Published online Dec 14, 2025. doi: 10.3748/wjg.v31.i46.111631
Revised: September 21, 2025
Accepted: October 24, 2025
Published online: December 14, 2025
Processing time: 158 Days and 6.8 Hours
N6-methyladenosine (m6A) exerts a pro-carcinogenic effect in diverse cancers. The relationship between m6A-reading protein IGF2BP3 and gastric cancer (GC) has not yet been fully elucidated.
To investigate the molecular mechanisms of IGF2BP3 in GC carcinogenesis and progression and thus provide a rationale for novel therapeutic strategies.
Expression levels of IGF2BP3 in GC were determined using quantitative reverse transcription polymerase chain reaction (qRT-PCR), western blot (WB), and immunohistochemistry, and their associations with patients’ clinicopathological characteristics were analyzed. The role of IGF2BP3 in GC was investigated using cellular functional assays and subcutaneous xenograft models, and its downstream targets and signaling pathways were identified using high-throughput sequencing, bioinformatics analysis, RNA immunoprecipitation qPCR, dual luciferase reporter assay, qRT-PCR, and WB. The mechanism of IGF2BP3 in GC was validated via WB and rescue and inhibition experiments.
IGF2BP3 was highly expressed in GC and associated with diffuse-type GC, incidence of lymph node metastasis, advanced tumor node metastasis stage, and deeper tumor invasion depth. In vitro experiments demonstrated that IGF2BP3 promoted proliferation, migration, and invasiveness of GC cells, while inhibiting apoptosis and augmenting intracellular levels of glucose metabolism. In vivo experiments revealed that IGF2BP3 contributes to the growth of GC. Mechanistically, IGF2BP3 recognized and bound to the m6A site at position 1427 on FBXO32 messenger RNA, thereby increasing protein expression of FBXO32, and further activated the downstream cyclic guanosine monophosphate-protein kinase G (cGMP-PKG) signaling pathway to modulate various biological functions of GC cells and promote progression of GC. Furthermore, treatment with a selective PKG inhibitor KT5823 significantly suppressed the proliferative capacity of GC cells.
IGF2BP3 increases FBXO32 protein expression in an m6A-dependent manner, activates the cGMP-PKG signaling pathway, and promotes GC progression. Targeting of the IGF2BP3/FBXO32/cGMP-PKG axis could thus represent a promising therapeutic modality for GC.
Core Tip: IGF2BP3 is highly expressed in gastric cancer and is closely associated with poor patient prognosis. IGF2BP3 promotes gastric cancer growth and progression both in vivo and in vitro. Mechanistically, IGF2BP3 recognizes and binds to N6-methyladenosine-modified site on FBXO32 messenger RNA, thereby upregulating FBXO32 protein expression. This activates the downstream cyclic guanosine monophosphate-protein kinase G (cGMP-PKG) signaling pathway, thereby regulating multiple biological functions of gastric cancer cells and promoting tumor progression. We propose that targeting of the IGF2BP3/FBXO32/cGMP-PKG axis could represent a promising therapeutic modality for gastric cancer.
- Citation: Si Y, Tian B, Zhang R, Xuan MD, Liu KY, Jiao J, Han SS, Li HF, Hu YH, Zhao HY, He WJ, Wang J, Liu T, Yu WF. IGF2BP3 binds to FBXO32 to activate the cyclic guanosine monophosphate-protein kinase G pathway, promoting gastric cancer progression. World J Gastroenterol 2025; 31(46): 111631
- URL: https://www.wjgnet.com/1007-9327/full/v31/i46/111631.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i46.111631
Gastric cancer (GC) is among the most prevalent malignant tumor types worldwide and is characterized by a significant disease burden and high mortality rate[1]. It ranks among the top malignancies in terms of both incidence and mortality globally, particularly in China, and poses a serious threat to public health[2,3]. Although recent studies have demonstrated that systemic therapies can alleviate symptoms, improve survival, and enhance quality of life in patients with locally advanced or metastatic GC, the overall prognosis remains poor[4]. To date, the precise mechanisms underlying gastric carcinogenesis have not been fully elucidated. Previous studies have identified several risk factors contributing to the development of GC, including family history, dietary habits, alcohol consumption, and smoking, as well as infections with Helicobacter pylori and Epstein-Barr virus[5]. In addition, epigenetic alterations have been recognized as fundamental mechanisms in oncogenesis[6]. Therefore, further elucidation of the molecular mechanisms involved in the initiation and progression of GC could provide novel therapeutic targets and have substantial clinical implications for the treatment of this disease.
In recent years, the role of epigenetics in the pathogenesis of GC has attracted extensive research attention worldwide. Epigenetics refers to chemical modifications to DNA and RNA, the main forms of which are DNA and RNA methylation, histone modification, non-coding RNA modification, and chromatin rearrangement[7]. The N6-methyladenosine (m6A) modification is of particular interest, as it represents the most common internal modification of eukaryotic messenger RNA (mRNA) and regulates the stability, translation efficiency, alternative splicing, and localization of RNA at the post-transcriptional level. It also participates in various biological processes including initiation, proliferation, differentiation, metastasis, and metabolic reprogramming of tumor cells[8,9]. The majority of m6A-related proteins are methyltransferases, demethylases, or ‘readers’, encoded by 20 genes[10]. The main function of m6A readers is to interpret RNA methylation modifications and regulate the subsequent translation and degradation of the RNA[11,12]. IGF2BP3, an m6A reader, is located on human chromosome 7p15.3 and contains two RNA recognition motifs and four K homologous domains[13]. Its primary role is to recognize m6A binding sites through the K homology domain, thereby increasing the stability and translation efficiency of target mRNAs[14]. Overexpression of IGF2BP3 was first observed in pancreatic cancer[15] and has since been implicated in the development of various cancers, with distinct roles in each. For instance, IGF2BP3 enhances the stability of TMBIM6 mRNA by binding to its m6A site, thereby promoting the progression of laryngeal squamous cell carcinoma[16]. Similarly, in hepatocellular carcinoma, it stabilizes and increases the expression of lnc-CTHCC (cancer-testis associated long non-coding RNA) by recognizing its m6A site, facilitating the progression of the disease[17]. In colorectal cancer, IGF2BP3 stabilizes CCND1 mRNA via its m6A site, promoting CCND1 protein expression, which in turn affects regulation of the cell cycle and promotes proliferation and growth of cancer cells[18]. Furthermore, Wang et al[19] found that m6A reader IGF2BP2 enhanced the mRNA stability of GLUT1, a key gene in aerobic glycolysis, in an m6A-dependent manner, thereby significantly promoting aerobic glycolysis of colorectal cancer cells. Taken together, these findings indicate that IGF2BP3 may be a critical oncoprotein.
Here, we observed that IGF2BP3 was highly expressed in GC, and its expression was closely associated with poor prognosis in GC patients. IGF2BP3 was also found to enhance malignant phenotypes of GC cells and promote gastric tumor growth in vivo. Furthermore, through omics analysis and subsequent experimental validation, we demonstrated for the first time that the FBXO32, is a downstream target of IGF2BP3, harboring m6A modification in GC. In addition, we observed high expression of FBXO32 in GC and found that IGF2BP3 upregulates protein expression of FBXO32 in an m6A-dependent manner. Together, IGF2BP3 and FBXO32 co-operatively activated the cyclic guanosine monophosphate-protein kinase G (cGMP-PKG) signaling pathway, thereby enhancing the proliferative, migratory, and invasive capacities of GC cells. These effects were inhibited by application of selective cGMP-dependent PKG inhibitor KT5823.
Sixty-three pairs of GC tissues and adjacent normal tissues were obtained from the First Hospital of Hebei Medical University. None of the patients had received radiation or chemotherapy prior to surgery, and all patients signed informed consent forms. The tissue samples were promptly placed in liquid nitrogen after isolation and then transferred to the Biological Sample Bank of the First Hospital of Hebei Medical University for storage. The cancer tissue samples used in this study were pathologically confirmed to consist of gastric adenocarcinoma/signet-ring cell carcinoma tissues. The study was approved by the Ethics Committee of the First Hospital of Hebei Medical University (approval No. S00112) and adheres to the principles of the Declaration of Helsinki.
Human gastric mucosa cells (GES-1) and GC cells (HGC-27, AGS, MKN7, and MKN74) were obtained from Wuhan Pricella Biotechnology Co., Ltd., and were verified by short tandem repeat analysis. The cells were cultured in RPMI 1640 medium (Gibco, Gaithersburg, MD, United States) supplemented with 10% fetal bovine serum (FBS) (Gibco, Gaithersburg, MD, United States) and 1% penicillin-streptomycin solution (Solarbio Sciences and Technology Co. Ltd., Beijing, China) and maintained in an incubator at constant temperature (37 °C) and humidity with 5% carbon dioxide. Routine mycoplasma detection was performed monthly using polymerase chain reaction (PCR). In all experiments, cells were used for a maximum of 25 passages.
Transfection was performed using lipofectamine 3000 (Invitrogen, Carlsbad, CA, United States) when cell confluence reached 80%-90% in the cell culture flask or dish. A small interfering RNA (siRNA) targeting IGF2BP3 and a negative control (NC) siRNA (GenePharma Co., Ltd., Shanghai, China) were transfected into HGC-27 and MKN74 cells, whereas an IGF2BP3-overexpression plasmid and a NC plasmid (GenePharma) were transfected into MKN7 and GES-1 cells. In addition, an IGF2BP3 short hairpin RNA (shRNA-IGF2BP3) and a NC (shRNA-NC) (GenePharma) were transfected into HGC-27 cells. For FBXO32, a targeted siRNA and NC (GenePharma) were transfected into MKN7 cells, and an FBXO32-overexpression plasmid and NC (GenePharma) were transfected into HGC-27 cells. Follow-up experiments were conducted 48 hours after transfection. To establish HGC-27 cell lines stably transfected with shRNA-NC and shRNA-IGF2BP3 (designated HGC-27-shNC and HGC-27-shIGF2BP3, respectively), 48 hours after transfection, the cell culture medium was replaced with medium including neomycin at a final concentration of 600 μg/mL (no other antibiotics were present). Once clones had formed, the neomycin concentration was adjusted to 300 μg/mL, and cell culture was continued until day 10. Selective PKG inhibitor KT5823 (126643-37-6, MedChemExpress, United States) was reconstituted according to the manufacturer’s instructions and stored under recommended conditions.
Total RNA was extracted from cells and tissues using RNA-Easy Isolation Reagent (Vazyme Biotech Co., Ltd., Nanjing, Jiangsu Province, China). The RNA was then reverse-transcribed into complementary DNA (cDNA) following the instructions of PrimeScript RT reagent kit (Takara, Beijing, China). Quantitative reverse transcription PCR (qRT-PCR) was performed using AceQ Universal SYBR qPCR master mix (Vazyme), with β-actin serving as the endogenous control. Each sample was tested in triplicate. The mRNA expression level was determined based on the cyclic threshold value of each sample, and the relative expression level of each sample was calculated using the 2-∆∆CT method. The sequences of primers are detailed in Supplementary Table 1.
