Published online Oct 28, 2025. doi: 10.3748/wjg.v31.i40.111951
Revised: July 29, 2025
Accepted: September 16, 2025
Published online: October 28, 2025
Processing time: 105 Days and 14.7 Hours
Histone deacetylase 10 (HDAC10) is emerging as a critical modulator of tumor immunity, chemoresistance, and transcriptional plasticity in colorectal cancer. Its suppression has been linked to altered CD8+ T cell activity, increased p53 expres
Core Tip: Histone deacetylase 10 (HDAC10) plays a multifaceted role in colorectal cancer by modulating immune evasion, chemoresistance, and transcriptional regulation. Its suppression influences CD8⁺ T cell activation, enhances p53 expression, and reduces programmed death ligand 1 levels, suggesting critical involvement in tumor-immune interactions. Selective HDAC10 inhibition, unlike pan-HDAC blockade, may offer targeted therapeutic benefit, particularly in microsatellite-stable tumors. Further in vivo studies and mechanistic dissection of HDAC10-p53-programmed death ligand 1 signaling are essential to harness its full potential in reshaping the tumor microenvironment and improving immunotherapy outcomes.
- Citation: Afzal A, Muanprasat C, Khawar MB. Prognostic role of histone deacetylase 10 in colorectal cancer: Strengths and gaps. World J Gastroenterol 2025; 31(40): 111951
- URL: https://www.wjgnet.com/1007-9327/full/v31/i40/111951.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i40.111951
We read with great interest the recent study by Nie et al[1] published in World Journal of Gastroenterology. This study has provided valuable and appealing evidence on the role of histone deacetylase 10 (HDAC10) in shaping the tumor microenvironment (TME) and mediating immunotherapy resistance in colorectal cancer (CRC). The depth of the multi-modal approach opted in current study, and integrating transcriptomic analyses with functional assays are appreciable, and we wish to offer some of critical reflections and suggestions for further consideration.
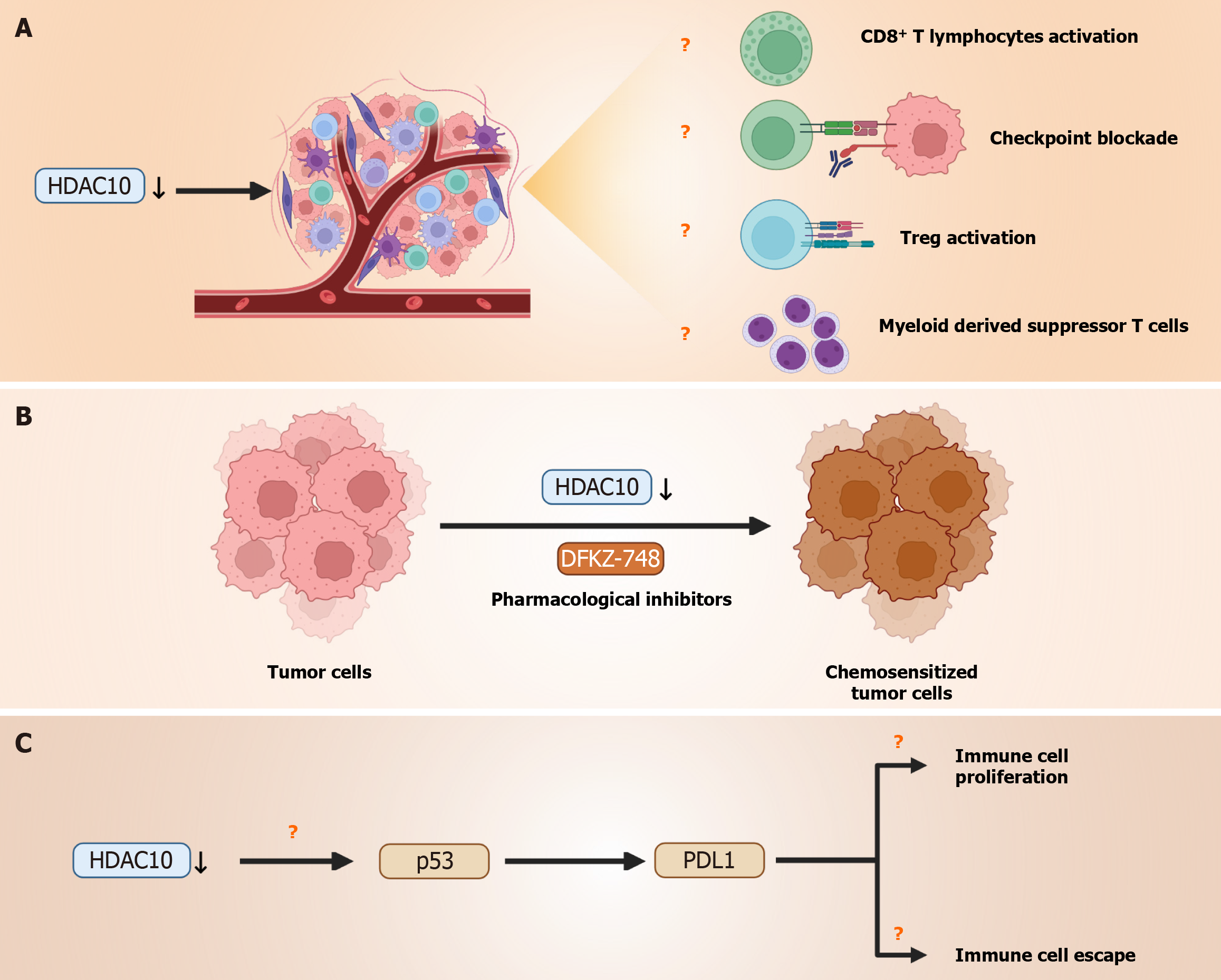
