©The Author(s) 2021.
World J Stem Cells. Jun 26, 2021; 13(6): 485-502
Published online Jun 26, 2021. doi: 10.4252/wjsc.v13.i6.485
Published online Jun 26, 2021. doi: 10.4252/wjsc.v13.i6.485
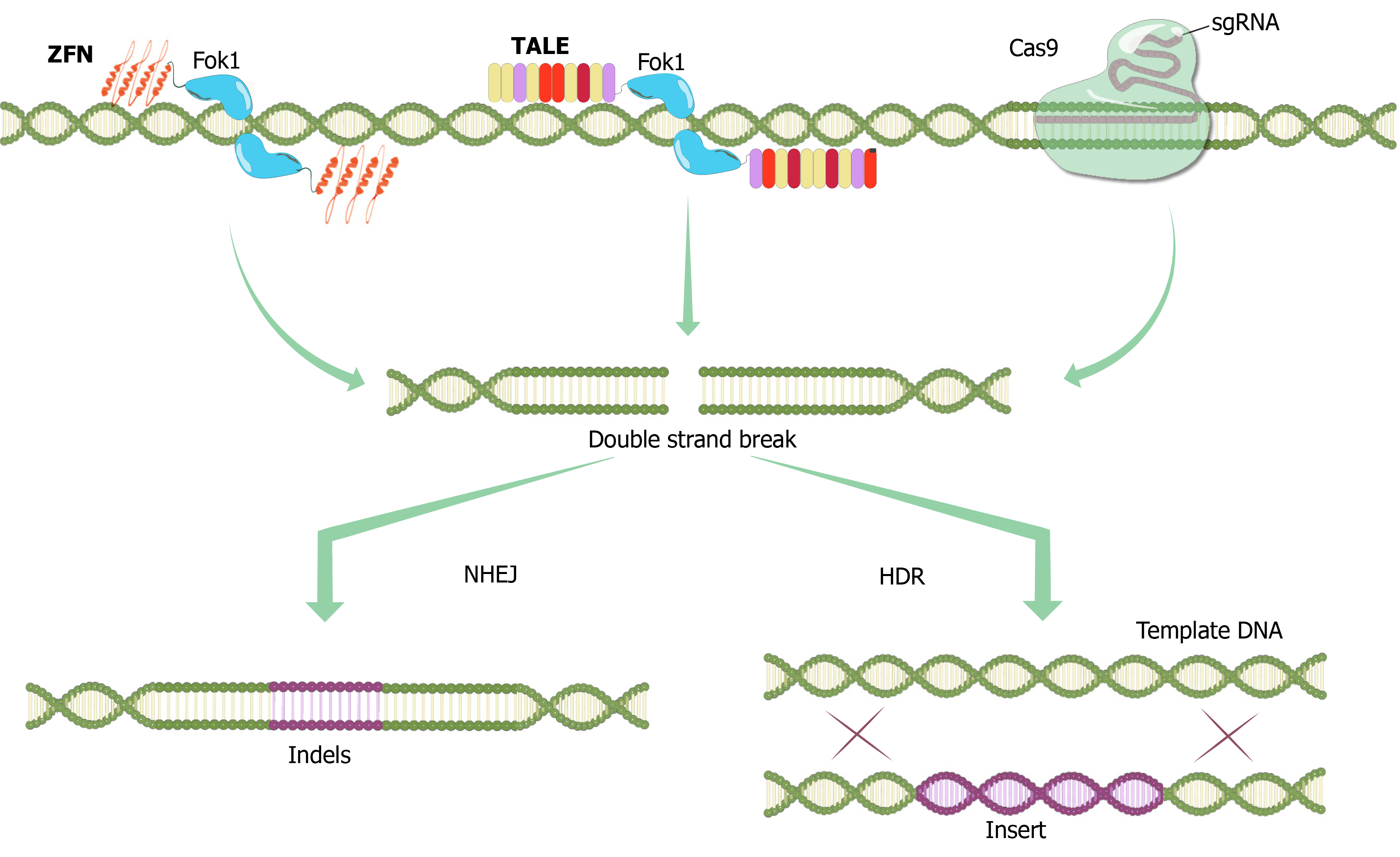
Figure 1 Programmable nucleases.
