Published online Oct 14, 2025. doi: 10.3748/wjg.v31.i38.110999
Revised: July 14, 2025
Accepted: September 8, 2025
Published online: October 14, 2025
Processing time: 116 Days and 12.8 Hours
Eosinophilic esophagitis (EoE) is a chronic, immune-mediated condition leading to esophageal inflammation and a range of symptomatic complications if inade
Core Tip: The integration of artificial intelligence (AI) and deep learning models into the diagnosis and management of eosinophilic esophagitis shows great promise in enhancing diagnostic accuracy, reducing human variability in interpretation, and facilitating timely interventions. By addressing the limitations of traditional diagnostic methods, AI technologies can streamline the clinical workflow, improve patient outcomes, and potentially transform the approach to managing this complex, chronic condition. However, ongoing research is necessary to validate these tools across diverse clinical settings and ensure their ethical application.
- Citation: Issa IA, Youssef O, Issa T. Can artificial intelligence improve the diagnosis and management of patients with eosinophilic esophagitis? World J Gastroenterol 2025; 31(38): 110999
- URL: https://www.wjgnet.com/1007-9327/full/v31/i38/110999.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i38.110999
Eosinophilic esophagitis (EoE) is a chronic, immune-mediated disease characterized by inflammation of the esophagus that can lead to symptoms such as dysphagia, food impaction, and long-term tissue remodeling if not properly managed[1,2]. The rising prevalence and clinical complexity of EoE have important implications for both patient care and healthcare systems, as the disease often requires multifaceted diagnostic and therapeutic approaches[3,4]. EoE is prima
Recent advances in artificial intelligence (AI) and deep learning (DL) models have begun to influence various domains of medicine, offering new possibilities for enhancing diagnostic accuracy, automating image analysis, and supporting individualized clinical decision-making[9,10]. Within the field of gastroenterology, the application of AI to EoE is a developing area of research, aiming to address some of the inherent limitations of current practices[11]. AI-driven tools have shown high concordance with manual histopathological assessment and have demonstrated diagnostic performance in endoscopic image analysis that compares favorably to traditional clinical evaluation[12,13]. These approaches have the potential to reduce the burden on medical personnel, increase consistency in diagnostic criteria application, and facilitate earlier detection, all of which are critical for optimizing patient care[14]. However, challenges remain in terms of integrating these technologies into clinical workflows, addressing data heterogeneity, ensuring robust validation across diverse populations, and managing ethical concerns such as algorithmic transparency and patient data privacy[15,16]. Furthermore, deployment of AI enabled products or schemes requires a strict regulatory framework to ensure benefit while protecting from harm.
The primary objective of this research paper is to provide a comprehensive review of the current and emerging roles of AI and DL in the diagnosis and management of EoE. The central research question addressed is how AI-based technologies can improve diagnostic precision, facilitate timely identification, and support personalized management strategies for EoE, as well as what barriers and limitations impede their implementation. This work is based on a critical synthesis of peer-reviewed studies, systematic reviews, and clinical trials, with a focus on interpreting and contextualizing existing evidence rather than generating new experimental data[3,17]. Key methodological approaches include evaluation of AI algorithms for histological and endoscopic diagnosis, analysis of the reliability and generalizability of these findings across different healthcare settings, and source criticism with attention to potential biases and gaps in the literature[18,19].
To comprehensively grasp the current state of EoE, it is imperative to gain a holistic understanding of its clinical mani
EoE has experienced a dramatic rise in its epidemiological burden over the past few decades, with recent studies acknowledging an 800% increase in prevalence compared to data from the late 20th century. This surge can be attributed to a dual phenomenon of enhanced recognition due to advancements in diagnostic tools and physician awareness, and an actual rise in disease incidence[20,21]. Researchers have also documented EoE incidence rates ranging from 3.7 to 10 new cases per 100000 annually, with prevalence rates spanning from 22.7 per 100000 to over 100 per 100000 in high-prevalence nations such as Spain and the United States[22,23]. Furthermore, estimates suggest that up to 1 in 2000 individuals in the United States may be affected, emphasizing the substantial burden of this condition[23]. The possibility of regional disparities or underdiagnosis is evidenced by the wide geographic variability in reported cases, necessitating robust epidemiological surveillance systems to better capture the true scope of the disease[20]. Beyond these figures, con
The burden of EoE is substantial, with the condition responsible for 2%-7% of all endoscopies performed for esophageal symptoms, positioning it prominently among frequent esophageal disorders encountered in clinical practice[22,24]. In adults, EoE significantly contributes to dysphagia, accounting for 12%-22% of cases[22]. Moreover, it plays a critical role in esophageal food impaction, because it underlies up to 50% of such cases in this demographic[23]. These statistics not only underline the clinical significance of EoE but also emphasize its impact on healthcare systems, which must address the growing number of affected individuals. EoE’s prominence in acute care settings further reinforces the importance of timely and accurate diagnosis, since delays could exacerbate complications and increase resource utilization. The increasing patient burden calls for optimized diagnostic protocols and the exploration of emerging tools such as AI to complement traditional methods.
The strong association between EoE and atopic disorders is a hallmark of its type 2 immune-mediated pathogenesis. Up to 75% of individuals diagnosed with EoE have coexisting atopic conditions such as asthma, eczema, allergic rhinitis, or food allergies[22]. This overlap highlights shared immunologic pathways, that could open avenues for therapeutic strategies targeting both EoE and broader allergic profiles[23]. The emphasis on multidisciplinary care becomes evident, since managing concurrent conditions can improve long-term outcomes for individuals with EoE. Furthermore, the high prevalence of comorbid allergic conditions supports the case for systematic screening for EoE in populations with complex allergic profiles. Such targeted approaches may identify at-risk individuals earlier, providing opportunities for timely interventions that could mitigate disease progression.
Delayed diagnosis remains a major concern, because the risk of developing esophageal strictures rises each year EoE is left undiagnosed. Studies have consistently shown that this risk increases by 9% annually[22,23]. The link between diagnostic delay and disease progression illustrates the shift from reversible inflammatory stages to irreversible esophageal remodeling. Chronic untreated EoE leads to subepithelial fibrosis, esophageal narrowing, and an elevated risk of food impaction, adding to the condition’s significant morbidity[22]. In addition to degrading quality of life, these complications place a strain on healthcare resources since patients require recurrent medical interventions[25]. Early diagnosis and intervention, supported by epidemiological modeling, could prevent much of this stricture-related morbidity. This finding lends further credence to the importance of initiatives aimed at raising awareness among primary care and gastroenterology professionals, as well as the integration of advanced diagnostic tools such as AI to reduce diagnostic delays and improve patient outcomes.
