©The Author(s) 2023.
World J Stem Cells. May 26, 2023; 15(5): 302-322
Published online May 26, 2023. doi: 10.4252/wjsc.v15.i5.302
Published online May 26, 2023. doi: 10.4252/wjsc.v15.i5.302
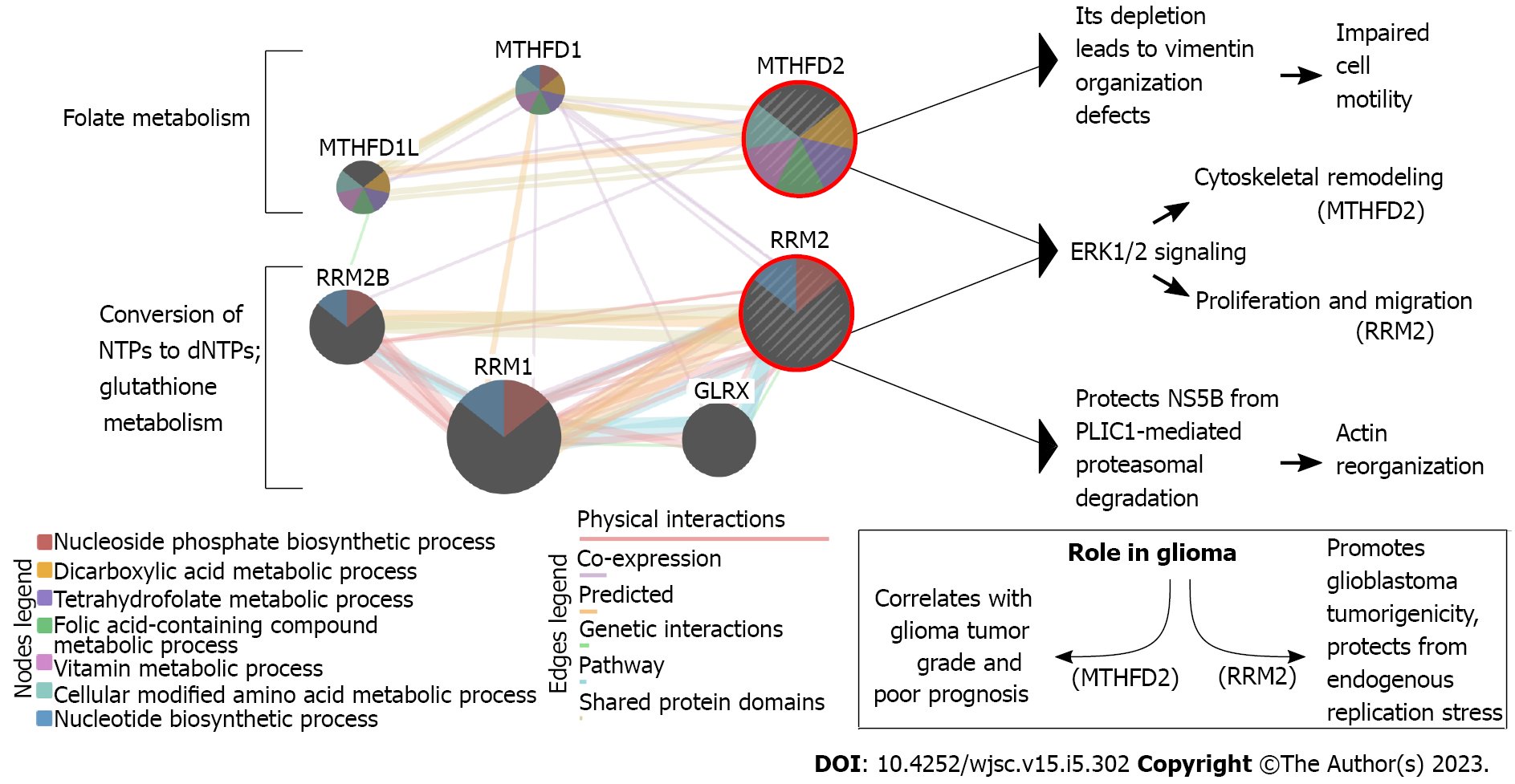
Figure 1 Example of the interplay between cytoskeleton and metabolism using the biological function of methylenetetrahydrofolate dehydrogenase 2 and ribonucleotide reductase subunit M2 enzymes.
Typically, methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) dehydrogenase is known for its activity in folate metabolism, whereas ribonucleotide reductase subunit M2 (RRM2) reductase is known for the conversion of ribonucleotide triphosphates to deoxyribonucleotide triphosphates which requires metabolic resources supplied by reduced glutathione. However, these two enzymes (encircled in red) are also involved in cytoskeletal rearrangements that are summarized on the right side of the figure. Literature data indicate that they also affect the same pathway (i.e., ERK1/2 signaling) but render various outcomes. Moreover, their role in glioma has already been proposed (bottom-right panel). Figure created using Inkscape and GeneMania (MTHFD2 and RRM2 as query genes; five “resultant” genes included to highlight interconnectivity; exemplary metabolism-related processes included from the built-in functional analysis). NTP: Ribonucleotide triphosphates; dNTPs: Deoxyribonucleotide triphosphates; MTHFD: Methylenetetrahydrofolate dehydrogenase; RRM2: Reductase subunit M2.