Zinc finger nucleases contain a specially designed N-terminal DNA-binding domain and a C-terminal endonuclease domain (bacterial Fok1 endonucleases) that dimerize to form a double-strand break (DSB) in DNA[111]. DNA-binding domain is a sequence of multiple zinc finger domains, each of which recognizes a specific three base pair sequence of DNA. Then, four to six zinc finger regions recognize specific 12-18 base pair nucleotide sequences. Transcription activator-like effector nucleases contain a C-terminal Fok1 endonuclease with an N-terminal DNA binding site. The DNA binding domain consists of highly conserved 33-35 amino acids with a variation at the 12th and 13th positions that ensure specificity against the target sequence. CRISPR/Cas9system; crRNA-tracrRNA (sgRNA) and Cas9 protein bind to DNA by forming a complex. The Cas9 enzyme has two nuclease sites. These are the HNH region (which creates a break in the complementary chain) and the RuvC region (which creates a break in the non-complementary chain). In the presence of the Protospacer adjacent motif (PAM) sequence, the sgRNA-Cas9 complex binds and cuts DNA generating DSBs. ZFN: Zinc Finger Nuclease; TALE: Transcription Activator-Like Effector; sgRNA: single guide RNA; NHEJ: Non-homologous end joining; HDR: Homology directed repair.
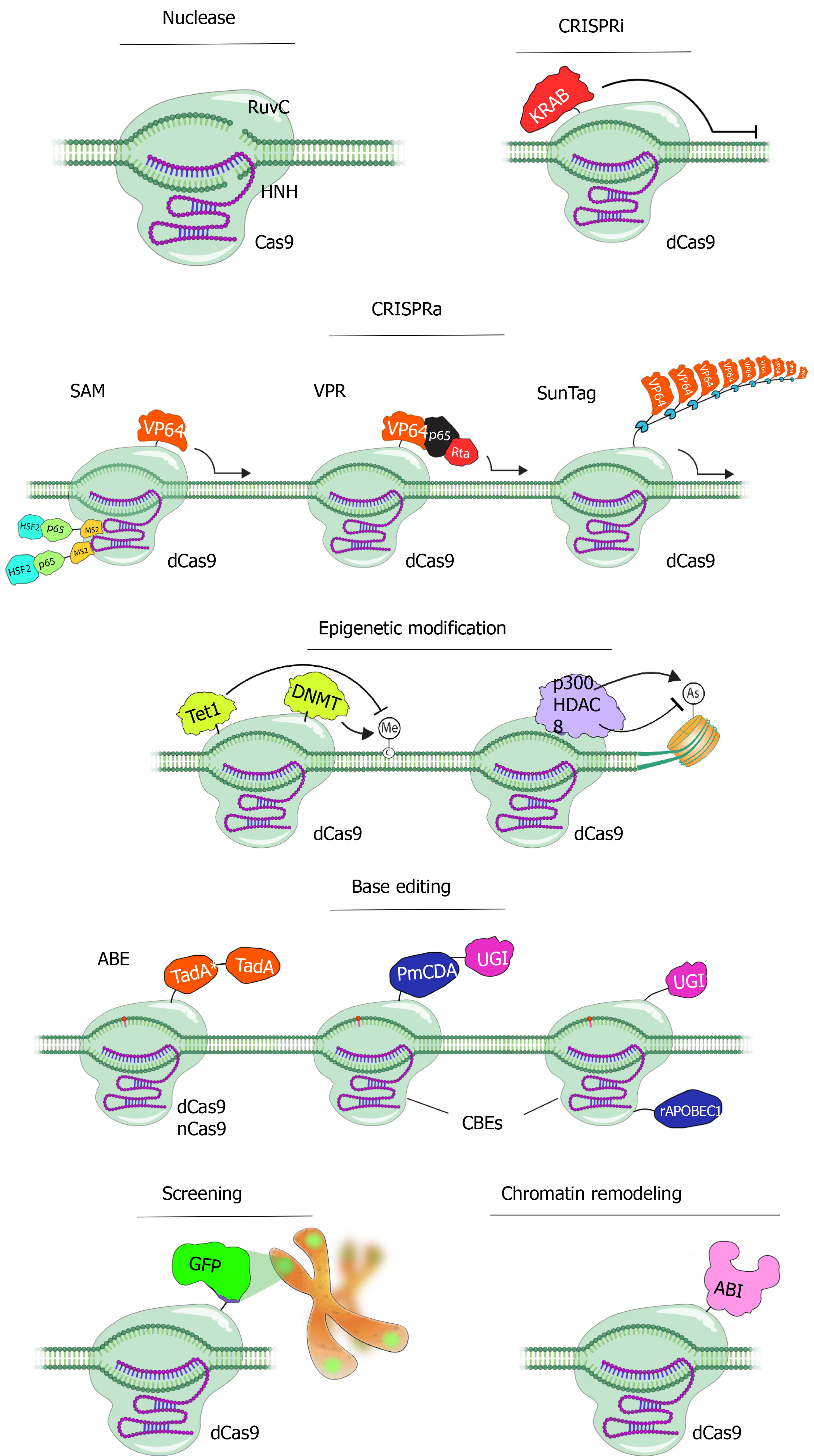
Figure 2 Applications of CRISPR/Cas9 technology.