The heterogeneous presentation of EoE further complicates its detection, with up to 12% of patients exhibiting normal findings on endoscopy despite confirmed disease[20,25]. This phenomenon is particularly evident in early or less active cases, where the absence of hallmark endoscopic abnormalities may hinder accurate identification. Pediatric studies have shown only moderate correlation (r = 0.61) between endoscopic severity scores and histologic activity, highlighting the insufficiency of endoscopy alone for diagnosing EoE[25]. Combining endoscopic assessment with histopathology and other advanced modalities is thus critical to ensure diagnostic accuracy. Missing subtle disease features during endo
Certain endoscopic signs, such as trachealization, exudates, and longitudinal furrows, are recognized as being more common in EoE than in non-EoE populations. However, their absence in some individuals complicates diagnosis. Liacouras et al[24] report that trachealization was observed in 25% of pediatric EoE patients but was absent in non-EoE controls. Additionally, the concurrent presence of exudates, trachealization, and longitudinal furrows was documented in 16% of EoE cases but in none of the control group. These findings support the necessity of developing disease-specific scoring systems and emphasize the potential of computer-aided diagnostic platforms in improving the recognition of such features. AI applications in endoscopic pattern recognition can not only enhance diagnostic accuracy but also reduce inter-observer variability, paving the way for more consistent outcomes.
Optimal diagnostic protocols for EoE are increasingly seen as a synthesis of clinical history, comprehensive atopic assessments, and detailed analyses of endoscopic and histologic findings. Advanced technologies, including AI and DL models, hold promise for integrating these diverse data points to facilitate real-time diagnostic accuracy. Recent innovations have demonstrated tangible benefits in this area; for instance, the application of AI to advanced imaging modalities like optical coherence tomography has achieved 97% diagnostic accuracy in detecting EoE-related basal layer thickening[26,27]. Such approaches can overcome some of the limitations of traditional diagnostic methods and provide a foundation for more effective disease management strategies. As the discussion progresses toward standard diagnostic approaches, the need for collaboration across technological and clinical domains to refine EoE detection and monitoring becomes evident. Table 1 summarizes the current diagnostic and endoscopic criteria for eosinophilic esophagitis.
| Feature | Description | Relevance |
| Definitive diagnosis | Histological identification of 15 or more eosinophils per HPF[65] | Gold standard for diagnosis |
| Endoscopic findings | Rings, linear furrows, exudates, edema[65] | Key visual indicators |
| Associated atopic conditions | Asthma, eczema, allergic rhinitis, food allergies[1] | Common co-morbidities, important for management |
| Symptoms | Dysphagia, food impaction, chest pain, reflux-like symptoms[1] | Impact on quality of life |
| Risk of complications | Increased risk of esophageal strictures and remodeling[1] | Importance of early diagnosis |
The endoscopic evaluation is a crucial initial step in the diagnosis of eosinophilic EoE, providing key visual hints of prevalent mucosal alterations, including rings (trachealization), linear furrows, white exudates, and edema. In spite of its diagnostic utility in contexts, the endoscopic process on its own is taxed by very real limitations. As discussed previously, in a minority of instances, up to 12% of EoE patients can have normal-appearing endoscopy, particularly in early or mild cases, and thus visual inspection alone is a dubious diagnostic criterion[28,29]. Moreover, infiltrates in EoE are often patchy in distribution; thus, focused and large biopsy sampling is needed in order to optimize sensitivity for diagnosis. Current recommendations are to obtain a minimum of six biopsies from various esophageal segments in an attempt to minimize sampling errors and maximize yield[30]. This requirement underlines the need for procedural rigor and identifies a significant limitation: Patient discomfort and procedural burden, particularly in high-prevalence areas where the demand for endoscopy is intensive. These limitations identify the reason why endoscopy, although being an absolute necessity, would preferably be complemented by adjunctive diagnostic methods in an effort to alleviate its limitations.
Histopathologic examination of esophageal biopsies, specifically identifying at least 15 eosinophils per high-power field (HPF), remains the gold standard for diagnosing EoE. This criterion is rooted in consensus guidelines and supported by extensive pathologic studies, offering an objective measure for diagnosis[24,31]. However, this threshold is not without challenges. Interobserver variability in counting eosinophils can lead to inconsistencies in diagnostic decisions, particularly in borderline cases. The specificity of this cutoff is further questioned by its overlap with conditions like proton pump inhibitor-responsive esophageal eosinophilia, where histological eosinophilia mimics EoE but resolves with acid suppression therapy[24]. Furthermore, the heterogeneous and patchy nature of eosinophilic infiltration introduces the risk of sampling error, especially when biopsies are insufficient or not adequately obtained from affected areas. Thus, reliance on eosinophil counts alone may oversimplify disease characterization, neglecting the dynamic interplay of tissue inflammation and remodeling that defines disease progression. This highlights the necessity for revised or supplemental histologic criteria that encompass broader pathologic changes beyond peak eosinophil counts.
While endoscopy and histopathological analysis represent the cornerstone of EoE diagnosis, these traditional approaches are invasive, posing procedural risks and discomfort for patients. The need for repeated endoscopies, often required for both diagnosis confirmation and monitoring of treatment response especially in pediatric and medically complex patients exacerbates these limitations[32]. Furthermore, delays in diagnosis, whether due to under-recognition of subtle endoscopic findings or unrepresentative biopsies, extend the disease course and contribute to complications like esophageal strictures. The risk of stricture formation increases by approximately 9% annually if diagnosis is delayed[22]. Beyond the impact on patient outcomes, the reliance on tissue sampling imposes a considerable burden on histopatho
In response to the shortcomings of standard diagnostic practices, several ancillary tools and emerging modalities have been explored for their potential to enhance EoE detection. Among these, magnifying endoscopy with narrow-band imaging and endoscopic ultrasonography have been investigated for their ability to provide better visualization of subtle mucosal abnormalities, which may be overlooked during standard endoscopy[31]. Another promising tool is the cytosponge-a minimally invasive device that collects a significant number of cells from the esophageal mucosa and has demonstrated an 83% accuracy rate in identifying active EoE in early studies[32,33]. This device offers unique advantages in sampling large surface areas, aiding in detecting active inflammation and potentially identifying molecular bio
In short, while traditional diagnostic approaches endoscopy and histopathology remain foundational to EoE diagnosis, their limitations stress the need for supplemental and innovative strategies. Emerging tools and AI-driven solutions hold great promise for addressing these gaps, potentially transforming the diagnostic landscape of this challenging condition.
