133. | Rattan R, Ali Fehmi R, Munkarah A. Metformin: an emerging new therapeutic option for targeting cancer stem cells and metastasis. J Oncol. 2012;2012:928127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 93] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 134. | Zhang R, Zhang P, Wang H, Hou D, Li W, Xiao G, Li C. Inhibitory effects of metformin at low concentration on epithelial-mesenchymal transition of CD44(+)CD117(+) ovarian cancer stem cells. Stem Cell Res Ther. 2015;6:262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 72] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 135. | Honjo S, Ajani JA, Scott AW, Chen Q, Skinner HD, Stroehlein J, Johnson RL, Song S. Metformin sensitizes chemotherapy by targeting cancer stem cells and the mTOR pathway in esophageal cancer. Int J Oncol. 2014;45:567-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 136. | Fasih A, Elbaz HA, Hüttemann M, Konski AA, Zielske SP. Radiosensitization of pancreatic cancer cells by metformin through the AMPK pathway. Radiat Res. 2014;182:50-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 137. | Chiorean EG, LoRusso P, Strother RM, Diamond JR, Younger A, Messersmith WA, Adriaens L, Liu L, Kao RJ, DiCioccio AT. A Phase I First-in-Human Study of Enoticumab (REGN421), a Fully Human Delta-like Ligand 4 (Dll4) Monoclonal Antibody in Patients with Advanced Solid Tumors. Clin Cancer Res. 2015;21:2695-2703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 111] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 138. | Smith DC, Eisenberg PD, Manikhas G, Chugh R, Gubens MA, Stagg RJ, Kapoun AM, Xu L, Dupont J, Sikic B. A phase I dose escalation and expansion study of the anticancer stem cell agent demcizumab (anti-DLL4) in patients with previously treated solid tumors. Clin Cancer Res. 2014;20:6295-6303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 139. | Prieur A, Cappellini M, Habif G, Lefranc MP, Mazard T, Morency E, Pascussi JM, Flacelière M, Cahuzac N, Vire B. Targeting the Wnt Pathway and Cancer Stem Cells with Anti-progastrin Humanized Antibodies as a Potential Treatment for K-RAS-Mutated Colorectal Cancer. Clin Cancer Res. 2017;23:5267-5280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 140. | Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12:1167-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 891] [Cited by in RCA: 913] [Article Influence: 45.7] [Reference Citation Analysis (0)] |
| 141. | Li L, Hao X, Qin J, Tang W, He F, Smith A, Zhang M, Simeone DM, Qiao XT, Chen ZN. Antibody against CD44s inhibits pancreatic tumor initiation and postradiation recurrence in mice. Gastroenterology. 2014;146:1108-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 125] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 142. | Naujokat C. Monoclonal antibodies against human cancer stem cells. Immunotherapy. 2014;6:290-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 143. | Pérez-Alea M, McGrail K, Sánchez-Redondo S, Ferrer B, Fournet G, Cortés J, Muñoz E, Hernandez-Losa J, Tenbaum S, Martin G. ALDH1A3 is epigenetically regulated during melanocyte transformation and is a target for melanoma treatment. Oncogene. 2017;36:5695-5708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 47] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 144. | Gudas LJ, Wagner JA. Retinoids regulate stem cell differentiation. J Cell Physiol. 2011;226:322-330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 329] [Cited by in RCA: 318] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 145. | Petrie K, Zelent A, Waxman S. Differentiation therapy of acute myeloid leukemia: past, present and future. Curr Opin Hematol. 2009;16:84-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 132] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 146. | Ginestier C, Wicinski J, Cervera N, Monville F, Finetti P, Bertucci F, Wicha MS, Birnbaum D, Charafe-Jauffret E. Retinoid signaling regulates breast cancer stem cell differentiation. Cell Cycle. 2009;8:3297-3302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 166] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 147. | Yan Y, Li Z, Xu X, Chen C, Wei W, Fan M, Chen X, Li JJ, Wang Y, Huang J. All-trans retinoic acids induce differentiation and sensitize a radioresistant breast cancer cells to chemotherapy. BMC Complement Altern Med. 2016;16:113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 51] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 148. | Nguyen PH, Giraud J, Staedel C, Chambonnier L, Dubus P, Chevret E, Bœuf H, Gauthereau X, Rousseau B, Fevre M. All-trans retinoic acid targets gastric cancer stem cells and inhibits patient-derived gastric carcinoma tumor growth. Oncogene. 2016;35:5619-5628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (1)] |