Tissues from patients and nude mice were fixed with 4% paraformaldehyde, followed by paraffin embedding (Shanghai YiYang Instrument Co., Ltd., Shanghai, China), sectioning, and staining. A rabbit/mouse two-step detection kit (Beijing ZSBG-Bio Co., Ltd., Beijing, China) was used for immunohistochemical (IHC) analysis according to the manufacturer’s instructions. Anti-IGF2BP3 (1:150, 14642-1-AP, Proteintech, China), anti-Ki67 (1:1000, 27309-1-AP, Proteintech, China), and anti-FBXO32 (1:500, 67172-1-Ig, Proteintech, China) antibodies were used for IHC staining, and the staining results were analyzed using the H-score method as follows. First, the percentage of positive cells within each high-magnification microscopic field of view as determined (ranging from 0% to 100%). The staining intensity was then graded, with no staining classified as grade 0, weak staining (pale yellow) as grade 1, moderate staining (brownish yellow) as grade 2, and strong staining (brown) as grade 3. The percentage of positive cells was multiplied by the corresponding staining intensity grade, and the sum of the products from all fields of view was divided by the total number of fields to obtain the average product, expressed as H-score = Σ [(% positive cells × staining intensity grade)]/total number of fields. Scores ≤ 100 were classified as negative group, whereas scores > 100 were classified as positive.
Proteins were extracted using radio immunoprecipitation assay lysis buffer (Solarbio) and protease/phosphatase inhibitors (Solarbio) at a ratio of 100:1, separated electrophoretically on a 10% sodium dodecyl sulfate polyacrylamide gel (Bio-Rad Laboratories Inc, Hercules, CA, United States), and transferred to a polyvinylidene fluoride membrane (Merck Millipore, Billerica, MA, United States). The membrane was then incubated with the following antibodies: Anti-IGF2BP3 (1:1000, ab177477, Abcam, United Kingdom), anti-β-actin (1:1000, TA-09, ZSBG-Bio, China), anti-FBXO32 (1:500, 67172-1-Ig, Proteintech, China), anti-glyceraldehyde-3-phosphate dehydrogenase (1:1000, AB-P-R 001, Goodhere Co. Ltd, China), anti-PKG1 (1:500, 21646-1-AP, Proteintech, China), anti-phospho-VASP (Ser239) (1:500, 3114, Cell Signaling Technology, United States), and anti-VASP (1:500, 13472-1-AP, Proteintech, China). Immunoreactive protein bands were detected using enhanced chemiluminescence (Millipore, Burlington, MA, United States).
For the cell counting kit-8 (CCK-8) assay, cells from each group were uniformly distributed in 96-well plates at a density of 1 × 103 cells per well and cultured with complete culture medium. At 24, 48, 72, and 96 hours after distribution, 10 μL of CCK-8 (Dojindo, Tokyo, Japan) reagent was added to each well, and the cells were incubated for an additional 2 hours. The absorbance of each well at 450 nm was measured using a Promega GloMax luminescence detector (Promega, Madison, WI, United States). For the colony formation assay, cells from each group were uniformly distributed in six-well plates at a density of 500 cells per well. Fresh culture medium was replaced every 3 days. Cells were cultured for 7-14 days, then washed twice with phosphate-buffered saline (PBS), treated with 4% paraformaldehyde for 30 minutes, and stained with 0.1% crystal violet for 20 minutes.
For the wound healing assay, cells from each group were uniformly distributed in six-well plates. When the cell confluence reached or was close to 100%, a 200 μL pipette tip was used to draw two straight lines on each well to simulate a wound. The cells were then washed twice with PBS and cultured in medium without FBS. Each well was photographed at 0 hour and 48 hours after the wound was created, and the migration rate was expressed as the ratio of the gap width measured at 48 hours to the gap width measured at 0 hour.
For the transwell migration assay, 3 × 104 (HGC-27, GES-1) or 1 × 105 (MKN7, MKN74) cells were added to the upper chamber of a transwell apparatus (Corning Inc., Corning, NY, United States) in 200 μL FBS-free medium. To induce downward migration, 700 μL of complete medium was added to the lower chamber. After 48 hours of culture, cells on the upper-chamber side of the polycarbonate membrane were removed using a swab. The chamber was washed twice with PBS, then treated with 4% paraformaldehyde for 30 minutes, followed by 0.1% crystal violet for 20 minutes. Excess crystal violet was removed by washing again with PBS. Five visual fields were randomly selected for photography, and the stained cells were counted using ImageJ software (National Institutes of Health, Bethesda, MD, United States).
For the transwell invasion assay, cells were resuspended in 100 μL FBS-free medium and added to the upper chamber, and 500 μL of complete medium was added to the lower chamber to induce downward invasion of the cells in the upper chamber. All other experimental procedures were the same as for the transwell migration assay.
Cells from each group were collected 48 hours after transfection and stained using an Annexin V-fluorescein isothiocyanate/propidium iodine apoptosis assay kit (NeoBioscience, Shenzhen, China). Flow cytometry (BD Biosciences, San Jose, CA, United States) was used to analyze apoptosis in a sample of 1 × 105 cells, and FlowJo software was used to calculate the percentage of apoptotic cells.
MKN7 and HGC-27 cells with overexpression or knockdown of IGF2BP3 were collected and processed using a glucose assay kit (Abcam, ab65333), l-lactate assay kit (Abcam, ab65331), and adenosine triphosphate (ATP) assay kit (Abcam, ab83355) according to the kit instructions. In brief, standard solutions of glucose, lactic acid, and ATP were prepared in a gradient dilution. Reaction mixtures were then prepared and mixed with the diluted standard solutions, followed by incubation (at 37 °C or room temperature) for 30 minutes. A spectrophotometer was adjusted to the appropriate wavelength, absorbance values were read, and standard curves for glucose, lactic acid, and ATP were generated. The corresponding reagents from the kits were added to the lysed cells, and the cell lysate was deproteinized using deproteinizing sample preparation kit tricarboxylic acid (Abcam, ab204708). The deproteinized cell lysates were then mixed with the reaction mixtures, followed by incubation (at 37 °C or room temperature) for 30 minutes, and absorbance values were measured and compared with the standard curves to calculate concentrations of glucose, lactic acid, and ATP.
Ten male BALB/c nude mice aged 4-6 weeks, weighing 16-20 g (procured from Beijing Huafukang Biotechnology Co., Ltd.) were randomly divided into two groups (five mice per group) and housed in a pathogen-free environment with sufficient water and food. The room was maintained at a temperature of approximately 22 °C and a 12-hour light/12-hour dark cycle. On a clean bench, the skin of the left hind limb flank of each nude mouse was disinfected, and HGC-27-shNC and HGC-27-shIGF2BP3 cells (5 × 106 cells per mouse) were subcutaneously injected using a sterile syringe. Starting from day 3 post-injection, tumor size was evaluated every 2 days using following formula: Volume = (long diameter × short diameter2)/2. When the tumor volume approached 1000 mm3, the nude mice were euthanized humanely. Tumor specimens were collected, photographed, weighed, and fixed in formalin. The tumor samples were then paraffin-embedded and sectioned for subsequent hematoxylin and eosin and IHC analysis. These experiments were approved by the Ethics Committee of the First Hospital of Hebei Medical University and carried out in accordance with the experimental animal care and use system.
Total RNA was extracted from HGC-27 cells transfected with siRNA-NC (normal control) and siRNA-IGF2BP3 (targeting IGF2BP3) for 24 hours. RNA library construction and sequencing analysis were performed by Beijing Novogene Technology Co., Ltd. In short, a total of six samples [siRNA-NC (siNC) × 3, IGF2BP3 siRNA (siIGF2BP3) × 3] were tested using Illumina’s NovaSeq 6000 platform. Quality control and filtering were carried out on the output data from each sample to ensure that high-quality data were obtained. HISAT software was used to align the high-quality data to reference genomes and to quantify gene or transcript expression levels in the sample. DESeq2 software was used for differential expression analysis, with differential genes defined as those with log2|fold change| > 1.0 and P < 0.05.
Total RNA was extracted from HGC-27 cells transfected with siRNA-NC and siRNA-IGF2BP3 for 24 hours. RNA library construction and sequencing analysis were performed by Novogene. In short, the original data were processed using fastp (version 0.19.11) to yield high-quality data. The reference genome index was constructed using Burrows-Wheeler Aligner (BWA) (version 0.7.12), and the high-quality data were aligned with the reference genome using BWA mem (version 0.7.12). After reads had been mapped to the reference genome, the m6A peaks within each group were identified using the exomePeak R package (version 2.16.0). The genes corresponding to each peak were defined as peak-related genes. The m6A enrichment motif of each group was identified with HOMER (version 4.9.1). The peak distribution in the 5’ untranslated region, coding DNA sequence, 3’ untranslated region, and other functional regions on mRNA transcripts was statistically analyzed.
According to the instructions provided with the RNA immunoprecipitation (RIP) kit (BersinBio, Guangzhou, Guangdong Province, China; catalog number: Bes5101), HGC-27 cells transfected with siRNA-NC and siRNA-IGF2BP3 for 48 hours were collected and lysed. An IGF2BP3-specific antibody (10 μg; Proteintech; catalog number: 14642-1-AP) was used to bind endogenous expression of IGF2BP3 protein in the cells, precipitate the target protein-RNA complex, and subsequently isolate and purify the RNA within the complex. Novogene was commissioned to perform ribosomal RNA removal, RNA library construction, and high-throughput sequencing analysis. In brief, RNA fragment distribution and concentration after immunoprecipitation and ribosomal RNA removal were measured using an Agilent 2100 Bioanalyzer and a simpliNano spectrophotometer (GE Healthcare). Library quality was assessed using an Agilent Bioanalyzer 2100 system, and the library was then sequenced using the Illumina NovaSeq platform. fastp software was utilized to filter the original data and obtain high-quality data. The reference genome index was established using BWA (version 0.7.12), and the filtered data were compared with the reference genome using BWA mem (version 0.7.12). Following the comparison, the immunoprecipitation enrichment region was identified against the background using MACS2 (version 2.1.0) peak-calling software. After peak-calling, the results were analyzed with respect to chromosome distribution, peak width, fold abundance, significance level, and peak number per peak. HOMER (version 4.9.1) was used to identify m6A enrichment motifs in each group, and Peak Annotator was used to identify peak-related genes. Differential peak analysis was carried out based on the enrichment of the peaks, and genes related to the differential peaks were identified.
Dual-luciferase reporter vectors containing the wild-type coding sequences of FBXO32 and its mutant versions with m6A site substitutions [adenine (A) to cytosine (C)] at nucleotide positions 1427 (Mut-1), 1399 (Mut-2), and 1493 (Mut-3) were constructed by Saierbio (Tianjin, China). Subsequently, HEK-293T cells were co-transfected with an IGF2BP3-overexpressing plasmid and the FBXO32-wild-type or FBXO32 Mut-1, Mut-2 or Mut-3 luciferase reporter vector. After 48 hours of incubation, the cells were lysed, and luciferase activity was measured using a dual-luciferase reporter assay kit (RG009, Beyotime, China) and GloMax 96 microplate luminometer (Promega, United States). Firefly luciferase activity was normalized to Renilla luciferase activity.