AI is revolutionizing the diagnosis and management of EoE by enhancing accuracy, efficiency, and consistency across various diagnostic modalities. Its integration into endoscopic and histopathological evaluation holds promise for over
The term AI usually alludes to computer generated algorithms that are capable of learning and have problem solving abilities much like the human mind[34]. Machine learning (ML) is a field that can be taught to analyze data samples, discriminate their characteristics and then generate conclusions based on previously learned experiences[35]. Supervised ML with support vector machines involves researchers’ crafted algorithms in which they direct the system to recognize distinctive variables of concern to assess them and provide output[34]. DL is a subset of ML based on artificial neural networks capable of autonomously extracting data and generating analysis. They are often crafted as convolutional neural networks (CNNs)[34,36]. Algorithms trained in DL models can be further refined longitudinally using prospective feedback methods[37]. However, DL models are similar to black boxes where the output is reached and clear but the mechanism by which it is generated is not[38]. Much effort and research are currently invested to understand and elucidate the mystery behind these processes[39,40]. Another field progressing at a fast pace is that of “omics” including genomics, transcriptomics, epigenetic and others, where large sets of biological data offer new information about pathogenesis of diseases[41]. ML models can be quite useful in these instances where large sets of data need to be integrated, analyzed and studied[42]. Table 2 summarizes the various technical AI terms and their functions[34,35].
| AI methodology | Description | Applications in EoE |
| Supervised ML | Algorithms that rely on labeled training data to learn and predict outcomes | Classifying endoscopic images, predicting eosinophil counts |
| DL | Subset of ML utilizing neural networks for pattern recognition | Analyzing endoscopic and histopathological data |
| Convolutional neural networks | Special type of DL particularly adept at image recognition | Enhancing diagnostic accuracy in endoscopic images |
| Random decision forest | ML algorithm combining multiple data sources for improved decision making | Integrating clinical and endoscopic data for diagnosis |
| Natural language processing | AI field focused on interaction between computers and human language | Analyzing electronic health records for diagnostic insights |
AI models have already been validated in multiple medical disciplines. For example, in radiology modules have been created to detect diseases such as breast cancer or acute neurological events with high accuracy[43]. Furthermore, data suggests that AI significantly increases detection of colorectal neoplasia during lower endoscopy[44,45]. Similarly in upper endoscopy AI models have been shown beneficial in detecting subtle lesions and reducing variability related to an endoscopist’s experience[38]. For instance, one recent meta-analysis showed that the sensitivity and specificity in diagnosing Barrett’s neoplasia is 89% and 86% respectively[46]. In general, there is clear evidence that AI generated models perform well in gastrointestinal pathologies.
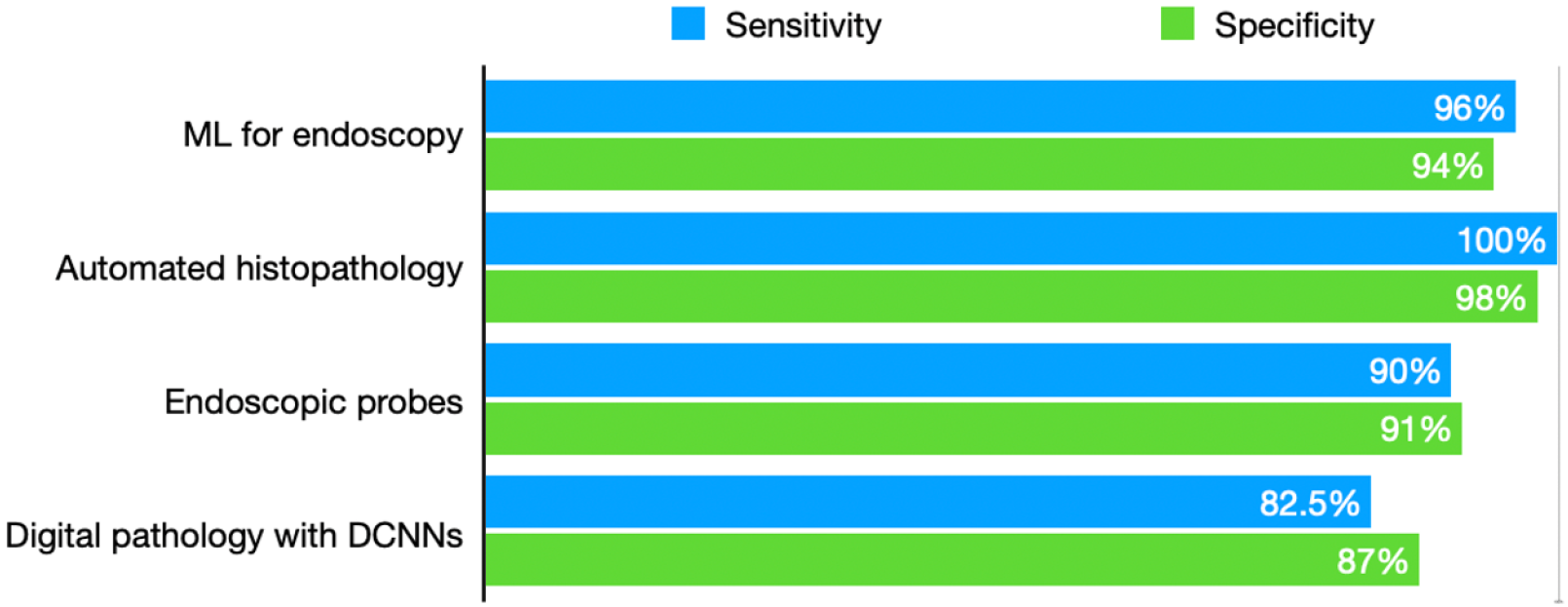
AI in improving endoscopy: The use of AI in diagnosing EoE has garnered considerable attention for its potential clinical impact, particularly through enhanced diagnostic methodologies. DL algorithms offer particular value by addressing the variability often seen in EoE endoscopic presentations. Subtle features, such as esophageal rings, linear furrows, white exudates, and edema, can sometimes be overlooked by endoscopists, particularly less experienced clinicians or in atypical cases. AI models trained to recognize these features consistently enhance diagnostic accuracy and reduce reliance on individual expertise[35]. While promising, these algorithms must undergo rigorous external validation to ensure robustness across varied clinical scenarios, particularly in resource-constrained environments where diagnostic expertise may vary. Integral to the success of AI-assisted diagnostic methods is the incorporation of established scoring systems, such as the EoE endoscopic reference score (EREFS), into AI algorithms. This approach improves diagnostic capacity and facilitates objective assessment of disease activity, which is crucial for monitoring treatment responses and disease progression[44]. The integration of EREFS reflects the potential of AI to standardize assessments, although questions remain regarding its applicability in pediatric populations and individuals with overlapping esophageal pathologies. AI has demonstrated high diagnostic accuracy in analyzing endoscopic images of individuals with EoE. For instance, DL models such as the AI-EoE-EREFS algorithm have achieved sensitivity, specificity, and accuracy levels of 0.96, 0.94, and 0.95, respectively, with an area under the curve (AUC) of 0.992[44]. These figures highlight the significant reliability and potential of AI tools as adjuncts in endoscopic settings. However, the validity of these outcomes necessitates critical consideration of data sources, diversity within training datasets, and the reproducibility of results across different populations and healthcare facilities.