Firstly, while the authors convincingly correlate higher expression of HDAC10 with poor prognosis, immunosuppressive TME, and chemoresistance, we are of the view that further functional in vivo validation will strengthen the findings of this study. We commend that the authors have acknowledged that the lack of animal models limits a comprehensive understanding of immunological dynamics in the TME. Prior studies have shown that CD8+ T cell infiltration is a critical determinant of response to immunotherapy in CRC, and HDAC modulation has been reported to influence cytotoxic T cell function via major histocompatibility class I upregulation and chemokine expression[2,3]. Future investigations should focus to explore how HDAC10 suppression influences CD8+ T cell activation and checkpoint blockade efficacy in vivo. Similarly, investigating specific immune cell subsets such as T-regulatory cells or myeloid-derived suppressor cells could clarify its impact on the TME (Figure 1A).
Secondly, even though the authors have reported associations between HDAC10 expression and chemotherapeutic sensitivity via in silico IC50 predictions using the “oncoPredict” R package, we believe investigating the direct pharmacological inhibition of HDAC10 using selective inhibitors such as DKFZ-748 (Figure 1B) and subsequent biological response assessments would elucidate role of HDAC10 in chemoresistance and strengthen the therapeutic rationale for its selective targeting in CRC. Recent efforts toward class- and isoform-selective HDAC inhibitors, such as HDAC6 and HDAC10-selective compounds, have demonstrated distinct transcriptional and immune-modulatory effects compared to pan-HDAC inhibition[4]. Isoform-selective such as DKFZ-748 offers significant therapeutic advantages over pan-HDAC inhibitors. They provide higher therapeutic precision to minimize off-target effects and systemic toxicity associated with pan-HDAC inhibitors[1]. This target approach may improve clinical applicability and enhance patient safety. Therefore, we suggest in vivo studies to validate the oncogenic function of HDAC10 and evaluate its potential as a therapeutic target, which would provide critical mechanistic insights into its role in tumor progression and therapy resistance.
