Published online Feb 15, 2003. doi: 10.3748/wjg.v9.i2.300
Revised: April 4, 2002
Accepted: April 20, 2002
Published online: February 15, 2003
AIM: To investigate the interaction between hepatitis C virus core protein and translin protein and its role in the pathogenensis of hepatocellular carcinoma and lymphoma.
METHODS: With the components of the yeast two hybrid system 3,“ bait” plasmids of HCV core the gene was constructed. After proving that hepatitis C virus core protein could be firmly expressed in AH109 yeast strains, yeast two- hybrid screening was performed by mating AH109 with Y187 that transformed with liver cDNA library plasmids - pACT2 and then plated on quadrople dropout (QDO) medium and then assayed for α-gal activity. Sequencing analysis of the genes of library plasmids in yeast colonies that could grow on QDO with α-gal activity was performed. The interaction between HCV core protein and the protein we obtained from positive colony was further confirmed by repeating yeast two - hybrid analysis and coimmunoprecipitation in vitro.
RESULTS: A gene from a positive colony was the gene of translin, a recombination hotspot binding protein. The interaction between HCV core protein and translin protein could be proved not only in yeast, but also in vitro.
CONCLUSION: The core protein of HCV can interact with translin protein. This can partly explain the molecular mechanism for hepatocellular carcinoma and lymphoma caused by HCV.
- Citation: Li K, Wang L, Cheng J, Lu YY, Zhang LX, Mu JS, Hong Y, Liu Y, Duan HJ, Wang G, Li L, Chen JM. Interaction between hepatitis C virus core protein and translin protein- a possible molecular mechanism for hepatocellular carcinoma and lymphoma caused by hepatitis C virus. World J Gastroenterol 2003; 9(2): 300-303
- URL: https://www.wjgnet.com/1007-9327/full/v9/i2/300.htm
- DOI: https://dx.doi.org/10.3748/wjg.v9.i2.300
The core protein of hepatitis C virus (HCV) is the structural protein of the virus[1-4]. However, some evidences suggested that this protein has a pleiotropic nature. In addition to having a packaging function, the core protein has been shown to act in trans on the viral and cellular promoters and it is also capable of transformation of rat embryonic fibroblasts through cooperation with the ras oncogene. Previous studies showed that the core protein could interact with several proteins such as lymphotoxin-β Receptor, heterogeneous nuclear ribonucleoprotein K, RNA helicase[5-8]. In order to understand the pathogenesis of HCV infection we examined the possibility that the HCV core protein interacts with cellular proteins.
Bacterial, yeast strains and Plasmids All yeast strains and plasmids for yeast two-hybrid experiments were obtained from Clontech (Palo Alto, Calif., USA) as components of the MATCHMAKER Two Hybrid System 3. Yeast strain AH109 (MATa, trp1-901, leu2-3,112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2 URA3::MEL1TATA-lacZ MEL1) containing pGBKT7-53,coding for DNA-BD/mouse p53 fusing protein and AH109 used for cloning of bait plasmids, yeast strain Y187 (MATa ura3-52, his3-200, Ade2-101, trp1-901, leu2-3, 112, gal4Δ, gal80Δ, met-, URA3::GAL1UAS-GAL1TATA-lacZ MEL1)containing pTD1-1, in which pACT2 coding for AD/SV40 large T antigen fusing protein and Y187 used for cloning of library plasmids. Pretransformed cDNA liver cell library Y187. Bacterial strain DH5α used for cloning of every shuttle plasmid. Yeast-Escherichia coli shuttle plasmids pGBKT7 DNA-BD cloning plasmid, pGADT7 AD cloning plasmid, pGBKT7-53 control plasmid, pGADT7, pGBKT7-Lam control plasmid, pCL1 plasmid from Clontech L.T.D Company (K1612-1). pGEM T vector from Promega Company, USA.