Okimoto et al[47] performed a very elegant study where they developed a CNN model and trained it to recognize EoE endoscopically using a set of images. The model performed very well with an accuracy of 95% when tested against patients and controls. Similarly, another trial compared EoE endoscopic appearance to normal controls and patients with esophageal candidiasis using another CNN model. The algorithm was trained on 484 endoscopic images from 134 patients. The results were compared and validated against 3 endoscopists, showing an accuracy of 91% vs 83% for the physicians[48]. Additionally, it seems that adding objective criteria to the model such as the EREFS score infers more benefit. German researchers concluded that the prediction of a CNN model improved when the endoscopic score was added to the algorithm increasing sensitivity to 85% and specificity to 95%[49]. The performance consistency of AI-EoE models, as demonstrated by high sensitivity, specificity, accuracy, and harmonic mean across different cohorts and image qualities, further supports their clinical utility. This robustness highlights the scalability of such systems; however, their long-term performance in real-world, multi-institutional settings remains a crucial area for further exploration. The extent to which these technologies can be seamlessly integrated into routine workflows, particularly in lower-resource envi
AI has also facilitated the development of random decision forest models that integrate both clinical and endoscopic data to enhance diagnostic precision. These models have achieved impressive AUCs of 0.90 with clinical data alone and 0.94 when combining clinical and endoscopic data, demonstrating their utility in improving patient selection and biopsy guidance in real-world settings[50]. However, the complexity of these multi-source systems may pose challenges to their seamless integration into routine practice. One major advantage is that the predictive accuracy of ML models improves with the inclusion of readily accessible clinical variables alongside endoscopic findings, enabling better prioritization for invasive procedures[50]. Such advancements could streamline patient management, however, disparities in healthcare infrastructure and diagnostic capacity must be acknowledged, as these factors could affect the performance of AI algorithms in underrepresented settings. Table 3 summarizes the main AI models applied to endoscopic diagnosis of EoE, their methodology and key limitations.
| Model | Training dataset | Population size | Validation status | Key limitations | Methodological rigor |
| AI-EoE-EREFS (DL/CNN)[48] | Endoscopic images, EREFS-labeled, multi-center | 484 images from 134 patients | Retrospective, some external validation | Retrospective data, limited diversity, mainly binary classification, limited real-world scenarios | High accuracy (AUC = 0.992), but generalizability and real-world robustness uncertain |
| CNN[47] | Endoscopic images (EoE vs controls) | 1192 characteristic endoscopic images of 108 patients | Tested against patients and controls | Dataset size not reported, unclear diversity, retrospective design | High accuracy (95%), but limited transparency and unclear reproducibility |
| CNN with EREFS integration[49] | Endoscopic images annotated with EREFS | 200 WLIs, including 100 WLIs from EoE patients and 100 WLIs of normal esophagus | Compared to human experts | Training/test data overlap possible, unclear external validation | Sensitivity improved to 85%, specificity 95% with EREFS |
| Random decision forest[50] | Clinical + endoscopic data | Not specified | Real-world data, AUC up to 0.94 | Complexity, integration challenges, population diversity not detailed | Robust with multi-source inputs, but needs more transparent validation |
| General endoscopic AI (DL/CNN) | Mixed datasets (WLI, NBI, CT images) | Varies; often small | Mostly retrospective, single/multi-center | Data heterogeneity, lack of multi-center, prospective validation | High reported accuracy, but shallow validation, risk of bias due to dataset curation |
AI in improving histology: AI has also shown remarkable promise in the automated histological evaluation of tissue biopsies. A DL approach achieved near-perfect diagnostic performance in one study, demonstrating 99.0% accuracy, 100% sensitivity, and 98.2% specificity in identifying ≥ 15 eosinophils per HPF in esophageal biopsies[24]. These results highlight the advanced precision of AI in meeting diagnostic thresholds.
AI-driven eosinophil counting has demonstrated excellent concordance with pathologist counts, achieving a Pearson correlation coefficient of 85% in gastrointestinal biopsy analyses[51]. This accuracy supports the broader adoption of AI as a quality control instrument in histopathology. Automated counting methods significantly reduce errors and alleviate fatigue-related inconsistencies in human reporting, fostering efficiency in high-volume pathology environments[51]. The faster turnaround times enabled by automated methods could lead to more prompt clinical decisions and interventions, further supporting the utility of AI tools for improving patient outcomes in EoE[51]. Nevertheless, ensuring consistent performance across diverse healthcare systems remains an ongoing challenge. Automated eosinophil quantification further enables comprehensive mapping of disease heterogeneity, an innovation that holds promise for distinguishing between phenotypic variants of EoE and predicting treatment responses[24]. This capability could help clinicians refine personalized management strategies. In general experts agree that CNN models can reliably take the place of manual eosinophils count.
Recent analytical advancements indicate that focusing solely on peak eosinophil counts may overlook critical diagnostic features of EoE. ML and DL analyses have offered innovative insights into global histopathologic changes, such as basal cell hyperplasia, spongiosis, and lamina propria fibrosis, which may help differentiate active EoE from non-EoE states[31]. This suggests a critical opportunity to expand histologic assessment frameworks by integrating both quantitative (eosinophil counts) and qualitative (global tissue features) analysis. Leveraging AI-driven models may enhance diagnostic consistency, decrease inter-observer variability, and allow for a more comprehensive assessment of disease activity. Importantly, adopting AI platforms for biopsy-based diagnostics may enable automated and reproducible analyses, addressing issues related to subjectivity. This advancement represents a significant step toward a more nuanced and technologically advanced approach to EoE evaluation. Looking further than eosinophilic count, pathologists have elaborated an EoE histologic scoring systems to include other important criteria such as fibroblasts in the laminated propria[52]. Additional characteristics beside eosinophil counts reliably predicted EoE in CNN models with an accuracy of 85%[53].