| 149. | Lim YC, Kang HJ, Kim YS, Choi EC. All-trans-retinoic acid inhibits growth of head and neck cancer stem cells by suppression of Wnt/β-catenin pathway. Eur J Cancer. 2012;48:3310-3318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 92] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 150. | Budd GT, Adamson PC, Gupta M, Homayoun P, Sandstrom SK, Murphy RF, McLain D, Tuason L, Peereboom D, Bukowski RM. Phase I/II trial of all-trans retinoic acid and tamoxifen in patients with advanced breast cancer. Clin Cancer Res. 1998;4:635-642. [PubMed] |
| 151. | Liu P, Kumar IS, Brown S, Kannappan V, Tawari PE, Tang JZ, Jiang W, Armesilla AL, Darling JL, Wang W. Disulfiram targets cancer stem-like cells and reverses resistance and cross-resistance in acquired paclitaxel-resistant triple-negative breast cancer cells. Br J Cancer. 2013;109:1876-1885. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 169] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 152. | Duan L, Shen H, Zhao G, Yang R, Cai X, Zhang L, Jin C, Huang Y. Inhibitory effect of Disulfiram/copper complex on non-small cell lung cancer cells. Biochem Biophys Res Commun. 2014;446:1010-1016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 80] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 153. | Ortiz RC, Lopes NM, Amôr NG, Ponce JB, Schmerling CK, Lara VS, Moyses RA, Rodini CO. CD44 and ALDH1 immunoexpression as prognostic indicators of invasion and metastasis in oral squamous cell carcinoma. J Oral Pathol Med. 2018;47:740-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 154. | Hu J, Li G, Zhang P, Zhuang X, Hu G. A CD44v+ subpopulation of breast cancer stem-like cells with enhanced lung metastasis capacity. Cell Death Dis. 2017;8:e2679. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 91] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 155. | Kanwal R, Shukla S, Walker E, Gupta S. Acquisition of tumorigenic potential and therapeutic resistance in CD133+ subpopulation of prostate cancer cells exhibiting stem-cell like characteristics. Cancer Lett. 2018;430:25-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 156. | Bigoni-Ordóñez GD, Ortiz-Sánchez E, Rosendo-Chalma P, Valencia-González HA, Aceves C, García-Carrancá A. Molecular iodine inhibits the expression of stemness markers on cancer stem-like cells of established cell lines derived from cervical cancer. BMC Cancer. 2018;18:928. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 157. | Zhang Y, Xu W, Guo H, Zhang Y, He Y, Lee SH, Song X, Li X, Guo Y, Zhao Y. NOTCH1 Signaling Regulates Self-Renewal and Platinum Chemoresistance of Cancer Stem-like Cells in Human Non-Small Cell Lung Cancer. Cancer Res. 2017;77:3082-3091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 158. | Durinikova E, Kozovska Z, Poturnajova M, Plava J, Cierna Z, Babelova A, Bohovic R, Schmidtova S, Tomas M, Kucerova L. ALDH1A3 upregulation and spontaneous metastasis formation is associated with acquired chemoresistance in colorectal cancer cells. BMC Cancer. 2018;18:848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 159. | Kozovska Z, Patsalias A, Bajzik V, Durinikova E, Demkova L, Jargasova S, Smolkova B, Plava J, Kucerova L, Matuskova M. ALDH1A inhibition sensitizes colon cancer cells to chemotherapy. BMC Cancer. 2018;18:656. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 160. | Pal D, Kolluru V, Chandrasekaran B, Baby BV, Aman M, Suman S, Sirimulla S, Sanders MA, Alatassi H, Ankem MK. Targeting aberrant expression of Notch-1 in ALDH+ cancer stem cells in breast cancer. Mol Carcinog. 2017;56:1127-1136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country of origin: Mexico
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Haider KH, He XH, Scuteri A, Zheng YW S- Editor: Dou Y L- Editor: Filipodia E- Editor: Song H