Chemical agents and cultural media Taq DNA polymerase purchased from MBI Company, T4 DNA ligase, EcoRI and BamHI restricton endonuclease from Takara. c-Myc monoclonal antibody secreted by 1-9E10.2 hybridoma (ATCC), goat anti-mouse IgG conjugated with horseradish peroxidase from Zhongshan Company,China. Lithium Acetate, semi-sulfate adenine, Acrylamide and N, N’-Bis-acrylamide from Sigma, TEMED from Boehringer Mannheim. Tryptone and yeast extracts from OXOID. X-α-Gal and Cultural media: YPDA, SD/-Trp SD/-Leu, SD/-Trp/-Leu, SD/-Trp/-Leu/-His, SD/-Trp/-Leu/-His/-Ade from Clontech L.T.D Company. protein-G agarose from Roche. RT-PCR kit and TNT®Coupled Reticulocyte Lysate Systems from Promega. [35S]-methionine (1000 Ci/mmol; 10 mCi/mL) from Isotope company of china. Amplify Fluorographic Reagent (#NAMP100) from Amersham Life Sciences. Others from Sigma company.
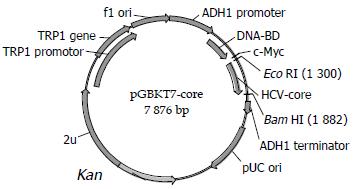
Construction of “bait” plasmid and expression of HCV core protein Plasmid pGBKT7-core (Figure 1) containing full-length HCV core gene was constructed by insertion of HCV core gene in-frame into EcoRI/BamHI site, which could direct expression of DNA binding domain, c-myc and core fusion protein. After the plasmid was transformed into yeast strain AH109 by using Lithium Acetate method[9]. Western blotting was performed to confirm the expression of the fusion protein by using c-myc monoclonal antibody. Transformed AH109 was cultured on quadrople dropout media to exclude the auto-activity.
Two-hybrid library screening using yeast mating One large (2-3-mm), fresh (< 2 mo old) colony of AH109[bait] was inoculated into 50 mL of SD/-Trp and incubated at 30 °C overnight (16-24 h) with shaking at 250-270 rpm. Then the cells were spinned down by centrifuging the entire 50-mL culture at 1000 × for 5 min and supernatant Decanted the cell pellet was resuspended in the residual liquid by vortexing. The entire AH109[bait] culture and the 1-mL library were combined and cultured in a 2-L sterile flask and Add 45 mL of 2X YPDA/Kan was added and swirl gently. After 20 h mating, the cells were spinned down and resuspened then spreaded on -50 large (150-mm) plates, containing 200 mL of SD/-Ade/-His/-Leu/-Trp (QDO). After 6-18 d grew, the yeast colonies were transferred onto the plates containing X-α-gal to check for expression of the MEL1 reporter gene (blue colonies).
Plasmid isolation from yeast and transforming E.coli with yeast plasmid Yeast plasmid was isolated with Lyticase method (provided by Clontech), and transformed into E. coli by using elctroporation[10], transformants were plated on ampicilin LB selection media, then, isolating plasmids from E.coli and sequencing analysis.
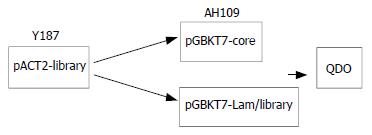
To confirm the true interaction in yeast To confirm the true protein-protein interaction and exclude false positives, the plasmids of positive colonies were transformed into yeast strain Y187, next mating experiments were carried out by mating with yeast strain AH109 containing pGBKT7-core or pGBKT7-Lam. After mating ,the diploids yeast were plated on QDO covered with X-α-gal (Figure 2).
Bioinformatic analysis After sequencing the positive colonies, the sequences blasted with GenBank to analogize the function of the genes.
RT-PCR In order to clone the full-length gene, RT-PCRs were conducted by using PCR primers the designing based on the information of GenBank. The genes amplified by RT-PCR were ligated into yeast plasmid pGADT7.
In vitro translation Mixture of TNT®reticulocyte 25 μL, TNT® reactio buffer 2 μL, T7 TNT®RNA polymerase 1 μL, amino acids mixture (minus methionine, 1 mM) 1 μL, [35S]methionine 2 μL, Rnasin®nuclease inhibitor (40u/μL) 1 μL, DNA template (pGBKT7-core or pGADT7-library gene) (0.5 μg/μL) 2 μL, ddH2O 50 μL, 30 °C incubated 90 min.