Dynamic CNNs have successfully classified esophageal tissue biopsies, achieving accuracy, sensitivity, and specificity rates of 85%, 82.5%, and 87%, respectively[54]. Notably, these systems incorporate global histopathologic features such as tissue architecture rather than solely relying on eosinophil counts, thus strengthening their diagnostic capacity. Down
Larey et al[56] developed an ML model incorporating not only eosinophilic counts of > 15/HPF but also basal cell fraction. They used 1066 biopsies to train their model. This algorithm predicted severity more accurately than the standard method (accuracy of 87% vs 79%). This carries additional importance in addressing treatment response and remission[56]. Similarly, Ricaurte Archila et al[57] presented a CNN model to evaluate histologic features of EoE and compared the results to five experienced pathologists. The AI model analyzed several pathologic features and faired well in comparison to the pathologists scores (94.5% vs 94.8%). Another important study researched an image classification model looking at multiple features other than the eosinophils count. It was able to predict the diagnosis of EoE in 99% of slide patches from treatment naive patients, the model was also useful in prediction of remission after follow up[58]. Even more impressive is the DL digital pathology model that incorporated novel features of EoE looking at the whole biopsy histology. It examined features such as tissue subregions, eosinophilic abscesses, cell types and even the spatial relationships between various cells[59]. These models offer the possibility of diagnosing difficult and unconventional patients such as those without elevated eosinophils or those maintained on chronic proton pump inhibitors.
Dunn et al[60] applied an AI clustering analysis on Th2 cytokine expression resulting in major breakthrough in EoE patients subtypes and classifications. They were able to delineate 5 groups of endotypes of patients. To note that all of these patients did not differ in their eosinophil count further confirming the theory peripheral eosinophil count may not be enough for diagnosing EoE patients. Group 1 showed the lowest interleukin-5 expression vs group 5 which showed the highest (suggesting an elevated inflammatory load). Groups 2 to 4 showed elevated levels of interleukin-13 and tendency for more fibrostenotic disease[60]. It is still unclear whether these subtypes result from a continuous progression in disease severity or distinct patients phenotypes.
The application of DL for histopathological analysis effectively mitigates longstanding issues of inter-observer subjectivity in manual eosinophil counting. Pathologist variability, particularly in borderline cases, has historically hindered consistent diagnoses. Automated systems not only provide reproducibility but also offer scalability in high-volume pathology labs, addressing critical workflow bottlenecks. The high specificity and sensitivity achieved with AI-based histopathological tools highlight their potential in supporting overburdened histopathologists, who face increasing demand for esophageal biopsies[24]. Nevertheless, implementing such technology requires significant investment, and its adaptability to small-scale or underfunded pathology centers is yet to be fully assessed. Employing sufficiently large and diverse training datasets has significantly improved the adaptability of AI models to various clinical conditions and institutions[38]. Models that are generalizable across populations are essential for equitable healthcare delivery. While the reported AI performance metrics such as 99% accuracy appear impressive, it is important to interpret these results with caution. Such high figures may inadvertently signal potential overfitting, especially if the model has primarily been evaluated on limited or non-representative datasets. Overfitting occurs when a model performs exceedingly well on training data but fails to generalize effectively to new, real-world scenarios. Additionally, models trained and validated on small sample sizes or narrowly defined datasets may embed underlying biases, leading to performance gaps when exposed to broader populations or diverse input conditions. This underscores the need for thorough testing across varied, real-world datasets and the inclusion of robustness checks to detect and mitigate bias. Figure 1 illustrates the some of the available AI driven innovations and their performance in EoE diagnosis.
AI in improving biomarkers: Point-of-care decision support enabled by ML models could drastically reduce unnecessary biopsies while ensuring that high-risk individuals are not overlooked[36]. ML applications that utilize peripheral blood markers, such as eosinophil levels, have demonstrated strong diagnostic associations in EoE and other gastrointestinal conditions[53]. Expanding these capabilities could aid in streamlining the diagnostic process; although, their accuracy and reliability when applied to complex, multi-disease scenarios require further study. The inclusion of auxiliary markers, such as platelet-lymphocyte ratios and white blood cell counts, enhances the predictive power of diagnostic models[53]. By considering these multifactorial inputs, AI could offer better decision-making insights for clinicians managing complex cases. Additionally, non-invasive, AI-driven analysis of peripheral blood data could serve as an effective triage tool, reducing reliance on invasive procedures. This approach has significant implications for populations with limited access to endoscopic services; nonetheless, further validation is crucial to ensure that it does not compromise diagnostic accuracy.
Multiple studies have explored non-invasive AI facilitated diagnostic tools ranging from swallowed strings to sponges. They capture eosinophils-derived proteins, but they still seem to offer lower accuracy in diagnosis that the EREFS score[61,62]. A more innovative approach is the new swallowable capsule that can identify eosinophils in the esophagus through reflectance microscopy. It is swallowed and then retrieved using a string. A CNN model was created to analyze the generated images and resulted in an 86% accuracy in diagnosis[63]. Wechsler et al[64] used an AI random forest model analyzing data from a combined six eosinophils associated blood markers and two urine markers. It accurately predicted the diagnosis 84% of the times.
