Published online Nov 7, 2025. doi: 10.3748/wjg.v31.i41.110367
Revised: September 2, 2025
Accepted: September 28, 2025
Published online: November 7, 2025
Processing time: 151 Days and 18.6 Hours
High expression of pescadillo ribosomal biogenesis factor 1 (PES1) has been re
Core Tip: In gastric cancer and head and neck squamous cell carcinoma, pescadillo ribosomal biogenesis factor 1 (PES1) and programmed death-ligand 1 (PD-L1) show significant co-expression. Across multiple tumor types, elevated PES1 is linked to poor prognosis and aggressive tumor behavior. Our analysis further reveals that PES1 expression negatively correlates with CD8+ T cell infiltration and positively correlates with PD-L1 expression. These findings suggest opportunities for combination cancer immunotherapy, although the molecular basis of PES1-PD-L1 interactions and the therapeutic benefit of dual targeting require further experimental validation.
- Citation: Yu J, Yu B, Ge FL, Ren ZG. Pescadillo ribosomal biogenesis factor 1 as a therapeutic target in tumor immunotherapy. World J Gastroenterol 2025; 31(41): 110367
- URL: https://www.wjgnet.com/1007-9327/full/v31/i41/110367.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i41.110367
Pescadillo ribosomal biogenesis factor 1 (PES1) has emerged as a potential prognostic biomarker and therapeutic target across multiple malignancies, yet its role in tumor immune evasion remains unclear. Hu et al[1] recently reported significant co-expression of PES1 and programmed death-ligand 1 (PD-L1) in gastric cancer (GC) and head and neck squamous cell carcinoma (HNSCC), with correlations to adverse clinical features. While their study was limited by methodological constraints, its findings nevertheless provide important insights into immune evasion mechanisms. To further explore this relationship, we analyzed GC data from The Cancer Genome Atlas (TCGA, namely the gastric adenocarcinoma dataset) and observed a negative correlation between PES1 expression and CD8+ T cell infiltration, alongside a positive correlation with PD-L1 expression. These findings suggest functional interactions between PES1 and PD-L1 within the tumor microenvironment. We hypothesize that PES1 may regulate PD-L1 expression through phosphatidylinositol 3-kinase/protein kinase B signaling or cellular Myc-mediated mechanisms. Although these path
Hu et al[1] reported significant positive expression rates of PES1 (51.72%) and PD-L1 (58.62%) in GC and HNSCC. PES1 expression correlated with tumor-node-metastasis stage, lymph node metastasis, and depth of infiltration (P < 0.05), while PD-L1 expression correlated with tumor differentiation, lymph node metastasis, and infiltration depth (P < 0.05). These findings suggest that PES1 and PD-L1 may contribute to tumor progression and immune evasion. However, the study was limited by substantial methodological and analytical shortcomings.
The investigation enrolled only 58 patients, an insufficient sample size for a study spanning two distinct tumor entities. Case numbers for GC vs HNSCC were not reported, and the rationale for combining these biologically and clinically disparate tumor types remains unjustified. More critically, the absence of normal tissue controls or benign comparators prevents confirmation that PES1 and PD-L1 expression is truly tumor-specific. Additionally, because PES1 is a core ribosomal biogenesis factor broadly expressed in proliferating cells, the lack of analysis of other proliferation markers or related ribosomal genes represents a key oversight that limits interpretation of its biological significance.
The report of this study did not define the explicit criteria used for PES1 and PD-L1 positivity and omitted the scoring systems used, as well as the thresholds for staining intensity and the percentage of positive cells. Critical quality control measures were also lacking, such as the inclusion of positive and negative controls, blinded evaluation protocols, and inter-observer concordance assessments to reduce subjective bias. Although the authors stated that they used “highly specific and sensitive” monoclonal antibodies, key technical details, including antibody sources, clone designations, and working concentrations, were not reported. These omissions represent major methodological shortcomings that under
The study’s statistical approach contained several major flaws. Analyses were restricted to univariate methods and did not adjust for potential confounders such as tumor stage, grade, immune infiltration profiles, mutational burden, estab
PES1 is a highly conserved nucleolar protein essential for ribosome biogenesis, particularly in precursor rRNA processing and 60S ribosomal subunit assembly. As a core component of the PeBoW complex (composed of PES1, block of proli
Overexpression of PES1 has been documented in multiple malignancies, including hepatocellular carcinoma[4], thyroid cancer[5], colorectal cancer[6], breast cancer[7], esophageal squamous cell carcinoma[8], and pancreatic cancer[9]. In these cancers, elevated PES1 Levels are strongly associated with poor prognosis and aggressive tumor behavior. Mechanistically, PES1 promotes tumorigenesis through modulation of oncogenic signaling pathways: In hepatocellular carcinoma, it activates phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) signaling[4]; in breast cancer, it regulates the estrogen receptor alpha/estrogen receptor beta balance[7]; and in pancreatic cancer, it drives proliferation via cellular-Myc upregulation[9]. Collectively, these diverse functions underscore PES1 as a critical oncogenic driver and a promising therapeutic target.