Coimmunoprecipitation The following reactants were combined in a 1.5-mL microcentrifuge tube on ice: Five μL in vitro translated bait protein, 5 μL in vitro translated library protein. The control only added 10 μL pGBKT7-core plasmid. The mixtures were incubated at 30 °C for 1 h. Then, the following reagents added into the reaction tubes: 470 mL coimmunoprecipitation buffer (20 mM Tris-HCl (pH7.5), 150 mM NaCl, 1 mM DTT, 5 μg/mL aprotinin, 0.5 mM PMSF, 0.1% Tween 20), 10 μL Protein-G Agarose Beads, 10 μL c-Myc Monoclonal Antibody. Incubated at 4 for 2 h with continuous rocking. The tubes were centrifuged at 14000 rpm for 1-2 min. The supernatants were removed. 0.5 mL TBST added to the tubes. Rinse steps were repeated three times. 15 mL SDS-loading buffer were added. The samples was heated at 80 °C for 5 min. The tubes were placed on ice. Briefly centrifuged, and 10 μL loaded onto an SDS-PAGE minigel to begin the electrophoretic separation. After electrophoresis, the gel was transfered to a tray containing Gel Fixation Solution, and placed on a rotary shaker for 10 min at room temperature. Rinsed the gel with H2O, then Amplify Fluorographic Reagent was added shaked for 20 min at room, then dryed at 80 °C under constant vacuum. The gel was exposed to a X-ray film overnight at room temperature. The film was developed by using standard techniques.
Yeast strain AH109 transformed with pGBKT7-core could stably express the fusion protein at high level (Figure 3) and could only grow on SD/-Trp medium and could not grow on QDO medium. Thus, the transformed yeast could be used for yeast hybrid analysis.
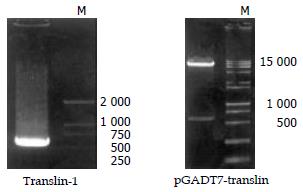
RT-PCR experiments The yeast two hybrid analysis showed 30 blue colonies grew on QDO plates containing X-α-gal. After confirming the true interaction in yeast, we isolated the plasmids from the blue colonies containing only pGBKT7-core and one library plasmid other than other plasmids. Sequencing the gene and blasted with the data from GenBank, a gene is translin. To further prove the interaction between HCV core protein and translin protein (Translin), a pair of primer were designed based on the gene of translin (Forward: 5'-GAA TTC ATG TCT GTG AGC GAG ATC TTC GTG G -3' down: 5'-GGA TCC CTA TTT TTC AAC ACA AGC TGC TGC C-3', up and down primers containing EcoRI and BamHI restriction endonuclease site, respectively. Total RNAs were prepared from HepG2, A 687bp fragment was amplified by using RT-PCR (Figure 4A). After cut by EcoRI/BamHI, the fragment was in-frame ligated into pGADT7 EcoRI/BamHI site (Figure 4B).
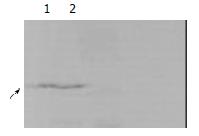
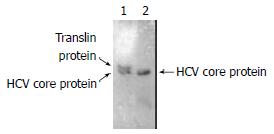
HCV core protein containing 192 aa, Mr = 20968, is smaller than Translin containing 228aa, Mr = 26182. Lane 1 showed two protein could interact with each other, lane 2 was only HCV core protein. (Figure 5).