This review aimed to assess the transformative potential of AI and DL models in the diagnosis and management of EoE, a chronic, immune-mediated esophageal disease characterized by its increasing prevalence and presenting significant diagnostic challenges. The primary objective was to determine whether AI-based methodologies can provide substantial improvements over conventional diagnostic paradigms and to evaluate the extent to which technological advancements overcome existing barriers in clinical practice. By synthesizing current literature and emerging evidence, the work systematically evaluated the limitations of conventional approaches, the efficacy of advanced computational tools, and the broader implications for disease detection and patient care. Standard diagnostic modalities, such as endoscopy and histopathology, are fundamental but are limited by significant shortcomings. Inter-observer variability, the invasiveness of procedures, and the potential for diagnostic delays substantially increase morbidity. Data presented demonstrate that AI-driven algorithms now attain high sensitivity and specificity in both endoscopic pattern recognition and histopathologic image analysis. Advanced models reliably identify subtle mucosal abnormalities and precisely quantify eosinophilic infiltration, resulting in outcomes that are comparable to or surpass those of expert human evaluators. Further
| 1. | Furuta GT, Katzka DA. Eosinophilic Esophagitis. N Engl J Med. 2015;373:1640-1648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 420] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 2. | Andersson ER, Chivukula IV, Hankeova S, Sjöqvist M, Tsoi YL, Ramsköld D, Masek J, Elmansuri A, Hoogendoorn A, Vazquez E, Storvall H, Netušilová J, Huch M, Fischler B, Ellis E, Contreras A, Nemeth A, Chien KC, Clevers H, Sandberg R, Bryja V, Lendahl U. Mouse Model of Alagille Syndrome and Mechanisms of Jagged1 Missense Mutations. Gastroenterology. 2018;154:1080-1095. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 94] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 3. | Holguin F, Bleecker ER, Busse WW, Calhoun WJ, Castro M, Erzurum SC, Fitzpatrick AM, Gaston B, Israel E, Jarjour NN, Moore WC, Peters SP, Yonas M, Teague WG, Wenzel SE. Obesity and asthma: an association modified by age of asthma onset. J Allergy Clin Immunol. 2011;127:1486-93.e2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 329] [Cited by in RCA: 311] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 4. | Plate J, Söderbergh T, Bergqvist J, Lingblom C, Bergquist H, Larsson H. Eosinophilic esophagitis prevalence, incidence, and presenting features: a 22-year population-based observational study from southwest Sweden. Dis Esophagus. 2025;38:doae025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 5. | Biedermann L, Straumann A, Greuter T, Schreiner P. Eosinophilic esophagitis-established facts and new horizons. Semin Immunopathol. 2021;43:319-335. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 6. | Thel HL, Anderson C, Xue AZ, Jensen ET, Dellon ES. Prevalence and Costs of Eosinophilic Esophagitis in the United States. Clin Gastroenterol Hepatol. 2025;23:272-280.e8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 45] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 7. | Rothenberg ME, Dellon ES, Collins MH, Hirano I, Chehade M, Bredenoord AJ, Lucendo AJ, Spergel JM, Sun X, Hamilton JD, Mortensen E, Laws E, Maloney J, Mannent LP, McCann E, Liu X, Glotfelty L, Shabbir A. Efficacy and safety of dupilumab up to 52 weeks in adults and adolescents with eosinophilic oesophagitis (LIBERTY EoE TREET study): a multicentre, double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Gastroenterol Hepatol. 2023;8:990-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 67] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 8. | Hirano I, Collins MH, Katzka DA, Mukkada VA, Falk GW, Morey R, Desai NK, Lan L, Williams J, Dellon ES; ORBIT1/SHP621-301 Investigators. Budesonide Oral Suspension Improves Outcomes in Patients With Eosinophilic Esophagitis: Results from a Phase 3 Trial. Clin Gastroenterol Hepatol. 2022;20:525-534.e10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 107] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 9. | Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S, Dean J. A guide to deep learning in healthcare. Nat Med. 2019;25:24-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1123] [Cited by in RCA: 1783] [Article Influence: 254.7] [Reference Citation Analysis (0)] |
| 10. | Topol E. The Patient Will See You Now: The Future of Medicine is in Your Hands. Basic Books, 2016. |
| 11. | Penrice DD, Rattan P, Simonetto DA. Artificial Intelligence and the Future of Gastroenterology and Hepatology. Gastro Hep Adv. 2022;1:581-595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Ali H, Muzammil MA, Dahiya DS, Ali F, Yasin S, Hanif W, Gangwani MK, Aziz M, Khalaf M, Basuli D, Al-Haddad M. Artificial intelligence in gastrointestinal endoscopy: a comprehensive review. Ann Gastroenterol. 2024;37:133-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 16] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 13. | Kim D, Pantanowitz L, Schüffler P, Yarlagadda DVK, Ardon O, Reuter VE, Hameed M, Klimstra DS, Hanna MG. (Re) Defining the High-Power Field for Digital Pathology. J Pathol Inform. 2020;11:33. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 14. | Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5683] [Cited by in RCA: 5713] [Article Influence: 634.8] [Reference Citation Analysis (0)] |
| 15. | Challen R, Denny J, Pitt M, Gompels L, Edwards T, Tsaneva-Atanasova K. Artificial intelligence, bias and clinical safety. BMJ Qual Saf. 2019;28:231-237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 268] [Cited by in RCA: 426] [Article Influence: 60.9] [Reference Citation Analysis (0)] |
| 16. | Visaggi P, de Bortoli N, Barberio B, Savarino V, Oleas R, Rosi EM, Marchi S, Ribolsi M, Savarino E. Artificial Intelligence in the Diagnosis of Upper Gastrointestinal Diseases. J Clin Gastroenterol. 2022;56:23-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 17. | Yuriy R, Tatarina O, Kaminskyy V, Silina T, Bashkirova L. Modern Methods and Prospects for Using Artificial Intelligence in Disease Diagnostics: A Narrative Review. Futur Med. 2024;3. [DOI] [Full Text] |
| 18. | Castagnaro E, Felici E, Spaccapelo R, Amoroso M, Moretti D, Saab JP, Stassaldi A, Tassone A, Borrelli O, De Bortoli N, Corso GD, Gaynor E, Goh L, Oliva S, Rea F, Renzo S, Romano C, Savarino EV, Visaggi P. Systematic Review: Use of Artificial Intelligence and Unmet Needs in Eosinophilic Oesophagitis. Aliment Pharmacol Ther. 2025;62:110-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 19. | Koo TK, Li MY. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J Chiropr Med. 2016;15:155-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9979] [Cited by in RCA: 17999] [Article Influence: 1799.9] [Reference Citation Analysis (1)] |