CRISPR/Cas9 system, as a result of rapidly developing technology, has become the most commonly used tool in almost all gene editing fields today. The Cas9 enzyme is used for many different purposes by subjecting it to various changes and fusing it with a series of effective proteins[112]. Catalytically inactive Cas9 (dCas9) can be attached to numerous transcription-modifiable enzymes to achieve epigenetic and transcriptional control of targeted genes. Transcription activators are recruited to control gene expression using the CRISPR-based activators SunTag, viral protein regulatory (VPR), and synergistic activator mediator (SAM). Conventional peptide linkers are used by VPR to attach the tripartite VP64, p65, and Rta effector to dCas9. MCPs linked to a p65-HSF1 domain are directed by the SAM using the MS2 RNA aptamer to induce transcription. In the SunTag system, a GCN4-epitope array is used to recruit VP64 activators to transcription start sites. Transcription can be inhibited by the dCas9-KRAB via deploying similar strategies. CpG dinucleotides can be de-novo methylated by the DNA methyltransferase 3A (DNMT3A) attached to dCas9 in a programmable manner. Methylated CpG sites can be demethylated via ten-eleven translocation’s (TET1) catalytic domain linked to dCas9. By aiming CpG-containing promoter regions for epigenetic alteration, dCas9-DNMT3A/TET1 can successfully control gene transcription. The human p300 acetyltransferase (p300core) I or histone deacetylase 8 linked to dCas9 can regulate the acetylation status of histone 3 Lysine 27 residues to control transcription from promoters and enhancers. Moreover, in