Expression levels of IGF2BP3 mRNA in cancer and adjacent tissues of patients with GC were obtained from The Cancer Genome Atlas (TCGA) (https://www.aclbi.com/static/index.html#/tcga) and Gene Expression Omnibus (GEO) (https://www.aclbi.com/static/index.html#/geo). Kaplan-Meier plotter (https://kmplot.com/analysis/) was used to analyze the influence of gene expression levels on the prognosis of GC patients. Gene Expression Profiling Interactive Analysis (http://gepia2.cancer-pku.cn/#index) was used to analyze gene expression levels in GC and adjacent normal tissues, and GeneCards (https://www.genecards.org/) was used to gain insight into gene sublocalization within cells. PrimerBank (https://pga.mgh.harvard.edu/primerbank/) and the Primer-BLAST tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) were used for primer design, and catRAPID omics (http://s.tartaglialab.com/page/catrapid_group) was used to predict binding site motifs between the IGF2BP3 protein and FBXO32 mRNA. The Integrative Genomics Viewer software was utilized for analysis of genomic m6A methylation levels. STRING (https://cn.string-db.org/) and Cytoscape were used to predict and visualize the protein–protein interaction network, and BioRender (https://www.biorender.com/) facilitated the creation of scientific schematic diagrams.
Three independent replicates were performed for each group of experiments. Results in the form of normally distributed data were presented as the mean ± SD, whereas non-normally distributed data were presented as the median and interquartile distance. Statistical analyses were performed using GraphPad Prism 9.5 (GraphPad Software, La Jolla, CA, United States) and SPSS 26.0 (IBM, Armonk, NY, United States). Statistical methods used included Student’s t-test and one-way and two-way analyses of variance. P < 0.05 was considered to indicate statistical significance.
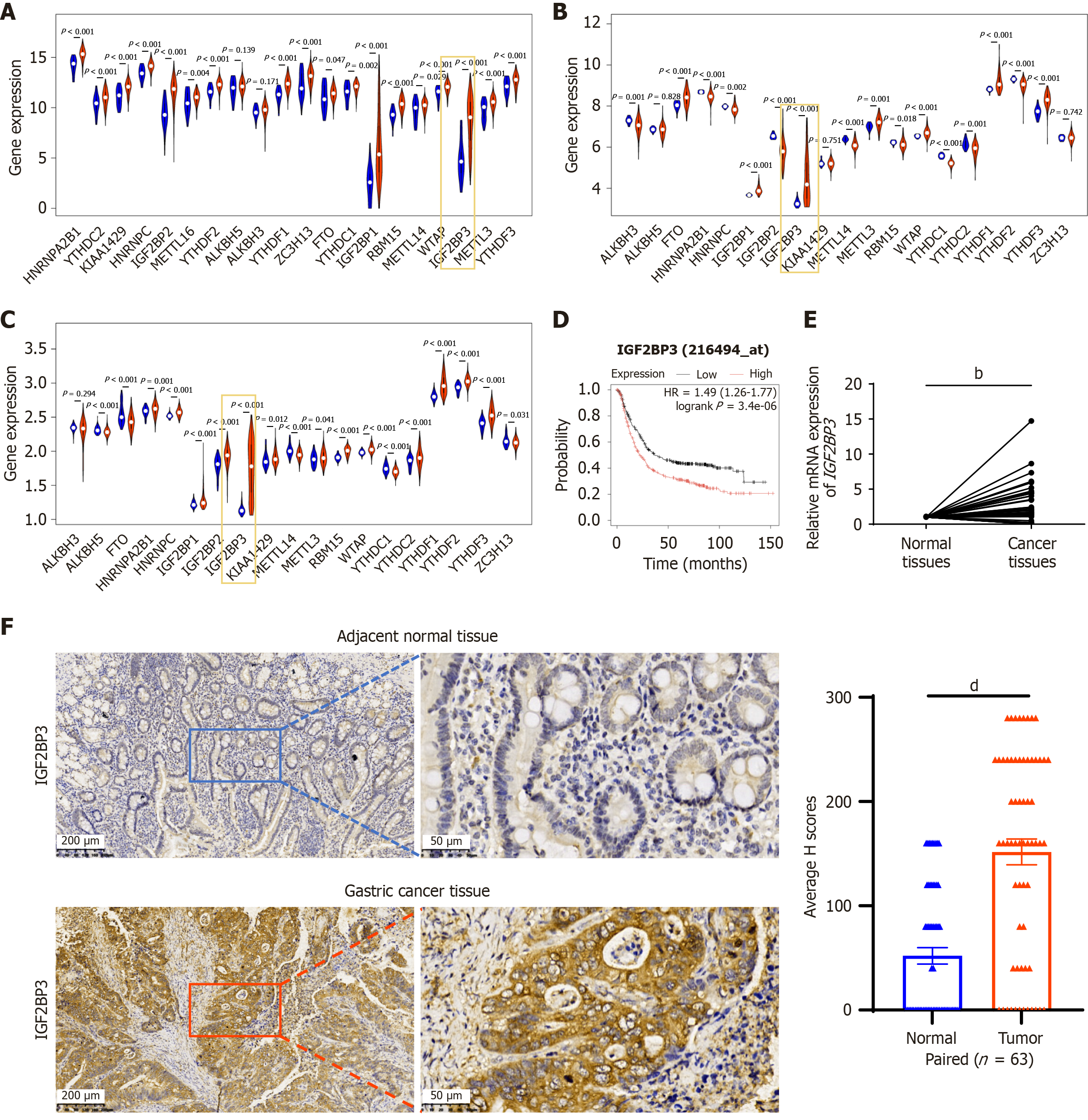
To investigate the role of m6A-related genes in the pathogenesis of GC, we first analyzed the expression of m6A related genes in GC and adjacent normal tissues in the TCGA database and the GSE54129 and GSE66229 datasets (Figure 1A-C). We identified four m6A reader proteins (YTHDF1, YTHDF3, IGF2BP1, and IGF2BP3) with higher expression levels in GC than in adjacent normal tissues, among which IGF2BP3 exhibited the most significant differential expression. There has also been limited research on the mechanism of this protein in GC. Therefore, we selected IGF2BP3 as our research focus for the present work.
Kaplan-Meier survival analysis indicated that GC patients with high IGF2BP3 expression had worse overall survival than those with low IGF2BP3 expression (Figure 1D). We then assessed IGF2BP3 mRNA expression in 50 pairs of fresh frozen GC tissues and adjacent normal tissues using qRT-PCR. Consistent with the TCGA and GEO data, the expression of IGF2BP3 was higher in GC tissues compared with adjacent normal tissues, and the difference was statistically significant (P < 0.01) (Figure 1E). Next, we used IHC to detect IGF2BP3 protein expression in these samples and found that IGF2BP3 was primarily localized in the cytoplasm of GC cells and expressed at significantly higher levels than in adjacent normal tissues (χ2 = 22.415; P < 0.001) (Figure 1F and Table 1). Finally, we analyzed the correlations between IGF2BP3 expression and clinical characteristics of GC patients. IGF2BP3 expression was significantly associated with diffuse-type GC (P < 0.05), lymph node metastasis (P < 0.001), tumor node metastasis (TNM) stage (P = 0.001), and invasion range (P = 0.001), whereas it showed no association with gender, age, tumor size, or tumor differentiation (Table 2). Taken together, these findings suggest that m6A reader protein IGF2BP3 is both highly expressed in GC and closely associated with poor prognosis in GC patients.
| Group | Cases | IGF2BP3 expression | Positive rate (%) | χ2 value | P value | |
| High | Low | |||||
| Gastric cancer tissues | 63 | 38 | 25 | 60 | 22.415 | < 0.001c |
| Adjacent normal tissues | 63 | 12 | 51 | 19 | ||
| Group | Cases | IGF2BP3 expression | χ2 value | P value | |
| High | Low | ||||
| Gender | |||||
| Male | 48 | 33 | 15 | 2.404 | 0.121 |
| Female | 15 | 7 | 8 | ||
| Age (years) | |||||
| ≤ 60 | 16 | 11 | 5 | 0.256 | 0.613 |
| > 60 | 47 | 29 | 18 | ||
| Tumor size (cm) | |||||
| < 5 | 34 | 20 | 14 | 1.550 | 0.2131 |
| ≥ 5 | 27 | 20 | 7 | ||
| Differentiation | |||||
| Well | 27 | 14 | 13 | 2.762 | 0.097 |
| Poorly | 36 | 26 | 10 | ||
| Lauren classification | |||||
| Intestinal type | 33 | 18 | 15 | 3.873 | 0.049a,1 |
| Diffuse type | 28 | 22 | 6 | ||
| Lymph node metastasis | |||||
| Negative | 18 | 5 | 13 | 13.867 | < 0.001c |
| Positive | 45 | 35 | 10 | ||
| TNM staging | |||||
| 0 | 2 | 0 | 2 | 17.037 | 0.001b |
| I | 4 | 0 | 4 | ||
| II | 17 | 9 | 8 | ||
| III | 30 | 21 | 9 | ||
| IV | 10 | 10 | 0 | ||
| Invasion range | |||||
| Tis | 2 | 0 | 2 | 14.165 | 0.001b |
| T1 | 3 | 0 | 3 | ||
| T2 | 8 | 5 | 3 | ||
| T3 | 5 | 1 | 4 | ||
| T4 | 45 | 34 | 11 | ||
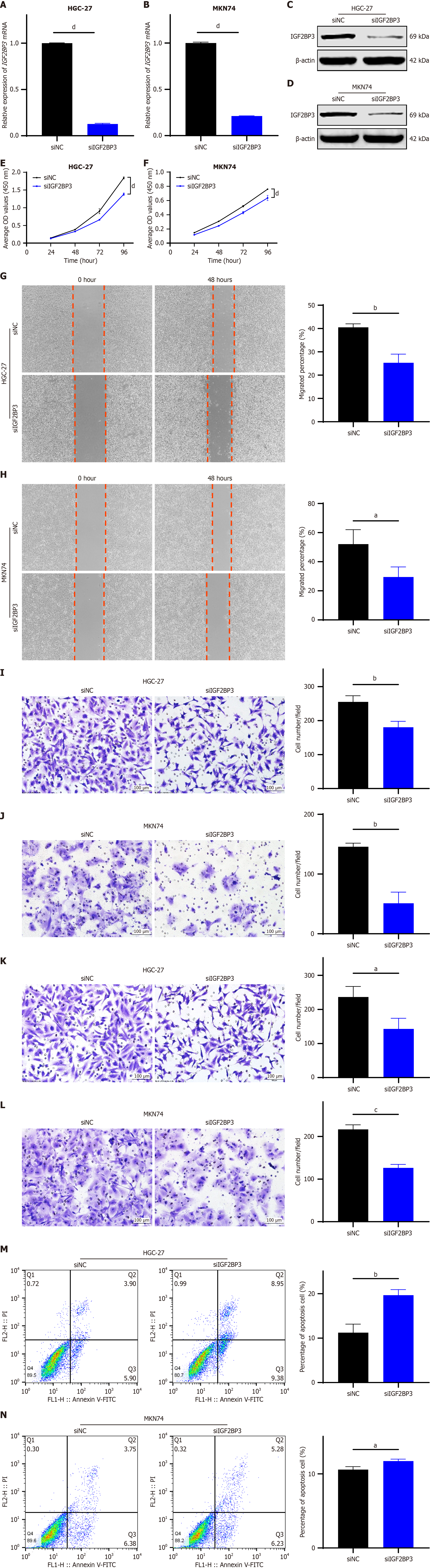
To investigate the mechanism of action of IGF2BP3 in GC, we first assessed the mRNA (Supplementary Figure 1A) and protein (Supplementary Figure 1B) expression levels of IGF2BP3 in GES-1, HGC-27, AGS, MKN74, and MKN7 cells. Results indicated that the expression levels of IGF2BP3 mRNA and protein were relatively high in both HGC-27 and MKN74 cells. Therefore, we further performed knockdown of IGF2BP3 in these cell types to determine its impact on cellular function. Two IGF2BP3 siRNAs were transfected simultaneously into HGC-27 and MKN74 cells, and the one with significantly better knockdown efficiency (siRNA-IGF2BP3-2) was selected for subsequent experiments (Supplementary Figure 1C and D). Following transfection with siRNA-IGF2BP3-2 (siIGF2BP3) and a siNC, both mRNA (Figure 2A and B) and protein (Figure 2C and D) levels of IGF2BP3 were significantly reduced. The CCK-8 assay revealed marked inhibition of cell proliferation after knockdown of IGF2BP3 (Figure 2E and F), and wound healing (Figure 2G and H) and transwell migration (Figure 2I and J) assays demonstrated that knockdown of IGF2BP3 significantly suppressed cell migration. Furthermore, the transwell invasion assay showed that the reduction in IGF2BP3 expression significantly impaired the invasion capability of the cells (Figure 2K and L), and flow cytometry analysis showed that silencing IGF2BP3 increased apoptosis of cells of both types (Figure 2M and N). To minimize the potential for off-target effects, an additional siRNA targeting IGF2BP3 was designed and transfected into HGC-27 and MKN74 cells for subsequent functional assays (Supplementary Figure 2). Based on the above results, it can be concluded that downregulation of IGF2BP3 seems to inhibit proliferation, migration, and invasion of GC cells and promote their apoptosis.