| 20. | Chang JW, Jensen ET. Epidemiologic and Clinical Clues to the Etiology of Eosinophilic Esophagitis. Immunol Allergy Clin North Am. 2024;44:145-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 21. | Mona R, Hruz P. Epidemiology of Eosinophilic Esophagitis: Really a Novel and Evolving Disease? Inflamm Intest Dis. 2025;10:34-40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 22. | Rothenberg ME. Biology and treatment of eosinophilic esophagitis. Gastroenterology. 2009;137:1238-1249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 195] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 23. | Furuta GT, Liacouras CA, Collins MH, Gupta SK, Justinich C, Putnam PE, Bonis P, Hassall E, Straumann A, Rothenberg ME; First International Gastrointestinal Eosinophil Research Symposium (FIGERS) Subcommittees. Eosinophilic esophagitis in children and adults: a systematic review and consensus recommendations for diagnosis and treatment. Gastroenterology. 2007;133:1342-1363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1400] [Cited by in RCA: 1174] [Article Influence: 61.8] [Reference Citation Analysis (0)] |
| 24. | Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, Burks AW, Chehade M, Collins MH, Dellon ES, Dohil R, Falk GW, Gonsalves N, Gupta SK, Katzka DA, Lucendo AJ, Markowitz JE, Noel RJ, Odze RD, Putnam PE, Richter JE, Romero Y, Ruchelli E, Sampson HA, Schoepfer A, Shaheen NJ, Sicherer SH, Spechler S, Spergel JM, Straumann A, Wershil BK, Rothenberg ME, Aceves SS. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3-20.e6; quiz 21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1605] [Cited by in RCA: 1518] [Article Influence: 101.2] [Reference Citation Analysis (1)] |
| 25. | González-Cervera J, Arias Á, Redondo-González O, Cano-Mollinedo MM, Terreehorst I, Lucendo AJ. Association between atopic manifestations and eosinophilic esophagitis: A systematic review and meta-analysis. Ann Allergy Asthma Immunol. 2017;118:582-590.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 124] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 26. | Wang C, Gan M, Zhang M, Li D. Adversarial convolutional network for esophageal tissue segmentation on OCT images. Biomed Opt Express. 2020;11:3095-3110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 27. | Santacroce G, Rossi CM, Lenti MV, Ghosh S, Iacucci M, Di Sabatino A. Understanding tissue injury and remodelling in eosinophilic oesophagitis: development towards personalised medicine. Gut. 2025;74:996-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 28. | Dellon ES. Eosinophilic esophagitis. Gastroenterol Clin North Am. 2013;42:133-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 29. | Kim HP, Vance RB, Shaheen NJ, Dellon ES. The prevalence and diagnostic utility of endoscopic features of eosinophilic esophagitis: a meta-analysis. Clin Gastroenterol Hepatol. 2012;10:988-96.e5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 264] [Cited by in RCA: 264] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 30. | Dhar A, Haboubi HN, Attwood SE, Auth MKH, Dunn JM, Sweis R, Morris D, Epstein J, Novelli MR, Hunter H, Cordell A, Hall S, Hayat JO, Kapur K, Moore AR, Read C, Sami SS, Turner PJ, Trudgill NJ. British Society of Gastroenterology (BSG) and British Society of Paediatric Gastroenterology, Hepatology and Nutrition (BSPGHAN) joint consensus guidelines on the diagnosis and management of eosinophilic oesophagitis in children and adults. Gut. 2022;71:1459-1487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 74] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 31. | Yao K. Clinical Application of Magnifying Endoscopy with Narrow-Band Imaging in the Stomach. Clin Endosc. 2015;48:481-490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 32. | Godwin B, Wilkins B, Muir AB. EoE disease monitoring: Where we are and where we are going. Ann Allergy Asthma Immunol. 2020;124:240-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 33. | Katzka DA, Smyrk TC, Alexander JA, Geno DM, Beitia RA, Chang AO, Shaheen NJ, Fitzgerald RC, Dellon ES. Accuracy and Safety of the Cytosponge for Assessing Histologic Activity in Eosinophilic Esophagitis: A Two-Center Study. Am J Gastroenterol. 2017;112:1538-1544. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 84] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 34. | Le Berre C, Sandborn WJ, Aridhi S, Devignes MD, Fournier L, Smaïl-Tabbone M, Danese S, Peyrin-Biroulet L. Application of Artificial Intelligence to Gastroenterology and Hepatology. Gastroenterology. 2020;158:76-94.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 355] [Article Influence: 59.2] [Reference Citation Analysis (3)] |
| 35. | Ebigbo A, Palm C, Probst A, Mendel R, Manzeneder J, Prinz F, de Souza LA, Papa JP, Siersema P, Messmann H. A technical review of artificial intelligence as applied to gastrointestinal endoscopy: clarifying the terminology. Endosc Int Open. 2019;7:E1616-E1623. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 36. | Sana MK, Hussain ZM, Shah PA, Maqsood MH. Artificial intelligence in celiac disease. Comput Biol Med. 2020;125:103996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 37. | Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. N Engl J Med. 2019;380:1347-1358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1274] [Cited by in RCA: 1890] [Article Influence: 270.0] [Reference Citation Analysis (3)] |
| 38. | Mori Y, Kudo SE, Mohmed HEN, Misawa M, Ogata N, Itoh H, Oda M, Mori K. Artificial intelligence and upper gastrointestinal endoscopy: Current status and future perspective. Dig Endosc. 2019;31:378-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 39. | Glover B, Teare J, Patel N. A systematic review of the role of non-magnified endoscopy for the assessment of H. pylori infection. Endosc Int Open. 2020;8:E105-E114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 23] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 40. | Nakashima H, Kawahira H, Kawachi H, Sakaki N. Artificial intelligence diagnosis of Helicobacter pylori infection using blue laser imaging-bright and linked color imaging: a single-center prospective study. Ann Gastroenterol. 2018;31:462-468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 67] [Article Influence: 8.4] [Reference Citation Analysis (1)] |
| 41. | Matsuyama K, Yamada S, Sato H, Zhan J, Shoda T. Advances in omics data for eosinophilic esophagitis: moving towards multi-omics analyses. J Gastroenterol. 2024;59:963-978. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 42. | Banerjee J, Taroni JN, Allaway RJ, Prasad DV, Guinney J, Greene C. Machine learning in rare disease. Nat Methods. 2023;20:803-814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 55] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 43. | Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2376] [Cited by in RCA: 3595] [Article Influence: 513.6] [Reference Citation Analysis (5)] |
| 44. | Hassan C, Spadaccini M, Iannone A, Maselli R, Jovani M, Chandrasekar VT, Antonelli G, Yu H, Areia M, Dinis-Ribeiro M, Bhandari P, Sharma P, Rex DK, Rösch T, Wallace M, Repici A. Performance of artificial intelligence in colonoscopy for adenoma and polyp detection: a systematic review and meta-analysis. Gastrointest Endosc. 2021;93:77-85.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 355] [Article Influence: 71.0] [Reference Citation Analysis (5)] |