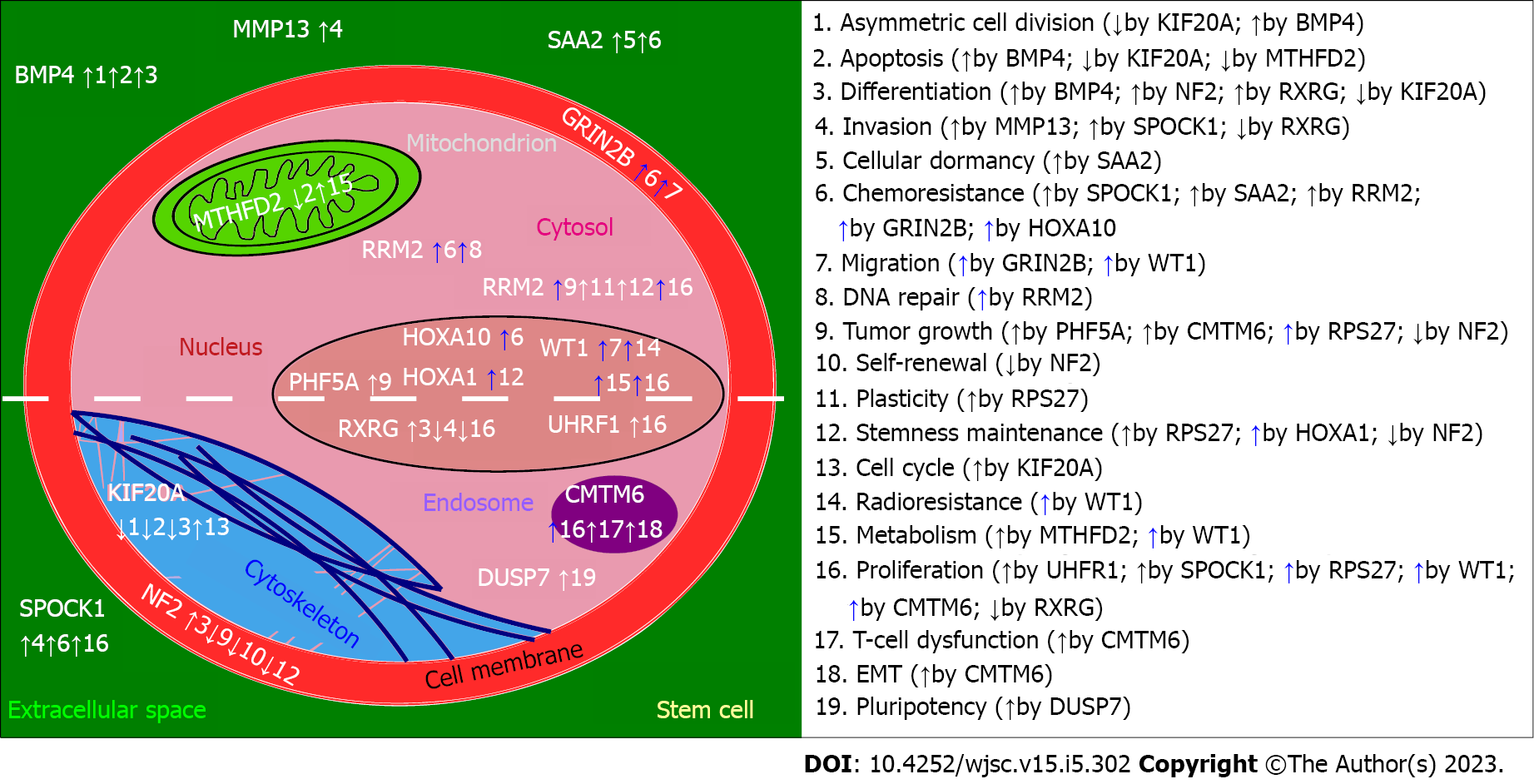
Figure 2 Impact of described genes on biological processes related to stem cells.
The “↑” or “↑” (blue) symbol indicates activation of the process while “↓“denotes inhibition. The impact of genes on processes (numbered from 1 to 19) is either directly confirmed (solid arrow next to the number) or recapitulated based on available data from various literature sources (dashed arrow next to the number). The “↓” (blue) symbol was not required as any gene inhibited the given process in an indirect manner. The white dashed line dividing the stem cell into two halves separates the genes with a confirmed role in glioblastoma stem cells (above the line) from those involved in cancer stemness outside the glioblastoma context (below the line). Figure created using Inkscape. NF2: Neurofibromatosis type 2 protein; BMP4: Bone morphogenetic protein 4; RXRG: Retinoid X receptor gamma; MMP13: Metalloproteinase 13; RRM2: Reductase subunit M2; SPOCK1: SPARC/Osteonectin; CWCV: Kazal-like domains 1; ECM: Extracellular matrix; CMTM: Chemokine-like factor superfamily.
- Citation: Kałuzińska-Kołat Ż, Kołat D, Kośla K, Płuciennik E, Bednarek AK. Delineating the glioblastoma stemness by genes involved in cytoskeletal rearrangements and metabolic alterations. World J Stem Cells 2023; 15(5): 302-322
- URL: https://www.wjgnet.com/1948-0210/full/v15/i5/302.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v15.i5.302