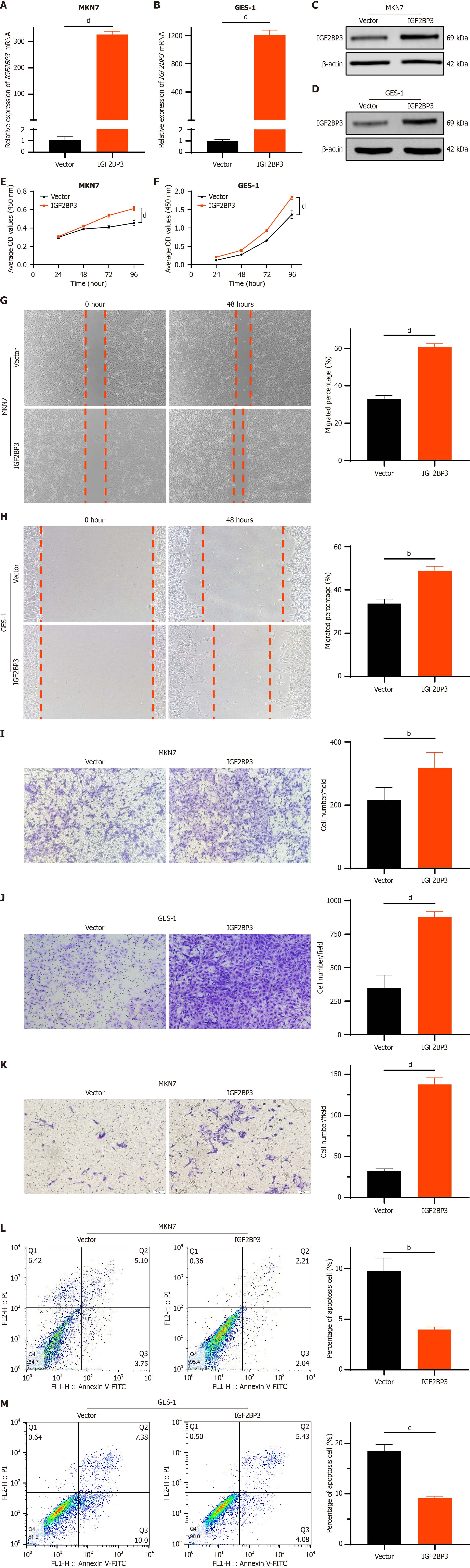
To further investigate the impact of IGF2BP3 on cellular biological functions, we overexpressed it in MKN7 and GES-1 cells, which exhibit relatively low endogenous expression levels. After transfecting cells with pcDNA3.1-IGF2BP3 (IGF2BP3-overexpression) and pcDNA3.1 (control) vectors, we observed significant increases in IGF2BP3 mRNA (Figure 3A and B) and protein (Figure 3C and D) levels. CCK-8 assay showed that cell proliferation was notably enhanced following overexpression of IGF2BP3 (Figure 3E and F), and wound healing (Figure 3G and H) and transwell migration (Figure 3I and J) assays demonstrated significantly increased migration capability. Furthermore, according to the transwell invasion assay, elevated IGF2BP3 expression significantly enhanced the invasion ability of cells (Figure 3K) (although GES-1 cells were not able to penetrate the matrix glue at the base of the transwell chambers, owing to the inherently weaker invasiveness of normal epithelial cells). Flow cytometry analysis demonstrated reduced apoptosis levels among both MKN7 and GES-1 cells with IGF2BP3 overexpression (Figure 3L and M). These results show that overexpression of IGF2BP3 can promote proliferation, migration, and invasion of GC cells and inhibit their apoptosis, suggesting that IGF2BP3 may function as an oncogene in GC.
Tumor cells rapidly produce large amounts of energy through a unique aerobic glycolysis pathway to meet their high metabolic demands while maintaining normal cellular functions. We monitored levels of glucose, lactic acid and ATP in HGC-27 and MKN7 GC cells after modulating expression levels of IGF2BP3 to explore its role in glucose metabolism. Downregulation and upregulation of IGF2BP3 expression led to decreased and increased intracellular levels of glucose (Supplementary Figure 3A), lactic acid (Supplementary Figure 3B), and ATP (Supplementary Figure 3C), respectively, suggesting that IGF2BP3 may have a significant role in regulation of glucose metabolism in GC cells. In future work, we will examine changes in the oxygen consumption rate and extracellular acidification rate in GC cells with altered IGF2BP3 expression, to further explore the effects of IGF2BP3 on the glycolytic capability of these cells.
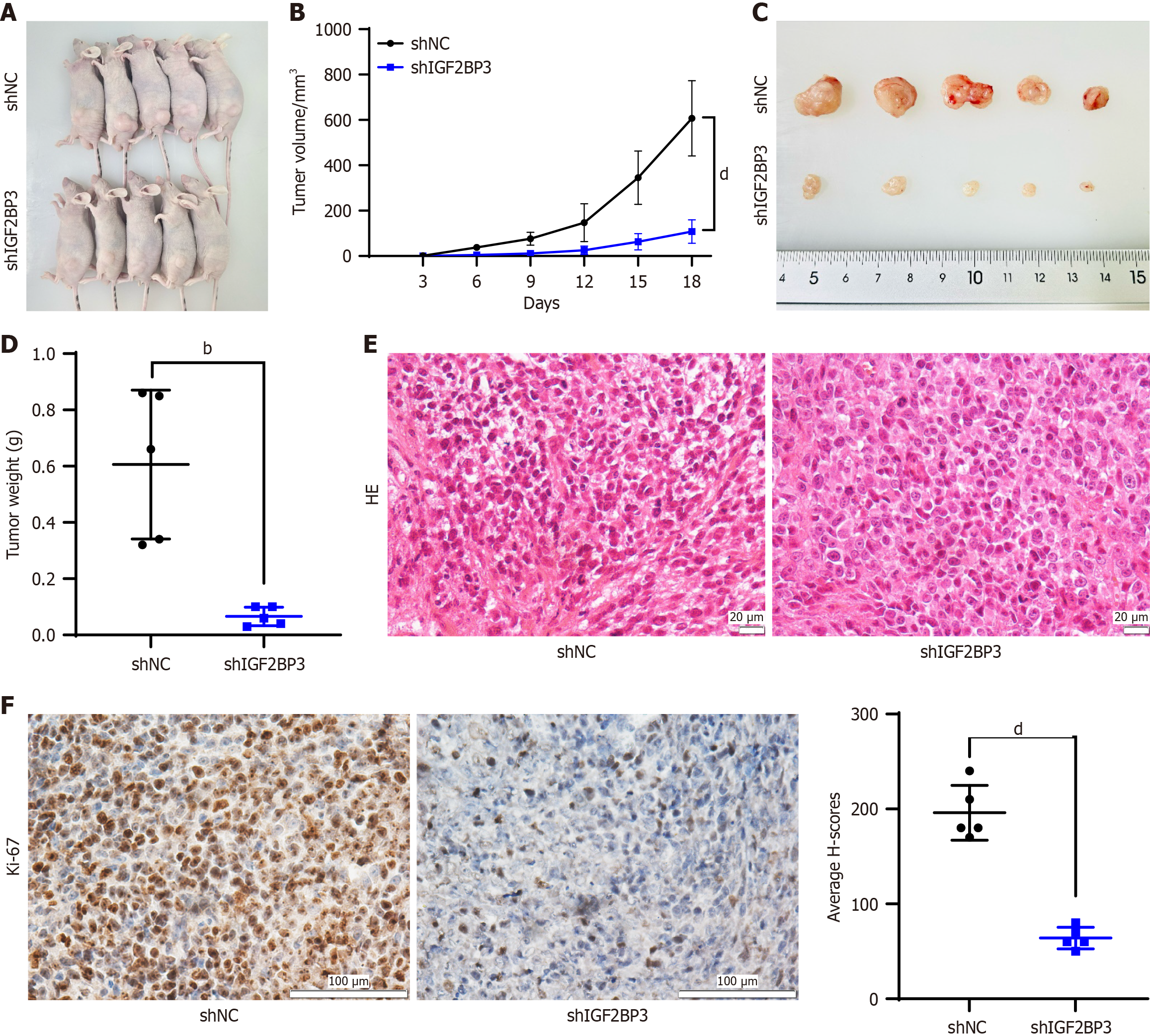
To further investigate the mechanism of IGF2BP3 in GC in vivo, we established tumor-bearing nude mouse models with low expression of IGF2BP3 and corresponding NCs. Following PCR (Supplementary Figure 4A) and Western blot (Supplementary Figure 4B) assays, we successfully established stable HGC-27-shIGF2BP3 and HGC-27-shNC cell lines. These cells were then injected subcutaneously into two groups of BALB/c nude mice. The resulting tumor-bearing models, together with tumor volume curves, photographs, and weights, are presented in Figure 4A-D. We observed significant reductions in tumor growth rate, volume, and weight in the shIGF2BP3 group compared with the shNC group, and PCR and Western blot assays showed that IGF2BP3 mRNA (Supplementary Figure 4C) and protein (Supplementary Figure 4D) expression levels in the xenograft tumors of nude mice in the shIGF2BP3 group were significantly lower than those in the shNC group. Hematoxylin and eosin staining was used to evaluate the morphological characteristics of the xenograft tissues, the results indicated that these tissues had similar cell morphology and structure to those of GC tissues (Figure 4E). Moreover, IHC staining for Ki67 revealed significantly inhibited cell proliferation in the shIGF2BP3 group compared with the shNC group (Figure 4F). These findings suggest that IGF2BP3 knockdown can inhibit the growth of GC in vivo.