| 45. | Barua I, Vinsard DG, Jodal HC, Løberg M, Kalager M, Holme Ø, Misawa M, Bretthauer M, Mori Y. Artificial intelligence for polyp detection during colonoscopy: a systematic review and meta-analysis. Endoscopy. 2021;53:277-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 178] [Article Influence: 35.6] [Reference Citation Analysis (1)] |
| 46. | Visaggi P, Barberio B, Gregori D, Azzolina D, Martinato M, Hassan C, Sharma P, Savarino E, de Bortoli N. Systematic review with meta-analysis: artificial intelligence in the diagnosis of oesophageal diseases. Aliment Pharmacol Ther. 2022;55:528-540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 47] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 47. | Okimoto E, Ishimura N, Adachi K, Kinoshita Y, Ishihara S, Tada T. Application of Convolutional Neural Networks for Diagnosis of Eosinophilic Esophagitis Based on Endoscopic Imaging. J Clin Med. 2022;11:2529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 48. | Guimarães P, Keller A, Fehlmann T, Lammert F, Casper M. Deep learning-based detection of eosinophilic esophagitis. Endoscopy. 2022;54:299-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 49. | Römmele C, Mendel R, Barrett C, Kiesl H, Rauber D, Rückert T, Kraus L, Heinkele J, Dhillon C, Grosser B, Prinz F, Wanzl J, Fleischmann C, Nagl S, Schnoy E, Schlottmann J, Dellon ES, Messmann H, Palm C, Ebigbo A. An artificial intelligence algorithm is highly accurate for detecting endoscopic features of eosinophilic esophagitis. Sci Rep. 2022;12:11115. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 50. | Mohsen F, Ali H, El Hajj N, Shah Z. Artificial intelligence-based methods for fusion of electronic health records and imaging data. Sci Rep. 2022;12:17981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 92] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 51. | Salo I, Nordlund L, Eklund L, Ho J, Soini M, Kumar D, Yeong J, Guan F, Metsälä E. Advancements and applications of AI technologies in pathology: a scoping review. Comput Methods Biomech Biomed Eng Imaging Vis. 2024;12. [DOI] [Full Text] |
| 52. | Collins MH, Martin LJ, Alexander ES, Boyd JT, Sheridan R, He H, Pentiuk S, Putnam PE, Abonia JP, Mukkada VA, Franciosi JP, Rothenberg ME. Newly developed and validated eosinophilic esophagitis histology scoring system and evidence that it outperforms peak eosinophil count for disease diagnosis and monitoring. Dis Esophagus. 2017;30:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 153] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 53. | Czyzewski T, Daniel N, Rochman M, Caldwell JM, Osswald GA, Collins MH, Rothenberg ME, Savir Y. Machine Learning Approach for Biopsy-Based Identification of Eosinophilic Esophagitis Reveals Importance of Global features. IEEE Open J Eng Med Biol. 2021;2:218-223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 54. | Tang D, Wang L, Ling T, Lv Y, Ni M, Zhan Q, Fu Y, Zhuang D, Guo H, Dou X, Zhang W, Xu G, Zou X. Development and validation of a real-time artificial intelligence-assisted system for detecting early gastric cancer: A multicentre retrospective diagnostic study. EBioMedicine. 2020;62:103146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 63] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 55. | Votto M, Rossi CM, Caimmi SME, De Filippo M, Di Sabatino A, Lenti MV, Raffaele A, Marseglia GL, Licari A. The State of the Art of Artificial Intelligence Applications in Eosinophilic Esophagitis: A Systematic Review. BDCC. 2024;8:76. [DOI] [Full Text] |
| 56. | Larey A, Aknin E, Daniel N, Osswald GA, Caldwell JM, Rochman M, Wasserman T, Collins MH, Arva NC, Yang GY, Rothenberg ME, Savir Y. Harnessing artificial intelligence to infer novel spatial biomarkers for the diagnosis of eosinophilic esophagitis. Front Med (Lausanne). 2022;9:950728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 57. | Ricaurte Archila L, Smith L, Sihvo HK, Koponen V, Jenkins SM, O'Sullivan DM, Cardenas Fernandez MC, Wang Y, Sivasubramaniam P, Patil A, Hopson PE, Absah I, Ravi K, Mounajjed T, Dellon ES, Bredenoord AJ, Pai R, Hartley CP, Graham RP, Moreira RK. Performance of an Artificial Intelligence Model for Recognition and Quantitation of Histologic Features of Eosinophilic Esophagitis on Biopsy Samples. Mod Pathol. 2023;36:100285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 58. | Javaid A, Fernandes P, Adorno W, Catalano A, Ehsan L, von Eckstaedt HV, Barnes B, Khan M, Raghavan SS, Mcgowan E, Collins MH, Rothenberg ME, Moskaluk CA, Brown DE, Syed S. Deep Learning Tissue Analysis Diagnoses and Predicts Treatment Response in Eosinophilic Esophagitis. 2021 Preprint. [DOI] [Full Text] |
| 59. | Archila LR, Smith L, Sihvo HK, Westerling-Bui T, Koponen V, O'Sullivan DM, Fernandez MCC, Alexander EE, Wang Y, Sivasubramaniam P, Patil A, Hopson PE, Absah I, Ravi K, Mounajjed T, Pai R, Hagen C, Hartley C, Graham RP, Moreira RK. Development and technical validation of an artificial intelligence model for quantitative analysis of histopathologic features of eosinophilic esophagitis. J Pathol Inform. 2022;13:100144. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 60. | Dunn JLM, Shoda T, Caldwell JM, Wen T, Aceves SS, Collins MH, Dellon ES, Falk GW, Leung J, Martin LJ, Menard-Katcher P, Rudman-Spergel AK, Spergel JM, Wechsler JB, Yang GY, Furuta GT, Rothenberg ME; Consortium of Eosinophilic Gastrointestinal Disease Researchers (CEGIR). Esophageal type 2 cytokine expression heterogeneity in eosinophilic esophagitis in a multisite cohort. J Allergy Clin Immunol. 2020;145:1629-1640.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 61. | Furuta GT, Kagalwalla AF, Lee JJ, Alumkal P, Maybruck BT, Fillon S, Masterson JC, Ochkur S, Protheroe C, Moore W, Pan Z, Amsden K, Robinson Z, Capocelli K, Mukkada V, Atkins D, Fleischer D, Hosford L, Kwatia MA, Schroeder S, Kelly C, Lovell M, Melin-Aldana H, Ackerman SJ. The oesophageal string test: a novel, minimally invasive method measures mucosal inflammation in eosinophilic oesophagitis. Gut. 2013;62:1395-1405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 210] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 62. | Katzka DA, Geno DM, Ravi A, Smyrk TC, Lao-Sirieix P, Miremadi A, Debiram I, O'Donovan M, Kita H, Kephart GM, Kryzer LA, Camilleri M, Alexander JA, Fitzgerald RC. Accuracy, safety, and tolerability of tissue collection by Cytosponge vs endoscopy for evaluation of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2015;13:77-83.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 114] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 63. | Kang D, Do D, Ryu J, Grant CN, Giddings SL, Rosenberg M, Hesterberg PE, Yuan Q, Garber JJ, Katz AJ, Tearney GJ. A miniaturized, tethered, spectrally-encoded confocal endomicroscopy capsule. Lasers Surg Med. 2019;51:452-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 64. | Wechsler JB, Ackerman SJ, Chehade M, Amsden K, Riffle ME, Wang MY, Du J, Kleinjan ML, Alumkal P, Gray E, Kim KA, Wershil BK, Kagalwalla AF. Noninvasive biomarkers identify eosinophilic esophagitis: A prospective longitudinal study in children. Allergy. 2021;76:3755-3765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 65. | Dellon ES. Diagnostics of eosinophilic esophagitis: clinical, endoscopic, and histologic pitfalls. Dig Dis. 2014;32:48-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/