Hepatitis C virus infects an estimated 170 million persons worldwide and thus represents a viral pandemic. Progression to chronic disease occurs in the majority of HCV-infected persons[11-17], and some patients can develop to hepatocellular carcinoma[18] and lymphoma. Infection with the virus has become the main indication for liver transplantation. Although research advances have been impeded by the inability to grow HCV easily in culture, there have been some insights into pathogenesis of the infection and improvements in treatment options. The core protein of HCV is a multifunctional protein involved in several processes; it is phosphorylated and has both cytoplasmic and nuclear localization and thus it may play multiple roles in the viral life cycle[3,4]. Several studies also suggested that it has regulatory roles for viral and cellular genes[19,20] and possesses transformation activity. More recent studies revealed that the core protein can interact with tumor necrosis factor receptor-1, lymphotoxin β-R and viral envelope protein 1 (E1) and also forms a complex with apolipoprotein AII of the lipid droplet[21]. A recent report showed that the HCV core protein can suppress the cisplatin- and c-Myc-mediated apoptotic effect, supporting its role in the establishment of persistent HCV infection[22]. But there have reverse results that that the core protein also has the ability to enhance cell death triggered by LT-βR ligand or anti-Fas monoclonal antibody. Some research considered that the core protein of HCV can induce steatosis and hepatocellular carcinoma in transgenic mice[23,24]. Thus, the molecular mechanism by which the HCV core protein and HCV induce hepatocellular carcinoma is not clear. Recent epidemical research indicated that a significant increase in the prevalence of HCV infection in a group of B-cell non-Hodgkin's lymphoma (NHL)[25-29] .
In this study, yeast two-hybrid system was used to clone oncogenic gene. Yeast two-hybrid system 3 based on the system originally designed by Fields and Song[30] is developed by Bendixen[31], which is commercially available from Clontech Company L.t.d. In this system, the promoters controlling HIS3, ADE2, and MEL1 expression in AH109 have significantly fewer false positives and the simple mating protocol significantly reduces the labor and time involved in performing a two-hybrid library screening and improves the chances of finding rare protein-protein interactions and leads to more reproducible results.
The “bait” plasmid pGBKT7-core was transformed into yeast strain, After mating with liver cDNA library yeast strain Y187, the diploid yeast cells were plated on QDO media containing X-α-gal, 30 true positives were obtained. Sequencing analysis of isolated library plasmids, we find one of the genes is translin[32] - a recombination hotspot binding protein. In order to further conform the interaction between the expressed protein and HCV core protein, we performed the experiment of coimmunoprecipitation of both proteins. A strong interaction between the HCV core protein and Translin protein in vitro was observed.
A number of studies have shown that chromosomal translocations either result in the activation of proto-oncogenes by joining them to immunoglobulin (Ig) or T-cell receptor genes or lead to the creation of tumor-specific fusion proteins. In man, such translocations consistently occur at particular sites in the genome. Translin protein, which specifically binds to the consensus sequences ATGCAG and GCCC (A/T) (G/C) (G/C) (A/T) found at the breakpoint junctions in many cases of chromosomal translocations, is a unique DNA binding protein[33]. The nuclear translocation of translin protein only happened in the time when the cells were treated with mutagen. translin protein may be a typical DNA end binding protein, which is in contrast with one of the other DNA binding proteins, the Ku antigen, that initially binds to DNA ends and then moves to internal positions within the DNA molecule[34]. Previous report showed that translin protein was not found in liver tumors. But in this study, we found the gene expressed in liver tumor cell HepG2 and in liver cDNA library. And the interaction between translin protein and HCV core protein not only existed in yeast, but also in vitro. The results suggested that translin protein may play a role in hepatocellular carcinoma. But there has had no report giving the evidence that the patients infected with HCV other than HBV have the chromosomal translocation, whether the hepatocellular carcinomas caused by infection of HCV have chromosomal translocation worthy of further studying.
The effects of translin protein on normal lymphocytes need the induction by some factors such as mutagens or biological factors (HCV infection). In lymphoproliferative disorder patients infected with HCV, some reports showed chromosomal translocation happened in B-cell[35,36]. Therefore, our report indicated a molecular mechnism that the interaction between HCV core protein and translin protein may trigger the B-cell progressing into lymphoma in patients infected with HCV. How the interaction between the HCV core protein and translin protein causes chromosomal translocation or rather, causes lymphoma, more experiments are necessary to elucidate it.