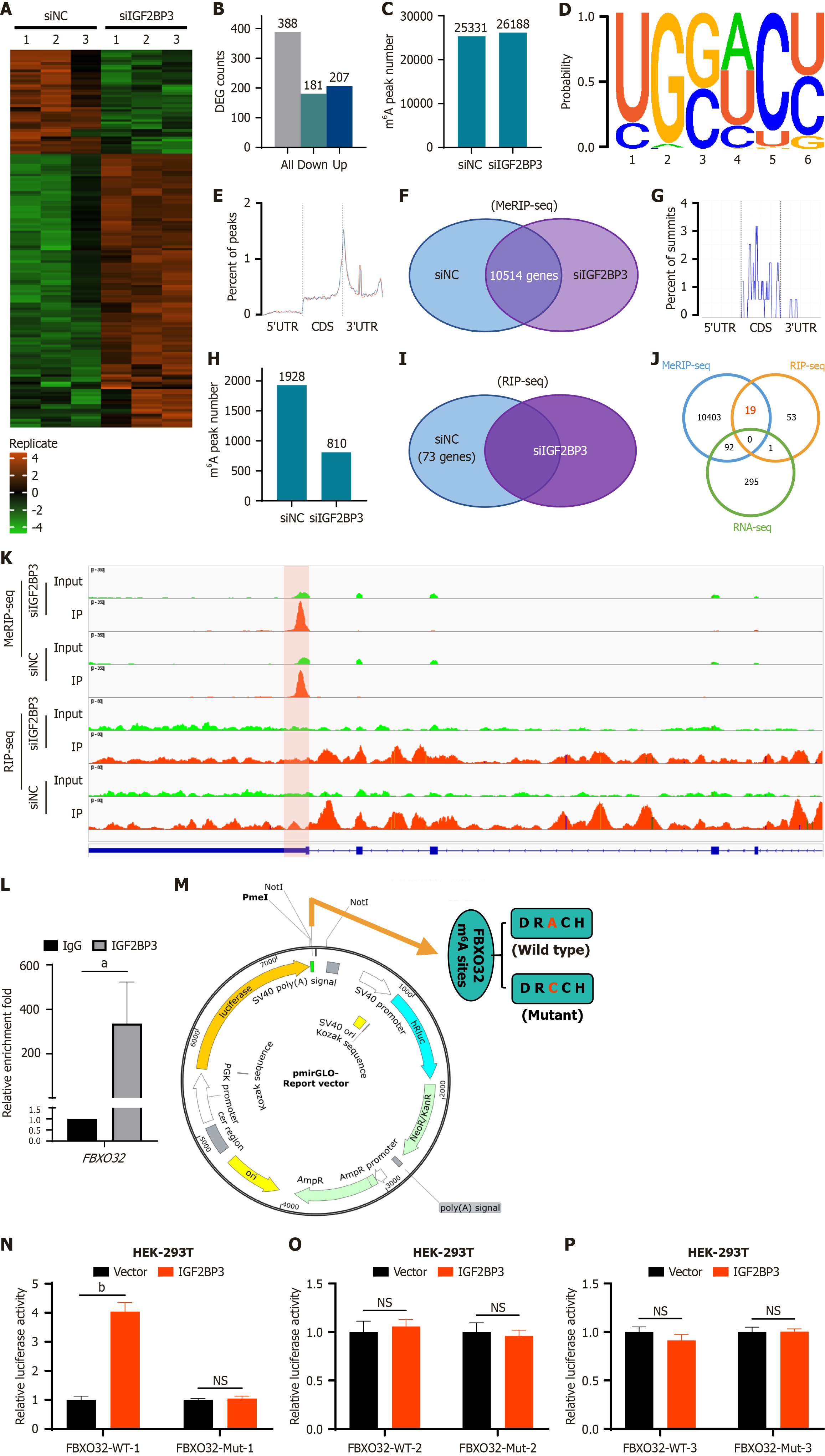
To further explore the potential mechanism of IGF2BP3 in the pathogenesis of GC, we performed RNA sequencing (RNA-seq), methylated RIP sequencing (MeRIP-seq), and RIP sequencing (RIP-seq) in HGC-27 cells of the siNC group and siIGF2BP3 group, respectively. The RNA-seq results showed significant changes in expression levels of 388 genes after IGF2BP3 knockdown, including 181 downregulated genes and 207 upregulated genes (Figure 5A and B). MeRIP-seq identified 25331 m6A peaks in the siNC group and 26188 m6A peaks in the siIGF2BP3 group (Figure 5C). When the m6A methyl group was mapped in HGC-27 cells, an m6A-consistent sequence motif (GGAC) could be identified, indicating successful enrichment of mRNA with m6A modifications (Figure 5D). In addition, m6A modification was mainly enriched in the coding DNA sequence and 3’ untranslated region of mRNA (Figure 5E). Intersection of the genes corresponding to the m6A peaks of the siNC group and siIGF2BP3 group yielded 10514 genes (Figure 5F). RIP-seq showed that m6A modification was again primarily enriched in the coding DNA sequence and 3’ untranslated region of mRNA (Figure 5G). Specifically, 1928 and 810 m6A peaks were identified in the siNC group and siIGF2BP3 group, respectively (Figure 5H). After removing the same genes from both groups, 73 unique genes were identified (Figure 5I). Subsequent analysis of the union of the genes identified by RNA-seq, MeRIP-seq, and RIP-seq showed that 19 genes bound to the IGF2BP3 protein were simultaneously labeled by m6A, and none of these genes showed changes in their transcription levels after IGF2BP3 knockdown (Figure 5J). This indicates that IGF2BP3 does not affect the abundance of its target RNAs; rather, it may modulate protein synthesis through its interactions with m6A-modified mRNAs.
Further analyses of these candidate genes included examination of their expression in GC tissues, their subcellular localization, their associations with survival of GC patients, and the abundance and loci of m6A on their mRNA. FBXO32 was found to be highly expressed in GC tissues based on TCGA data (Supplementary Figure 5A) and predominantly distributed in the cytoplasm and nucleus (Supplementary Figure 5B); moreover, the overall survival of patients with high expression of FBXO32 was poor (Supplementary Figure 5C), and the binding site of IGF2BP3 colocalize with m6A site on FBXO32 transcript (Figure 5K). These results suggest that IGF2BP3 may regulate FBXO32 in an m6A-dependent manner, thereby enhancing the malignant phenotype of GC. To verify this hypothesis, an RIP-qPCR assay was performed. The results indicated that IGF2BP3 protein could significantly enrich FBXO32 mRNA (Figure 5L). Subsequently, we used the catRAPID website to predict the presence of an m6A-binding site between IGF2BP3 protein and FBXO32 mRNA (Supplementary Figure 5D). Finally, to further elucidate the regulatory relationship between IGF2BP3 and FBXO32, we constructed luciferase reporter vectors containing either wild-type or m6A site-mutated sequences of FBXO32. The m6A sites on the FBXO32 transcript were predicted using the SRAMP database, and the top three high-scoring sites (1427, 1399, and 1493) were subjected to mutagenesis by substituting adenine with cytidine (Figure 5M). Overexpression of IGF2BP3 significantly enhanced luciferase activity in cells with the wild-type sequence site 1427, whereas no notable change was observed in the corresponding mutant group (Figure 5N). Notably, neither the wild-type nor mutant constructs for the other two sites resulted in significant alterations in luciferase activity (Figure 5O and P). These findings suggest that IGF2BP3 may exert its regulatory role by specifically targeting the m6A site at position 1427 on FBXO32 mRNA. Collectively, these data indicate that IGF2BP3 regulates FBXO32 in GC through an m6A-dependent mechanism.