Thirdly, the study also links HDAC10 suppression to augmented p53 expression. Since p53 is a crucial modulator of immunogenicity and programmed death ligand 1 (PD-L1) regulation, investigating the HDAC10-p53-PD-L1 axis could clarify how HDAC10 modulates both tumor proliferation and immune escape, particularly through transcriptional and epigenetic regulation of PDL1 (Figure 1C). HDAC-mediated regulation of PD-L1 via p53 and signal transducer and activator of transcription-1/interferon regulatory factor 1 has been described in multiple cancer types[5] which suggests a broader transcriptional control mechanism beyond direct promoter binding. Lastly, genes like REG1A and CLDN18 were downregulated in group with higher HDAC10 expression. Therefore, assessing whether REG1A and CLDN18 expression correlates with specific CRC subtypes in clinical samples would strengthen the link between HDAC10-driven transcriptional changes and tumor heterogeneity. Moreover, CRCs are further classified into microsatellite instability-high (MSI-H) and microsatellite stable (MSS) subtypes based on DNA mismatch repair status[6]. MSI-H tumor responds well than MSS inhibitors to immune checkpoint inhibitors due to high neoantigen load[7]. The distinct immune landscapes of MSS and MSI-H CRC subtypes would be worthwhile to explore whether HDAC10 inhibition offers great therapeutic advantages in MSS tumors. In conclusion, we commend the authors for highlighting HDAC10 as a promising therapeutic target and biomarker in CRC. We hope future studies will build upon this work by incorporating in vivo models, targeted HDAC10 inhibition as suggested, and deeper mechanistic insights into how HDAC10 regulates tumor immunity.
| 1. | Nie HH, Yang XY, Zhou JK, Gao GL, Ding L, Hong YT, Yu YL, Qiu PS, Zeng ZY, Lai J, Zheng T, Wang HZ, Zhao Q, Wang F. Histone deacetylases 10 as a prognostic biomarker correlates with tumor microenvironment and therapy response in colorectal cancer. World J Gastroenterol. 2025;31:108662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 2. | Huang Z, Li L, Cheng B, Li D. Small molecules targeting HDAC6 for cancer treatment: Current progress and novel strategies. Biomed Pharmacother. 2024;178:117218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 3. | Jin Z, Sinicrope FA. Mismatch Repair-Deficient Colorectal Cancer: Building on Checkpoint Blockade. J Clin Oncol. 2022;40:2735-2750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 110] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 4. | Li J, Wu C, Hu H, Qin G, Wu X, Bai F, Zhang J, Cai Y, Huang Y, Wang C, Yang J, Luan Y, Jiang Z, Ling J, Wu Z, Chen Y, Xie Z, Deng Y. Remodeling of the immune and stromal cell compartment by PD-1 blockade in mismatch repair-deficient colorectal cancer. Cancer Cell. 2023;41:1152-1169.e7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 221] [Reference Citation Analysis (0)] |
| 5. | Stewart TM, Foley JR, Holbert CE, Klinke G, Poschet G, Steimbach RR, Miller AK, Casero RA Jr. Histone deacetylase-10 liberates spermidine to support polyamine homeostasis and tumor cell growth. J Biol Chem. 2022;298:102407. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 6. | Wang H, Zhou Y, Zhang Y, Fang S, Zhang M, Li H, Xu F, Liu L, Liu J, Zhao Q, Wang F. Subtyping of microsatellite stability colorectal cancer reveals guanylate binding protein 2 (GBP2) as a potential immunotherapeutic target. J Immunother Cancer. 2022;10:e004302. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 7. | Zhou Q, Xu CR, Cheng Y, Liu YP, Chen GY, Cui JW, Yang N, Song Y, Li XL, Lu S, Zhou JY, Ma ZY, Yu SY, Huang C, Shu YQ, Wang Z, Yang JJ, Tu HY, Zhong WZ, Wu YL. Bevacizumab plus erlotinib in Chinese patients with untreated, EGFR-mutated, advanced NSCLC (ARTEMIS-CTONG1509): A multicenter phase 3 study. Cancer Cell. 2021;39:1279-1291.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 173] [Article Influence: 34.6] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/