| 1. | Harada S, Watanabe Y, Takeuchi K, Suzuki T, Katayama T, Takebe Y, Saito I, Miyamura T. Expression of processed core protein of hepatitis C virus in mammalian cells. J Virol. 1991;65:3015-3021. [PubMed] |
| 2. | Santolini E, Migliaccio G, La Monica N. Biosynthesis and biochemical properties of the hepatitis C virus core protein. J Virol. 1994;68:3631-3641. [PubMed] |
| 3. | Lo SY, Selby M, Tong M, Ou JH. Comparative studies of the core gene products of two different hepatitis C virus isolates: two alternative forms determined by a single amino acid substitution. Virology. 1994;199:124-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 58] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | Liu Q, Tackney C, Bhat RA, Prince AM, Zhang P. Regulated processing of hepatitis C virus core protein is linked to subcellular localization. J Virol. 1997;71:657-662. [PubMed] |
| 5. | Yasui K, Wakita T, Tsukiyama-Kohara K, Funahashi SI, Ichikawa M, Kajita T, Moradpour D, Wands JR, Kohara M. The native form and maturation process of hepatitis C virus core protein. J Virol. 1998;72:6048-6055. [PubMed] |
| 6. | Mamiya N, Worman HJ. Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem. 1999;274:15751-15756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 137] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 7. | You LR, Chen CM, Yeh TS, Tsai TY, Mai RT, Lin CH, Lee YH. Hepatitis C virus core protein interacts with cellular putative RNA helicase. J Virol. 1999;73:2841-2853. [PubMed] |
| 8. | Chen CM, You LR, Hwang LH, Lee YH. Direct interaction of hepatitis C virus core protein with the cellular lymphotoxin-beta receptor modulates the signal pathway of the lymphotoxin-beta receptor. J Virol. 1997;71:9417-9426. [PubMed] |
| 9. | Matsumoto M, Hsieh TY, Zhu N, VanArsdale T, Hwang SB, Jeng KS, Gorbalenya AE, Lo SY, Ou JH, Ware CF. Hepatitis C virus core protein interacts with the cytoplasmic tail of lymphotoxin-beta receptor. J Virol. 1997;71:1301-1309. [PubMed] |
| 10. | Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2506] [Cited by in RCA: 2821] [Article Influence: 83.0] [Reference Citation Analysis (0)] |
| 11. | Yan ZY, Wang HL. Short protocols in molecular biology. Beijing: Scientific press 1998; 23-24. |
| 12. | He Y, Liu W, Zeng L. [The effect of interferon in combination with ribavirin on the plus and minus strands of hepatitis C virus RNA in patients with hepatitis]. Zhonghua Neike Zazhi. 1996;35:32-35. [PubMed] |
| 13. | Wei L, Wang Y, Chen HS, Tao QM. Sequencing of hepatitis C virus cDNA with polymerase chain reaction directed sequencing. China Natl J New Gastroenterol. 1997;3:12-15. |
| 14. | Tang ZY, Qi JY, Shen HX, Yang DL, Hao LJ. Short_ and long_term effect of interferon therapy in chronic hepatitis C. China Natl J New Gastroenterol. 1997;3:77. |
| 15. | Zhou P, Cai Q, Chen YC, Zhang MS, Guan J, Li XJ. Hepatitis C virus RNA detection in serum and peripheral blood mononuclear cells of patients with hepatitis C. China Natl J New Gastroenterol. 1997;3:108-110. |
| 16. | Assy N, Minuk G. A comparison between previous and present histologic assessments of chronic hepatitis C viral infections in humans. World J Gastroenterol. 1999;5:107-110. [PubMed] |
| 17. | Huang F, Zhao GZ, Li Y. HCV genotypes in hepatitis C patients and their clinical significances. World J Gastroenterol. 1999;5:547-549. [PubMed] |
| 18. | Yan FM, Chen AS, Hao F, Zhao XP, Gu CH, Zhao LB, Yang DL, Hao LJ. Hepatitis C virus may infect extrahepatic tissues in patients with hepatitis C. World J Gastroenterol. 2000;6:805-811. [PubMed] |
| 19. | Zhang LF, Peng WW, Yao JL, Tang YH. Immunohistochemical detection of HCV infection in patients with hepatocellular carcinoma and other liver diseases. World J Gastroenterol. 1998;4:64-65. [PubMed] |
| 20. | Shih CM, Chen CM, Chen SY, Lee YH. Modulation of the trans-suppression activity of hepatitis C virus core protein by phosphorylation. J Virol. 1995;69:1160-1171. [PubMed] |