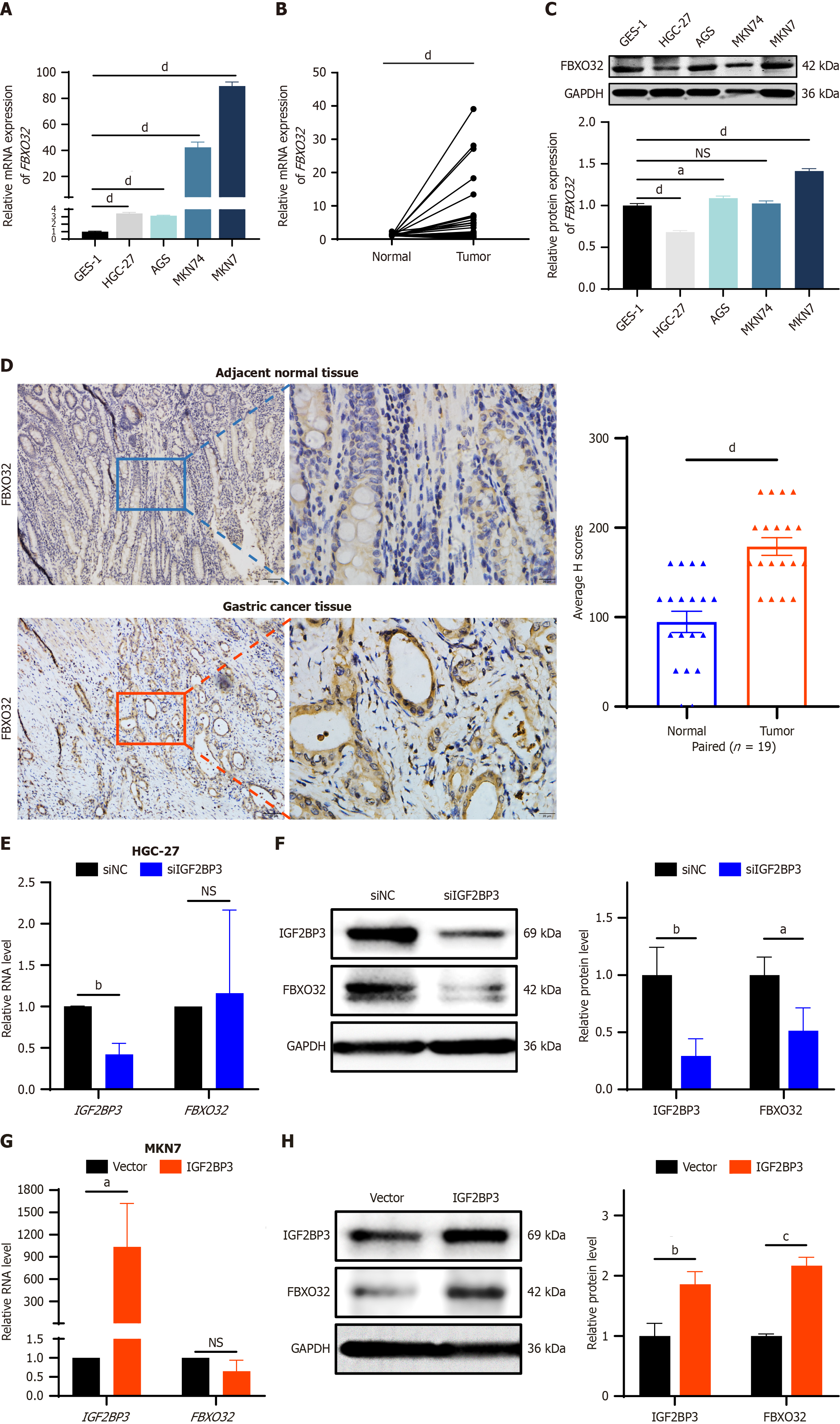
Next, we verified the expression of FBXO32 in GC cell lines and GC tissues. FBXO32 mRNA expression was higher in GC cell lines than in normal gastric mucosal cells (Figure 6A) and in tumor tissues compared to adjacent normal tissues (34 pairs of GC paired tissues) (Figure 6B). Moreover, the expression level of FBXO32 protein was also elevated in the majority of GC cell lines (Figure 6C) and was increased in tumor tissues relative to their matched adjacent normal tissues (19 pairs of GC paired tissues) (Figure 6D). These results suggest that FBXO32 may act as an oncogene in the pathogenesis of GC. To further confirm the regulation of FBXO32 expression by IGF2BP3 in GC cells, we examined transcriptional and translational changes in the expression of FBXO32 after IGF2BP3 knockdown or overexpression. After downregulation of IGF2BP3 expression in HGC-27 cells, there was no significant change in expression levels of FBXO32 mRNA (Figure 6E); however, FBXO32 protein expression was downregulated (Figure 6F). Similarly, when IGF2BP3 expression was upregulated in MKN7 cells, FBXO32 showed no significant change in terms of mRNA expression levels (Figure 6G), but its protein expression levels increased significantly (Figure 6H). These findings indicate that IGF2BP3 regulates the expression of FBXO32 at the translational rather than the transcriptional level, consistent with our initial hypothesis and with the mechanistic roles of other m6A reader proteins[20]. In conclusion, these results indicate that IGF2BP3 and FBXO32 interact with each other in GC, and IGF2BP3 can regulate the protein expression of FBXO32.
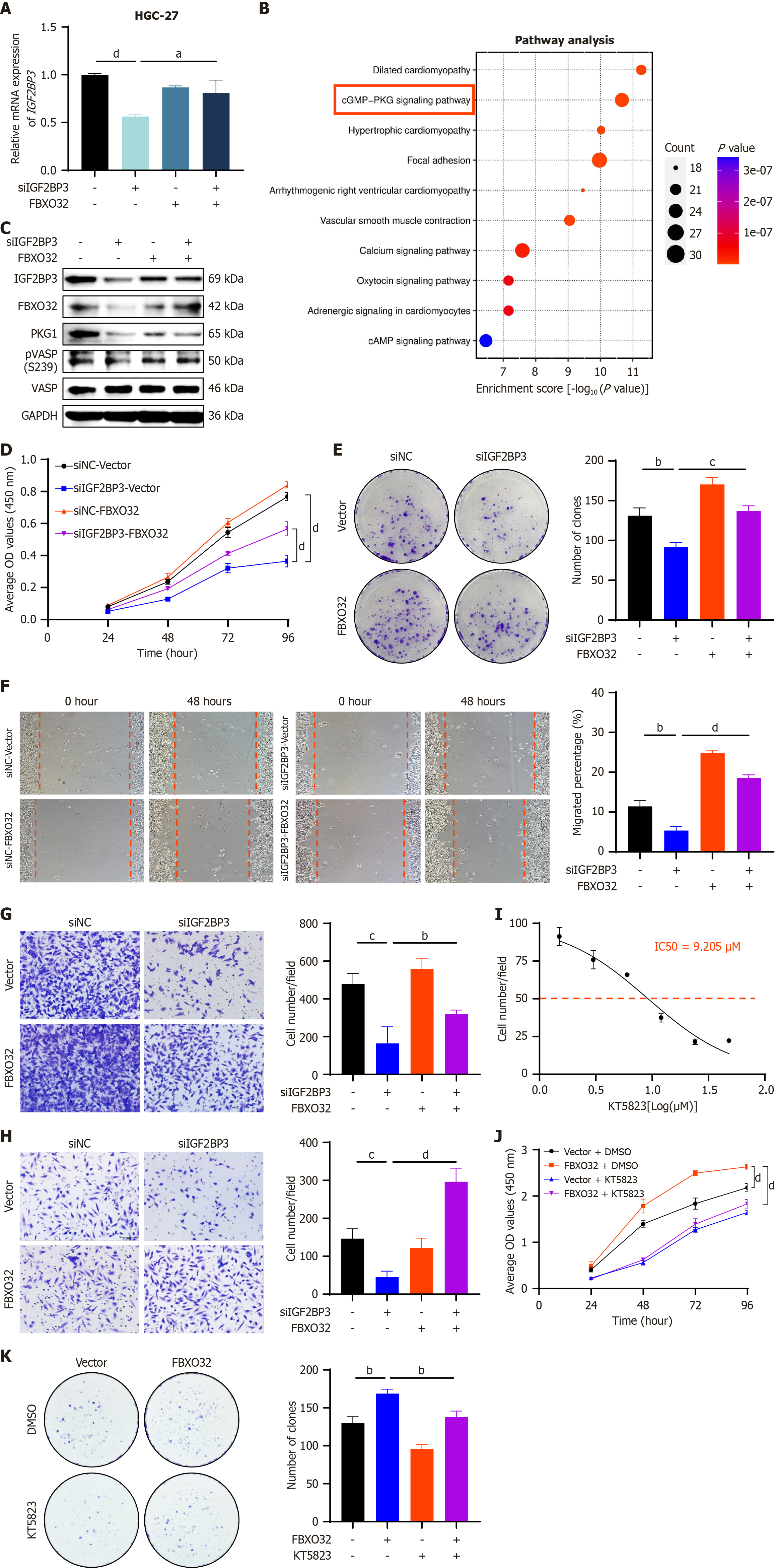
Four siRNA-FBXO32 sequences were transfected simultaneously into MKN7 cells, and the siRNA sequence with the most significant knockdown efficiency (si-FBXO32-910) was selected for subsequent experiments (Supplementary Figure 6A). Next, we transfected siNC or siRNA-IGF2BP3, and pcDNA3.1 or pcDNA3.1-FBXO32 into HGC-27 cells, and pcDNA3.1 or pcDNA3.1-IGF2BP3 and siRNA-NC or siRNA-FBXO32 into MKN7 cells, respectively. Changes of IGF2BP3 expression in each group were detected by qRT-PCR. The results suggested that the effects of IGF2BP3 knockdown in HGC-27 cells were attenuated by overexpression of FBXO32 (Figure 7A), and, conversely, the effects of IGF2BP3 overexpression in MKN7 cells were weakened by knockdown of FBXO32 (Supplementary Figure 6B). These findings indicate that FBXO32 may affect the expression of IGF2BP3 in GC cells. To further explore the downstream molecular mechanisms involving FBXO32, we selected the top 1000 genes co-expressed with FBXO32 in GC from TCGA and analyzed the signaling pathways that they controlled. Genes encoding proteins involved in the cGMP-PKG signaling pathway were significantly enriched (Figure 7B). Prediction using the STRING database indicated that both IGF2BP3 and FBXO32 had interactions with proteins in this pathway (Supplementary Figure 6C). We next investigated whether IGF2BP3 and FBXO32 could exert a synergistic effect on the activation of the cGMP-PKG signaling pathway. Knockdown of IGF2BP3 in HGC-27 cells led to marked downregulation of FBXO32 and downstream effectors of the cGMP-PKG signaling pathway (PKG1 and phospho-VASP). Moreover, overexpression of FBXO32 rescued the suppression of these effectors induced by IGF2BP3 depletion (Figure 7C). Conversely, IGF2BP3 overexpression in MKN-7 cells resulted in elevated levels of FBXO32 and cGMP-PKG pathway effectors (PKG1, phospho-VASP). This effect was attenuated by FBXO32 knockdown (Supplementary Figure 6D). These findings indicate that in GC, IGF2BP3 activates the cGMP-PKG signaling pathway through upregulation of FBXO32.
Next, we explored whether the effects of IGF2BP3 on GC cell function were influenced by FBXO32. The attenuation of cell proliferation, migration, and invasion induced by knockdown of IGF2BP3 in HGC-27 cells was rescued by upregulation of FBXO32 expression (Figure 7D-H), whereas the enhancement of cell proliferation, migration, and invasion caused by overexpression of IGF2BP3 in MKN7 cells was attenuated by downregulation of FBXO32 expression (Supplementary Figure 6E-I). These results demonstrate that IGF2BP3 relies on FBXO32 to promote the malignant phenotypes of GC.
Finally, we used KT5823, a selective inhibitor of PKG, to further investigate whether FBXO32 exerts its oncogenic effects through the cGMP-PKG signaling pathway. The half-maximal inhibitory concentration of KT5823 was determined to be 9.205 μM in HGC-27 cells via CCK-8 assay (Figure 7I). Subsequent CCK-8 and colony formation assays demonstrated that the pro-proliferative effect induced by FBXO32 overexpression in GC cells was attenuated by KT5823 treatment (Figure 7J and K). These results indicate that FBXO32 promotes tumor progression in a manner dependent on activation of the cGMP-PKG signaling pathway.