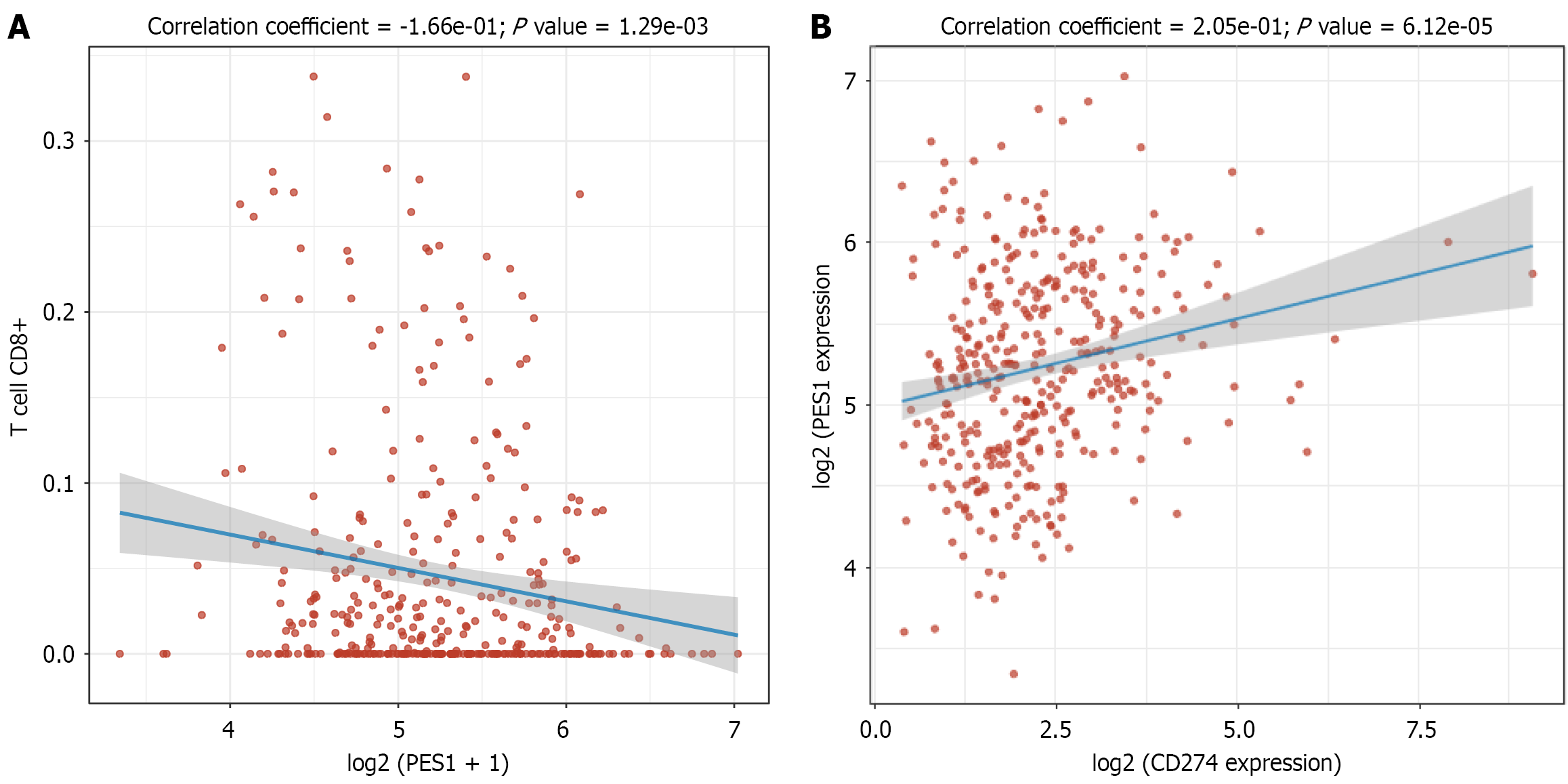
Research has shown that PES1 disrupts interleukin-enhancer binding factor 3-interleukin 15 complex formation in esophageal squamous cell carcinoma, thereby suppressing CD8+ T cell infiltration[8]. Although Hu et al[1] suggested a link between PES1 and tumor immunity in GC and HNSCC, the supporting evidence remains limited. To further evaluate this relationship in GC, we analyzed data from TCGA and observed a significant negative correlation between PES1 expression and CD8+ T cell infiltration (P = 1.29 × 10-3) (Figure 1A). These findings indicate that high PES1 expression is associated with reduced cytotoxic T lymphocyte presence in the tumor microenvironment, potentially facilitating immune evasion[10]. Consistent with the observations by Hu et al[1], however, we also detected a significant positive correlation between PES1 and PD-L1 expression in the TCGA cohort (P = 6.12 × 10-5) (Figure 1B). This suggests that PES1 and PD-L1 may act synergistically to promote immune evasion.
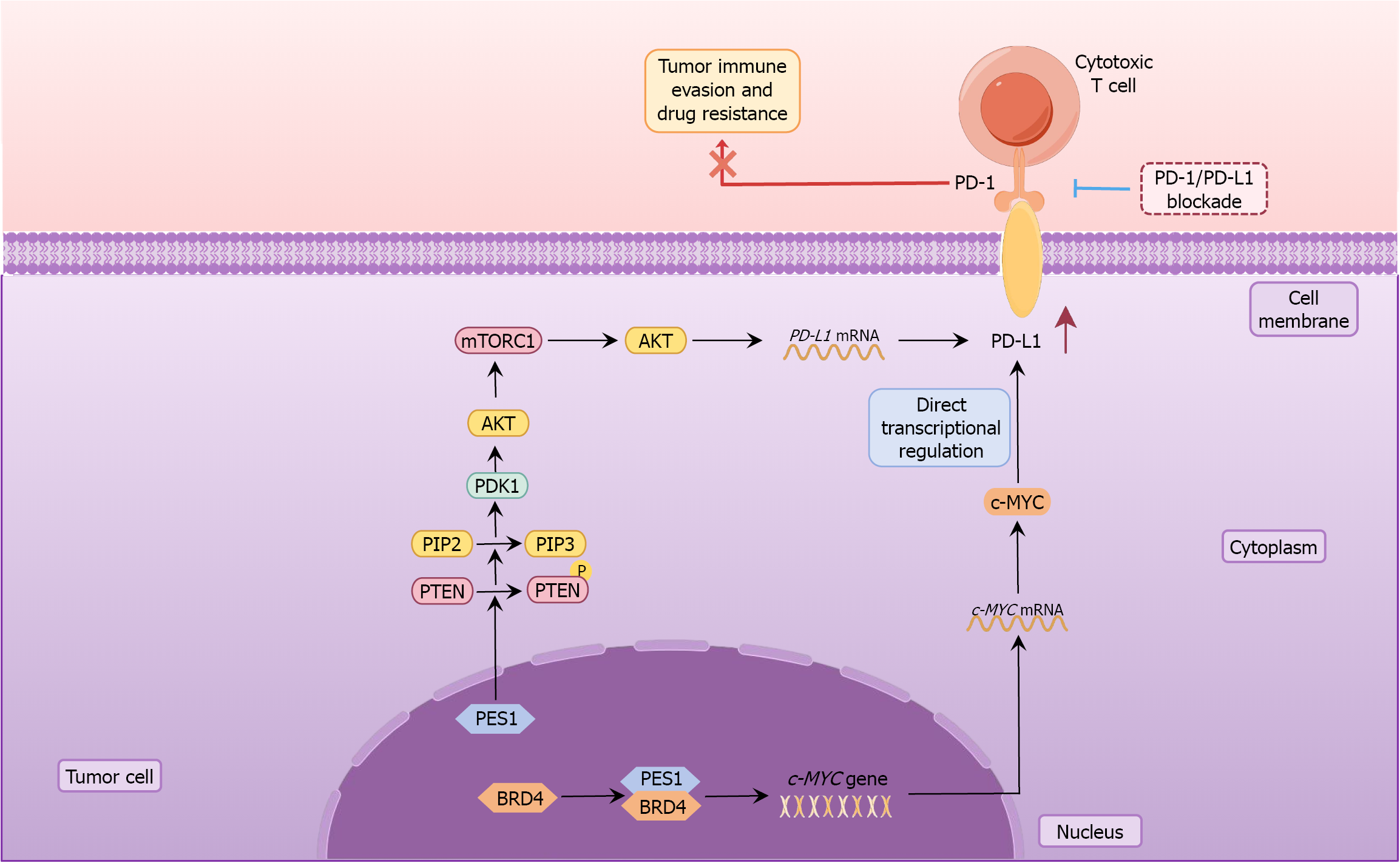