| 21. | Cho JW, Baek WK, Suh SI, Yang SH, Chang J, Sung YC, Suh MH. Hepatitis C virus core protein promotes cell proliferation through the upregulation of cyclin E expression levels. Liver. 2001;21:137-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 22. | Hahn CS, Cho YG, Kang BS, Lester IM, Hahn YS. The HCV core protein acts as a positive regulator of fas-mediated apoptosis in a human lymphoblastoid T cell line. Virology. 2000;276:127-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 95] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 23. | Sabile A, Perlemuter G, Bono F, Kohara K, Demaugre F, Kohara M, Matsuura Y, Miyamura T, Bréchot C, Barba G. Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology. 1999;30:1064-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 139] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 24. | Ray RB, Meyer K, Ray R. Suppression of apoptotic cell death by hepatitis C virus core protein. Virology. 1996;226:176-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 188] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 25. | Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 931] [Cited by in RCA: 912] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 26. | Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Ishibashi K, Matsuura Y, Miyamura T, Koike K. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J Gen Virol. 1997;78:1527-1531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 478] [Cited by in RCA: 470] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 27. | Ferri C, La Civita L, Longombardo G, Cecchetti R, Giannini C, Zignego AL. Type C chronic hepatitis complicated by B-cell lymphoma. Am J Gastroenterol. 1995;90:2071-2072. [PubMed] |
| 28. | Ferri C, Monti M, La Civita L, Careccia G, Mazzaro C, Longombardo G, Lombardini F, Greco F, Pasero G, Bombardieri S. Hepatitis C virus infection in non-Hodgkin's B-cell lymphoma complicating mixed cryoglobulinaemia. Eur J Clin Invest. 1994;24:781-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 56] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 29. | Brind AM, Watson JP, Burt A, Kestevan P, Wallis J, Proctor SJ, Bassendine MF. Non-Hodgkin's lymphoma and hepatitis C virus infection. Leuk Lymphoma. 1996;21:127-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 30. | Pioltelli P, Gargantini L, Cassi E, Santoleri L, Bellati G, Magliano EM, Morra E. Hepatitis C virus in non-Hodgkin's lymphoma. A reappraisal after a prospective case-control study of 300 patients. Lombart Study Group of HCV-Lymphoma. Am J Hematol. 2000;64:95-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 31. | Suriawinata A, Ye MQ, Emre S, Strauchen J, Thung SN. Hepatocellular carcinoma and non-Hodgkin lymphoma in a patient with chronic hepatitis C and cirrhosis. Arch Pathol Lab Med. 2000;124:1532-1534. [PubMed] |
| 32. | Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4257] [Cited by in RCA: 4358] [Article Influence: 117.8] [Reference Citation Analysis (5)] |
| 33. | Bendixen C, Gangloff S, Rothstein R. A yeast mating-selection scheme for detection of protein-protein interactions. Nucleic Acids Res. 1994;22:1778-1779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 91] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 34. | Aoki K, Suzuki K, Sugano T, Tasaka T, Nakahara K, Kuge O, Omori A, Kasai M. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat Genet. 1995;10:167-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 157] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 35. | Kasai M, Matsuzaki T, Katayanagi K, Omori A, Maziarz RT, Strominger JL, Aoki K, Suzuki K. The translin ring specifically recognizes DNA ends at recombination hot spots in the human genome. J Biol Chem. 1997;272:11402-11407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 79] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 36. | de Vries E, van Driel W, Bergsma WG, Arnberg AC, van der Vliet PC. HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J Mol Biol. 1989;208:65-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 206] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 37. | Zignego AL, Giannelli F, Marrocchi ME, Mazzocca A, Ferri C, Giannini C, Monti M, Caini P, Villa GL, Laffi G. T (14; 18) translocation in chronic hepatitis C virus infection. Hepatology. 2000;31:474-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 115] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 38. | Zuckerman E, Zuckerman T, Sahar D, Streichman S, Attias D, Sabo E, Yeshurun D, Rowe J. bcl-2 and immunoglobulin gene rearrangement in patients with hepatitis C virus infection. Br J Haematol. 2001;112:364-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
Supported partly by a grant from National Scientific Foundation of China, No.39970674
Edited by Zhang JZ