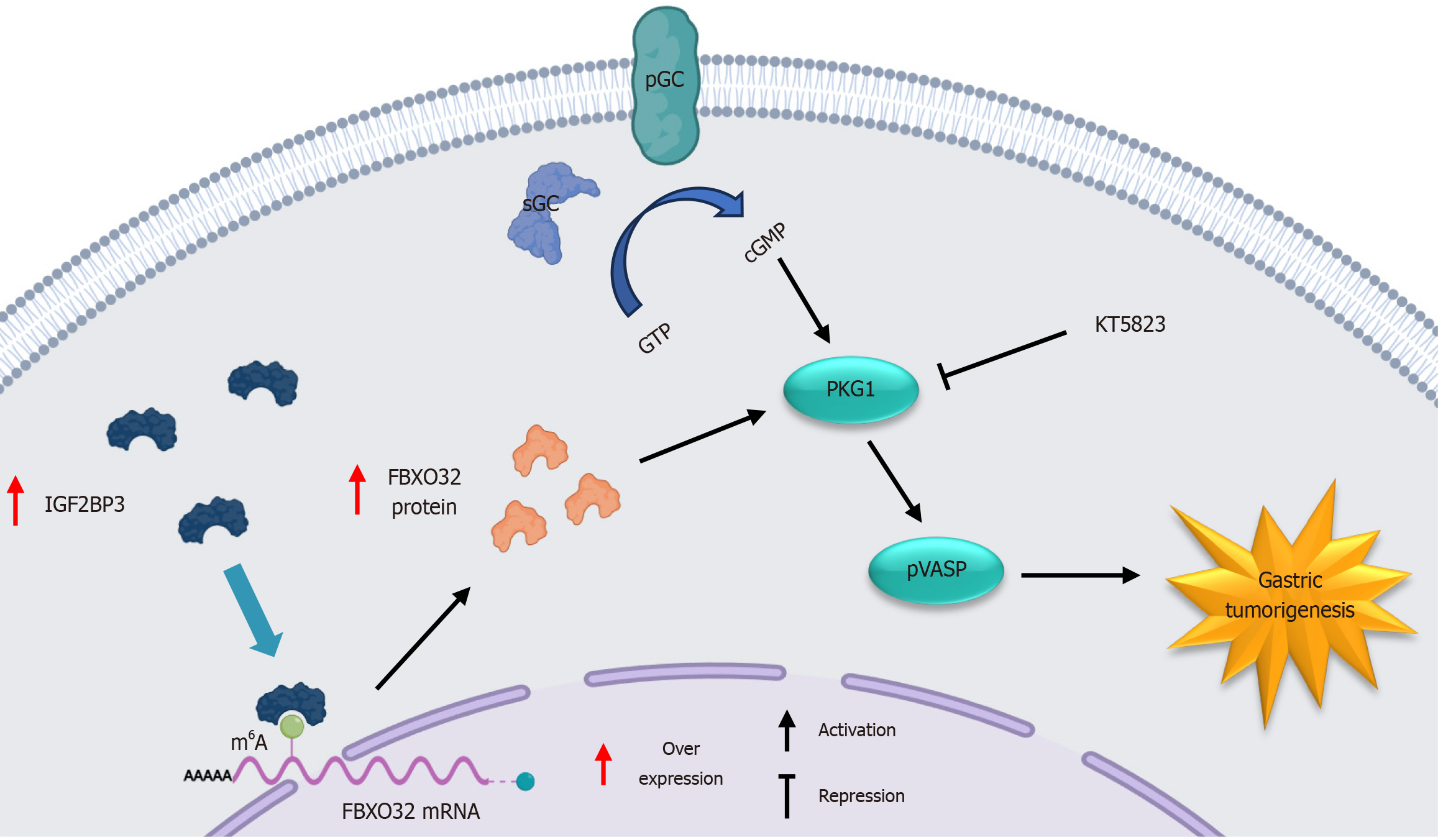
Taken together, these results indicate that IGF2BP3 promotes GC progression by m6A-dependently upregulating FBXO32 protein expression and subsequently activating the cGMP-PKG signaling pathway, and this oncogenic effect can be suppressed by the application of the selective PKG inhibitor KT5823 (Figure 8).
In this study, we report for the first time that IGF2BP3 binds to FBXO32 mRNA by recognizing the m6A site, resulting in increased intracellular levels of FBXO32 protein, which in turn lead to activation of the cGMP-PKG signaling pathway, ultimately promoting the progression of GC. Furthermore, we observed that selective PKG inhibitor KT5823 attenuated FBXO32-induced promotion of GC cell proliferation. Overall, our results demonstrate the clinical significance and potential therapeutic value of the IGF2BP3/FBXO32/cGMP-PKG axis in GC; this is of particular importance given the lack of effective biomarkers for GC to date.
Accumulating evidence indicates a significant oncogenic role for IGF2BP3 across various malignancies. For instance, it is notably upregulated in laryngeal squamous cell carcinoma relative to matched normal tissue[17] and is known to potentiate diverse malignant phenotypes in gallbladder cancer[21]. In colorectal cancer models, IGF2BP3 ablation has been demonstrated to induce cell cycle arrest, inhibit proliferation, and suppress tumor growth in vivo[18]. Consistent with these findings, we found that IGF2BP3 mRNA and protein expression levels in GC tissues were higher than those in adjacent normal tissues, and that this increased expression of IGF2BP3 was closely related to diffuse-type GC, lymph node metastasis, advanced TNM stage, and deeper invasion depth. Knockdown of IGF2BP3 in GC cells significantly inhibited cell proliferation, migration, and invasion and increased apoptosis, whereas upregulation of IGF2BP3 expression in GC cells had the opposite effect. Furthermore, IGF2BP3 deficiency inhibited the growth of GC in vivo. These findings suggest that IGF2BP3 is an important oncoprotein in GC. In addition, Wang et al[19] previously showed that IGF2BP2 enhanced the stability of mRNA transcribed from GLUT1, a key gene of aerobic glycolysis, in an m6A-dependent manner, significantly promoting aerobic glycolysis in colorectal cancer cells. We also found that upregulation and downregulation of IGF2BP3 expression in GC cells caused the glucose, lactic acid, and ATP contents of the cells to increase and decrease, respectively. Thus, the effects of IGF2BP3 on the biological function of GC cells may be closely related to the regulation of glucose metabolism in cells; this warrants further exploration in future work.
Through omics analyses and subsequent experimental validation, we identified FBXO32 with m6A modification as the target of IGF2BP3 in GC. Originally identified as a muscle-specific gene involved in muscle atrophy, FBXO32 encodes an E3 ubiquitin ligase and is located on chromosome 8q24.13[22,23]. FBXO32 has been implicated in diverse biological processes, including epithelial-mesenchymal transition and tumorigenesis[24]. Notably, FBXO32 has been demonstrated to exert oncogenic functions across multiple malignancies. For instance, its overexpression promotes proliferation and migration in melanoma cells, whereas its knockdown produces the opposite effects[25]. In pancreatic ductal adenocarcinoma, FBXO32 is aberrantly upregulated, enhancing cell migration and invasion in vitro and accelerating tumor growth and metastasis in vivo[26]. Furthermore, in lung adenocarcinoma, FBXO32 facilitates tumor progression by degrading PTEN, thereby augmenting the activity of the phosphatidylinositol 3-kinase/protein kinase B/mammalian target of rapamycin signaling pathway[27]. Similarly, we found that FBXO32 mRNA and protein levels were higher in GC tissues compared with adjacent normal tissues, and in GC cells compared with normal gastric mucosa cells. These findings suggest that FBXO32 may be an important oncogene in GC. However, Zhou et al[28] demonstrated that FBXO32 suppressed breast cancer cell colony formation in vitro and inhibited the initiation and growth of primary breast tumors in vivo through ubiquitination-mediated degradation of KLF4. Notably, the absence of the F-box domain abolished the ubiquitin-dependent degradation of KLF4 by FBXO32. These observations suggest a need to explore whether specific mutants or isoforms of FBXO32 could produce functionally opposing effects, a hypothesis that merits rigorous validation through extensive future studies.
Through bioinformatic predictions combined with RIP-qPCR and dual-luciferase reporter assays, we showed that IGF2BP3 protein specifically targets the m6A site located at position 1427 within FBXO32 mRNA. Furthermore, modulation of IGF2BP3 expression levels, followed by validation via qRT-PCR and western blot analyses, revealed that IGF2BP3 regulates FBXO32 at the translational rather than the transcriptional level. Previous studies have shown that IGF2BP3 enhances the translation efficiency of COPS7B mRNA, thereby promoting the growth and metastasis of colorectal cancer[29]. In addition, IGF2BP3 protein facilitates epidermal growth factor receptor mRNA translation through an m6A-dependent mechanism, contributing to colorectal cancer progression and conferring cetuximab resistance[30]. Based on these findings, we propose that IGF2BP3 protein binds to FBXO32 mRNA in an m6A-dependent manner and enhances its translation, ultimately driving the progression of GC. It is plausible that IGF2BP3 may also facilitate GC progression by modulating oncogenes in addition to FBXO32. Huang et al[31] demonstrated that insulin-like growth factor II mRNA-binding proteins enhanced the stability and promoted the translation of multiple transcripts, including MYC and cell-cycle-related genes, in an m6A-dependent manner, thereby exerting oncogenic functions across various malignancies. Therefore, our findings provide further insight into the oncogenic role of IGF2BP3 in gastric carcinogenesis.
To further elucidate the mechanism by which IGF2BP3 targets FBXO32 to promote GC progression, we analyzed the signaling pathways controlled by the top 1000 genes co-expressed with FBXO32 in gastric adenocarcinoma using TCGA data. These genes were significantly enriched in the cGMP-PKG signaling pathway, which was originally reported to participate in inhibition of platelet activation and regulates phosphorylation of VASP as an integral part of an efficient and sensitive signaling cascade[32]. Recent studies have shown that the cGMP-PKG signaling pathway is also involved in the development of various cancers. For example, cGMP can activate PKG and its downstream mitogen-activated protein kinase pathways, resulting in increased tumor cell stemness and metastasis[33]; DARS-AS1 accelerates the progression of cervical cancer by activating the cGMP-PKG pathway[34]; and nitric oxide-cGMP-PKG can increase the proliferation of colon cancer cells while inhibiting apoptosis via activation of extracellular regulated protein kinase-1/2 and activator protein-1[35]. Here, we demonstrated by western blot analysis that the inhibition or promotion of cGMP-PKG signaling activation resulting from IGF2BP3 knockdown or overexpression could be rescued or attenuated by FBXO32 upregulation or downregulation. Notably, the results of this analysis indicated that FBXO32 not only activates the cGMP-PKG pathway but also inversely regulates IGF2BP3 expression. This suggests a potential bidirectional regulatory or feedback mechanism between IGF2BP3 and FBXO32, a mechanism that warrants further elucidation in future studies. Rescue assays confirmed that the IGF2BP3/FBXO32/cGMP-PKG axis influences the proliferative, migratory, and invasive capacities of GC cells. Moreover, treatment of GC cells with selective PKG inhibitor KT5823 significantly suppressed their proliferation. These results indicate that the IGF2BP3/FBXO32/cGMP-PKG axis contributes to gastric carcinogenesis and progression and may thus represent a promising novel therapeutic target for clinical intervention in GC.
Our study had some limitations. For example, the human tissue samples used for clinical validation were all from the same hospital; future studies should therefore validate our results using multi-center clinical data. In addition, given our preliminary findings that changes in IGF2BP3 expression levels affect cellular glucose metabolism in GC cells, further exploration is warranted to more fully elucidate the mechanism underlying glucose metabolism in GC.
In conclusion, our study demonstrates that IGF2BP3 functions as an oncoprotein in GC, promoting the translation of FBXO32 in an m6A-dependent manner and activating the downstream cGMP-PKG signaling pathway. This, in turn, modulates multiple biological functions of GC cells and ultimately promotes the progression of GC. Consequently, targeting the IGF2BP3/FBXO32/cGMP-PKG axis may offer a novel approach to clinical treatment of GC.