recent years, it has been used with base regulatory proteins for base editing and fluorescent proteins such as GFP to monitor a specific region on the chromosome. In another application, the nuclear organization of chromosomes can be rearranged in 3D in a temporary and inducible manner (CRISPR GO). dCas9: dead Cas9; SAM: Synergistic activator mediator; DNMT: DNA methyltransferase; ABE: Adenine base editor; CBE: Cytosine base editor; GFP: Green fluorescent protein; VPR: Viral protein regulatory.
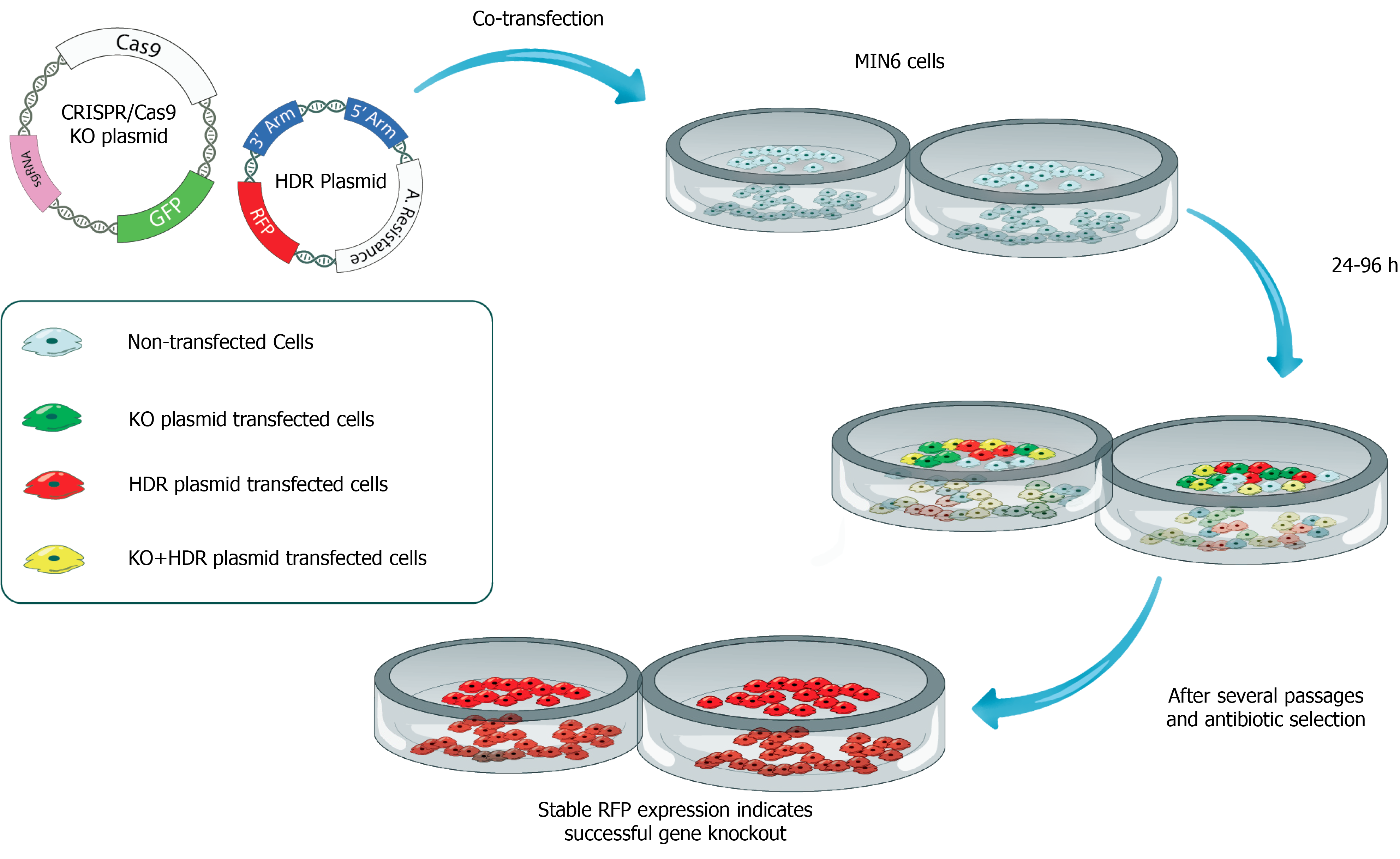
Figure 3 Production of CRISPR/Cas9-mediated insulin-deficient pancreatic beta cell line for testing the therapeutic efficacy of insulin gene therapy vectors.
In this scenario, the CRISPR/Cas9 knockout plasmid carries a Cas9 protein-encoding sequence, sgRNA, and green fluorescein protein as a reporter. Homology-directed repair (HDR) plasmid contains two genomic fragments of DNA complementary to insulin gene (3’ Arm and 5’ Arm) and red fluorescein protein-encoding DNA sequences. Both CRISPR/Cas9 knockout plasmid and HDR plasmid are transfected into a pancreatic beta-cell line such as MIN6 cells. After several passages and antibiotic selection, beta cells expressing red fluorescent protein remain alive suggesting successful knockout (KO) of the insulin (INS) gene. INS KO cells can be cloned by limited dilution and further expanded in cell culture. INS KO cells can be used in in vitro complementation assays as well as transplanted under the kidney capsule of diabetic rats to assess the therapeutic efficacy of gene therapy vectors. KO: Knockout; HDR: Homology-directed repair.