To investigate potential mechanisms linking PES1 and PD-L1, we conducted a comprehensive literature review. Prior studies have shown that PES1 promotes phosphatase and tensin homolog phosphorylation, leading to conversion of phosphatidylinositol 4,5-bisphosphate to phosphatidylinositol (3,4,5)-trisphosphate, subsequent 3-phosphoinositide dependent protein kinase-1 activation, and AKT phosphorylation[4]. Activation of the PI3K/AKT pathway is well established as a driver of PD-L1 upregulation across diverse tumor types. Specifically, PI3K/AKT signaling activates mammalian target of rapamycin[11], which enhances PD-L1 mRNA translational efficiency[12]. The mechanistic target of rapamycin complex 1 phosphorylates S6 kinase 1 and inactivates 4E-binding protein 1, thereby promoting PD-L1 trans
Based on these findings, we propose that PES1 may regulate PD-L1 expression via two complementary mechanisms: Modulation of the PI3K/AKT signaling pathway, and the PES1-BRD4-cellular-Myc axis (Figure 2). It is important to emphasize that these hypotheses are currently theoretical, derived from the published literature, and require rigorous functional and mechanistic validation. Moreover, the regulatory relationship between PES1 and PD-L1 may vary across tumor types, underscoring the need to investigate this mechanism in multiple cancer models to assess its generalizability.
Elevated PES1 expression is associated with poor prognosis and aggressive tumor behavior across multiple malignancies. However, the study by Hu et al[1] examining PES1 and PD-L1 co-expression in GC and HNSCC had substantial methodological limitations. Our analysis of TCGA data revealed a negative correlation between PES1 expression and CD8+ T cell infiltration, alongside a positive correlation with PD-L1 expression. Based on prior research, we further hypothesize that PES1 may regulate PD-L1 via the PI3K/AKT signaling pathway or the cellular-Myc-mediated axis. These observations provide novel insights into tumor immune evasion and establish a theoretical foundation for PES1-targeted therapeutic strategies. Future studies should aim to validate these regulatory mechanisms and evaluate the clinical potential of PES1 as a target in cancer immunotherapy.