The authors wish to acknowledge all those who provided technical support and constructive feedback throughout this study.
| 1. | López MJ, Carbajal J, Alfaro AL, Saravia LG, Zanabria D, Araujo JM, Quispe L, Zevallos A, Buleje JL, Cho CE, Sarmiento M, Pinto JA, Fajardo W. Characteristics of gastric cancer around the world. Crit Rev Oncol Hematol. 2023;181:103841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 233] [Reference Citation Analysis (3)] |
| 2. | Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5690] [Cited by in RCA: 12562] [Article Influence: 6281.0] [Reference Citation Analysis (6)] |
| 3. | Han B, Zheng R, Zeng H, Wang S, Sun K, Chen R, Li L, Wei W, He J. Cancer incidence and mortality in China, 2022. J Natl Cancer Cent. 2024;4:47-53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 1211] [Article Influence: 605.5] [Reference Citation Analysis (0)] |
| 4. | Ajani JA, D'Amico TA, Bentrem DJ, Chao J, Cooke D, Corvera C, Das P, Enzinger PC, Enzler T, Fanta P, Farjah F, Gerdes H, Gibson MK, Hochwald S, Hofstetter WL, Ilson DH, Keswani RN, Kim S, Kleinberg LR, Klempner SJ, Lacy J, Ly QP, Matkowskyj KA, McNamara M, Mulcahy MF, Outlaw D, Park H, Perry KA, Pimiento J, Poultsides GA, Reznik S, Roses RE, Strong VE, Su S, Wang HL, Wiesner G, Willett CG, Yakoub D, Yoon H, McMillian N, Pluchino LA. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2022;20:167-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 1163] [Article Influence: 290.8] [Reference Citation Analysis (0)] |
| 5. | Machlowska J, Baj J, Sitarz M, Maciejewski R, Sitarz R. Gastric Cancer: Epidemiology, Risk Factors, Classification, Genomic Characteristics and Treatment Strategies. Int J Mol Sci. 2020;21:4012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 897] [Cited by in RCA: 969] [Article Influence: 161.5] [Reference Citation Analysis (0)] |
| 6. | Grady WM, Yu M, Markowitz SD. Epigenetic Alterations in the Gastrointestinal Tract: Current and Emerging Use for Biomarkers of Cancer. Gastroenterology. 2021;160:690-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 164] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 7. | Tang SY, Zhou PJ, Meng Y, Zeng FR, Deng GT. Gastric cancer: An epigenetic view. World J Gastrointest Oncol. 2022;14:90-109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (2)] |
| 8. | Han X, Wang L, Han Q. Advances in the role of m(6)A RNA modification in cancer metabolic reprogramming. Cell Biosci. 2020;10:117. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 9. | Xie Q, Wu TP, Gimple RC, Li Z, Prager BC, Wu Q, Yu Y, Wang P, Wang Y, Gorkin DU, Zhang C, Dowiak AV, Lin K, Zeng C, Sui Y, Kim LJY, Miller TE, Jiang L, Lee-Poturalski C, Huang Z, Fang X, Zhai K, Mack SC, Sander M, Bao S, Kerstetter-Fogle AE, Sloan AE, Xiao AZ, Rich JN. N(6)-methyladenine DNA Modification in Glioblastoma. Cell. 2018;175:1228-1243.e20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 247] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 10. | An Y, Duan H. The role of m6A RNA methylation in cancer metabolism. Mol Cancer. 2022;21:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 642] [Article Influence: 160.5] [Reference Citation Analysis (0)] |
| 11. | Lee M, Kim B, Kim VN. Emerging roles of RNA modification: m(6)A and U-tail. Cell. 2014;158:980-987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 271] [Article Influence: 24.6] [Reference Citation Analysis (8)] |
| 12. | Jacobs BL, Lee CT, Montie JE. Bladder cancer in 2010: how far have we come? CA Cancer J Clin. 2010;60:244-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 249] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 13. | Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, Hüttelmaier S. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70:2657-2675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 366] [Cited by in RCA: 615] [Article Influence: 43.9] [Reference Citation Analysis (0)] |
| 14. | Müeller-Pillasch F, Lacher U, Wallrapp C, Micha A, Zimmerhackl F, Hameister H, Varga G, Friess H, Büchler M, Beger HG, Vila MR, Adler G, Gress TM. Cloning of a gene highly overexpressed in cancer coding for a novel KH-domain containing protein. Oncogene. 1997;14:2729-2733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 220] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 15. | Liu X, Chen J, Chen W, Xu Y, Shen Y, Xu X. Targeting IGF2BP3 in Cancer. Int J Mol Sci. 2023;24:9423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 46] [Reference Citation Analysis (0)] |
| 16. | Wang X, Tian L, Li Y, Wang J, Yan B, Yang L, Li Q, Zhao R, Liu M, Wang P, Sun Y. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J Exp Clin Cancer Res. 2021;40:80. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 192] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 17. | Xia A, Yuan W, Wang Q, Xu J, Gu Y, Zhang L, Chen C, Wang Z, Wu D, He Q, Yu W, Wang F, Xue C, Zhang Y, Bao G, Tao X, Liu S, Wang S, Hu Z, Sun B. The cancer-testis lncRNA lnc-CTHCC promotes hepatocellular carcinogenesis by binding hnRNP K and activating YAP1 transcription. Nat Cancer. 2022;3:203-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 18. | Yang Z, Wang T, Wu D, Min Z, Tan J, Yu B. RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and angiogenesis in colon cancer. J Exp Clin Cancer Res. 2020;39:203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 215] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 19. | Wang J, Zhu M, Zhu J, Li J, Zhu X, Wang K, Shen K, Yang K, Ni X, Liu X, Zhang G, Xi Q, Shi T, Chen W. HES1 promotes aerobic glycolysis and cancer progression of colorectal cancer via IGF2BP2-mediated GLUT1 m6A modification. Cell Death Discov. 2023;9:411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 20. | Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, Cheng C, Li L, Pi J, Si Y, Xiao H, Li L, Rao S, Wang F, Yu J, Yu J, Zou D, Yi P. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48:3816-3831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 548] [Article Influence: 91.3] [Reference Citation Analysis (0)] |
| 21. | Qin J, Cui Z, Zhou J, Zhang B, Lu R, Ding Y, Hu H, Cai J. IGF2BP3 drives gallbladder cancer progression by m6A-modified CLDN4 and inducing macrophage immunosuppressive polarization. Transl Oncol. 2023;37:101764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 22. | Sahu SK, Tiwari N, Pataskar A, Zhuang Y, Borisova M, Diken M, Strand S, Beli P, Tiwari VK. FBXO32 promotes microenvironment underlying epithelial-mesenchymal transition via CtBP1 during tumour metastasis and brain development. Nat Commun. 2017;8:1523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 23. | Mei Z, Zhang D, Hu B, Wang J, Shen X, Xiao W. FBXO32 Targets c-Myc for Proteasomal Degradation and Inhibits c-Myc Activity. J Biol Chem. 2015;290:16202-16214. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 24. | Wang H, Liu H, Zhao L, Luo S, Akinyemiju T, Hwang S, Yue Y, Wei Q. Association of genetic variants of FBXO32 and FOXO6 in the FOXO pathway with breast cancer risk. Mol Carcinog. 2021;60:661-670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 25. | Li F, Yu H, Zhang Y, Ma Y, Chen X, Zhang J, Sun L, Guo R, Wu Y, Zheng P, Wang X, Bie P, He F, Zhang L, Xie C, Xiong H. F-box protein FBXO32 ubiquitinates and stabilizes D-type cyclins to drive cancer progression. Nat Commun. 2025;16:4060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 26. | Su D, Wang R, Chen G, Ding C, Liu Y, Tao J, Wang Y, Qiu J, Luo W, Weng G, Yang G, Zhang T. FBXO32 Stimulates Protein Synthesis to Drive Pancreatic Cancer Progression and Metastasis. Cancer Res. 2024;84:2607-2625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 27. | Wu J, Wen T, Marzio A, Song D, Chen S, Yang C, Zhao F, Zhang B, Zhao G, Ferri A, Cheng H, Ma J, Ren H, Chen QY, Yang Y, Qin S. FBXO32-mediated degradation of PTEN promotes lung adenocarcinoma progression. Cell Death Dis. 2024;15:282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 14] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 28. | Zhou H, Liu Y, Zhu R, Ding F, Wan Y, Li Y, Liu Z. FBXO32 suppresses breast cancer tumorigenesis through targeting KLF4 to proteasomal degradation. Oncogene. 2017;36:3312-3321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 68] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 29. | Tang J, Wang S, Weng M, Guo Q, Ren L, He Y, Cui Z, Cong M, Qin M, Yu J, Su R, Li X. The IGF2BP3-COPS7B Axis Facilitates mRNA Translation to Drive Colorectal Cancer Progression. Cancer Res. 2023;83:3593-3610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 30. | Chen LJ, Liu HY, Xiao ZY, Qiu T, Zhang D, Zhang LJ, Han FY, Chen GJ, Xu XM, Zhu JH, Ding YQ, Wang SY, Ye YP, Jiao HL. IGF2BP3 promotes the progression of colorectal cancer and mediates cetuximab resistance by stabilizing EGFR mRNA in an m(6)A-dependent manner. Cell Death Dis. 2023;14:581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 36] [Reference Citation Analysis (0)] |
| 31. | Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, Hu YC, Hüttelmaier S, Skibbe JR, Su R, Deng X, Dong L, Sun M, Li C, Nachtergaele S, Wang Y, Hu C, Ferchen K, Greis KD, Jiang X, Wei M, Qu L, Guan JL, He C, Yang J, Chen J. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 928] [Cited by in RCA: 2242] [Article Influence: 280.3] [Reference Citation Analysis (0)] |
| 32. | Eigenthaler M, Nolte C, Halbrügge M, Walter U. Concentration and regulation of cyclic nucleotides, cyclic-nucleotide-dependent protein kinases and one of their major substrates in human platelets. Estimating the rate of cAMP-regulated and cGMP-regulated protein phosphorylation in intact cells. Eur J Biochem. 1992;205:471-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 124] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 33. | Lv Y, Wang X, Li X, Xu G, Bai Y, Wu J, Piao Y, Shi Y, Xiang R, Wang L. Nucleotide de novo synthesis increases breast cancer stemness and metastasis via cGMP-PKG-MAPK signaling pathway. PLoS Biol. 2020;18:e3000872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 91] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 34. | Kong X, Wang JS, Yang H. Upregulation of lncRNA DARS-AS1 accelerates tumor malignancy in cervical cancer by activating cGMP-PKG pathway. J Biochem Mol Toxicol. 2021;35:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 35. | Babykutty S, Suboj P, Srinivas P, Nair AS, Chandramohan K, Gopala S. Insidious role of nitric oxide in migration/invasion of colon cancer cells by upregulating MMP-2/9 via activation of cGMP-PKG-ERK signaling pathways. Clin Exp Metastasis. 2012;29:471-492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 78] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/