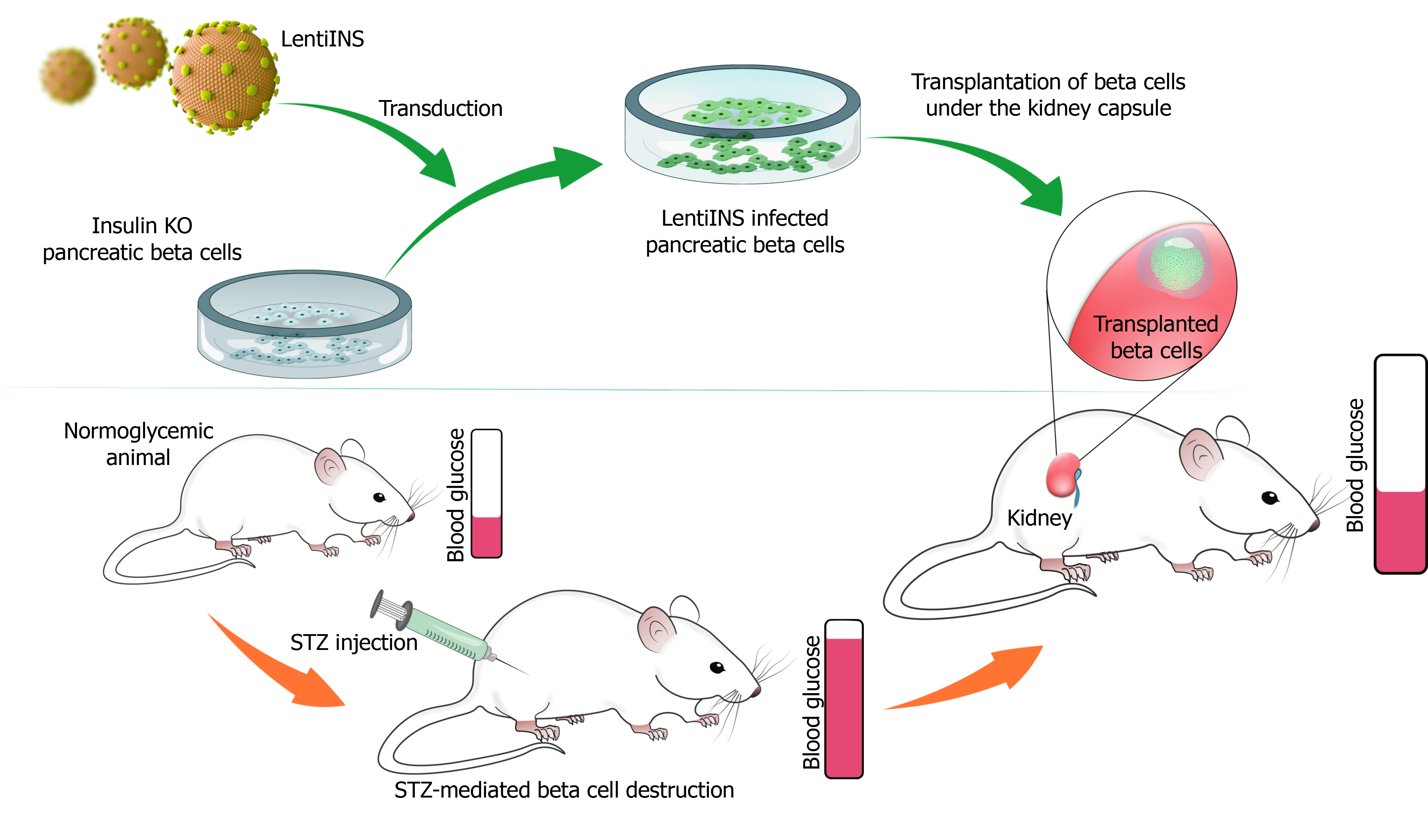
Figure 4 Testing the therapeutic efficacy of the lentivirus vector carrying insulin gene (LentiINS).
The chemotherapeutic agent Streptozotocin can be used to destroy endogenous pancreatic beta cells to induce an experimental animal model of diabetes. Insulin knockout (INS KO) pancreatic beta cells after transduction with the LentiINS can be transplanted under the kidney capsule of diabetic animals. Diabetic animals transplanted with the LentiINS-transduced INS KO pancreatic beta cells are expected to lower the blood glucose. On the other hand, hyperglycemia is anticipated in diabetic animals transplanted with INS KO pancreatic beta cells alone. STZ: Streptozotocin; KO: Knockout.
- Citation: Eksi YE, Sanlioglu AD, Akkaya B, Ozturk BE, Sanlioglu S. Genome engineering and disease modeling via programmable nucleases for insulin gene therapy: Promises of CRISPR/Cas9 technology . World J Stem Cells 2021; 13(6): 485-502
- URL: https://www.wjgnet.com/1948-0210/full/v13/i6/485.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i6.485