| 1. | Hu XN, Li CF, Huang SM, Nie CL, Pang R. Pescadillo ribosomal biogenesis factor 1 and programmed death-ligand 1 in gastric and head and neck squamous cell carcinoma. World J Gastroenterol. 2025;31:106644. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (1)] |
| 2. | Rohrmoser M, Hölzel M, Grimm T, Malamoussi A, Harasim T, Orban M, Pfisterer I, Gruber-Eber A, Kremmer E, Eick D. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol. 2007;27:3682-3694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 113] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 3. | Cheng L, Yuan B, Ying S, Niu C, Mai H, Guan X, Yang X, Teng Y, Lin J, Huang J, Jin R, Wu J, Liu B, Chang S, Wang E, Zhang C, Hou N, Cheng X, Xu D, Yang X, Gao S, Ye Q. PES1 is a critical component of telomerase assembly and regulates cellular senescence. Sci Adv. 2019;5:eaav1090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 4. | Wang J, Sun J, Zhang N, Yang R, Li H, Zhang Y, Chen K, Kong D. PES1 enhances proliferation and tumorigenesis in hepatocellular carcinoma via the PI3K/AKT pathway. Life Sci. 2019;219:182-189. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Qiu YB, Liao LY, Jiang R, Xu M, Xu LW, Chen GG, Liu ZM. PES1 promotes the occurrence and development of papillary thyroid cancer by upregulating the ERα/ERβ protein ratio. Sci Rep. 2019;9:1032. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 6. | Bian Z, Zhou M, Cui K, Yang F, Cao Y, Sun S, Liu B, Gong L, Li J, Wang X, Li C, Yao S, Yin Y, Huang S, Fei B, Huang Z. SNHG17 promotes colorectal tumorigenesis and metastasis via regulating Trim23-PES1 axis and miR-339-5p-FOSL2-SNHG17 positive feedback loop. J Exp Clin Cancer Res. 2021;40:360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 7. | Cheng L, Li J, Han Y, Lin J, Niu C, Zhou Z, Yuan B, Huang K, Li J, Jiang K, Zhang H, Ding L, Xu X, Ye Q. PES1 promotes breast cancer by differentially regulating ERα and ERβ. J Clin Invest. 2012;122:2857-2870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 8. | Ma N, Hua R, Yang Y, Liu ZC, Pan J, Yu BY, Sun YF, Xie D, Wang Y, Li ZG. PES1 reduces CD8(+) T cell infiltration and immunotherapy sensitivity via interrupting ILF3-IL15 complex in esophageal squamous cell carcinoma. J Biomed Sci. 2023;30:20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 9. | Jin X, Fang R, Fan P, Zeng L, Zhang B, Lu X, Liu T. PES1 promotes BET inhibitors resistance and cells proliferation through increasing c-Myc expression in pancreatic cancer. J Exp Clin Cancer Res. 2019;38:463. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 31] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 10. | He Y, Xiang J, Li Y, Huang W, Gu F, Wang Y, Chen R. PES1 is a biomarker of head and neck squamous cell carcinoma and is associated with the tumor microenvironment. Cancer Med. 2023;12:12622-12638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 11. | Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1178] [Cited by in RCA: 1235] [Article Influence: 51.5] [Reference Citation Analysis (0)] |
| 12. | Lastwika KJ, Wilson W 3rd, Li QK, Norris J, Xu H, Ghazarian SR, Kitagawa H, Kawabata S, Taube JM, Yao S, Liu LN, Gills JJ, Dennis PA. Control of PD-L1 Expression by Oncogenic Activation of the AKT-mTOR Pathway in Non-Small Cell Lung Cancer. Cancer Res. 2016;76:227-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 423] [Cited by in RCA: 633] [Article Influence: 57.5] [Reference Citation Analysis (0)] |
| 13. | Xu Y, Poggio M, Jin HY, Shi Z, Forester CM, Wang Y, Stumpf CR, Xue L, Devericks E, So L, Nguyen HG, Griselin A, Gordan JD, Umetsu SE, Reich SH, Worland ST, Asthana S, Barna M, Webster KR, Cunningham JT, Ruggero D. Translation control of the immune checkpoint in cancer and its therapeutic targeting. Nat Med. 2019;25:301-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 215] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 14. | Cerezo M, Guemiri R, Druillennec S, Girault I, Malka-Mahieu H, Shen S, Allard D, Martineau S, Welsch C, Agoussi S, Estrada C, Adam J, Libenciuc C, Routier E, Roy S, Désaubry L, Eggermont AM, Sonenberg N, Scoazec JY, Eychène A, Vagner S, Robert C. Translational control of tumor immune escape via the eIF4F-STAT1-PD-L1 axis in melanoma. Nat Med. 2018;24:1877-1886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 195] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 15. | Abdelhamed S, Ogura K, Yokoyama S, Saiki I, Hayakawa Y. AKT-STAT3 Pathway as a Downstream Target of EGFR Signaling to Regulate PD-L1 Expression on NSCLC cells. J Cancer. 2016;7:1579-1586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 103] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 16. | Chen S, Crabill GA, Pritchard TS, McMiller TL, Wei P, Pardoll DM, Pan F, Topalian SL. Mechanisms regulating PD-L1 expression on tumor and immune cells. J Immunother Cancer. 2019;7:305. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 281] [Cited by in RCA: 366] [Article Influence: 52.3] [Reference Citation Analysis (0)] |
| 17. | Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gütgemann I, Eilers M, Felsher DW. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 688] [Cited by in RCA: 1116] [Article Influence: 111.6] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/