Published online May 28, 2024. doi: 10.3748/wjg.v30.i20.2689
Revised: February 28, 2024
Accepted: April 19, 2024
Published online: May 28, 2024
Processing time: 164 Days and 6.5 Hours
The regulatory effects of KIF26B on gastric cancer (GC) have been confirmed, but the specific mechanism still needs further exploration. Pan-cancer analysis shows that the KIF26B expression is highly related to immune infiltration of cancer-associated fibroblasts (CAFs), and CAFs promote macrophage M2 polarization and affect cancers’ progression.
To investigate the regulatory functions of KIF26B on immune and metastasis of GC.
We analyzed genes’ mRNA levels by quantitative real-time polymerase chain reaction. Expression levels of target proteins were detected by immunohistochemistry, ELISA, and Western blotting. We injected AGS cells into nude mice for the establishment of a xenograft tumor model and observed the occurrence and metastasis of GC. The degree of inflammatory infiltration in pulmonary nodes was observed through hematoxylin-eosin staining. Transwell and wound healing assays were performed for the evaluation of cell invasion and migration ability. Tube formation assay was used for detecting angiogenesis. M2-polarized macrophages were estimated by immunofluorescence and flow cytometry.
KIF26B was significantly overexpressed in cells and tissues of GC, and the higher expression of KIF26B was related to GC metastasis and prognosis. According to in vivo experiments, KIF26B promoted tumor formation and metastasis of GC. KIF26B expression was positively associated with CAFs’ degree of infiltration. Moreover, CAFs could regulate M2-type polarization of macrophages, affecting GC cells’ migration, angiogenesis, invasion, and epithelial-mesenchymal transition process.
KIF26B regulated M2 polarization of macrophage through activating CAFs, regulating the occurrence and metastasis of GC.
Core Tip:KIF26B could promote cancer-associated fibroblast activation, mediating macrophage M2 polarization and affecting the occurrence, lung metastasis, and abdominal metastasis of gastric cancer (GC). This study provides useful insights for exploring new mechanisms of GC and suppressing its progression.
- Citation: Huang LM, Zhang MJ. Kinesin 26B modulates M2 polarization of macrophage by activating cancer-associated fibroblasts to aggravate gastric cancer occurrence and metastasis. World J Gastroenterol 2024; 30(20): 2689-2708
- URL: https://www.wjgnet.com/1007-9327/full/v30/i20/2689.htm
- DOI: https://dx.doi.org/10.3748/wjg.v30.i20.2689
Kinesin superfamily proteins (KIFs), known as molecular motor proteins, were dependent on microtubule or ATP and were used to transport membrane organelle[1-3]. KIFs have been identified as potential molecular targets in cancer treatment[4-6]. This study was based on the KIFs, and bioinformatics analysis was conducted to screen out the most likely potential gene (KIF26B) involved in gastric cancer (GC) progression and prognosis.
KIFs participate in the regulation of multiple cancers. Silencing of KIF15 can reduce cell proliferation and enhance cell apoptosis to retard the progression of osteosarcoma[7]. KIF11 aggravates the progression of breast cancer and results in poor prognosis[8]. KIF20B strengthens tumorigenesis in tongue cancer[9]. Suppression of KIF15 weakens cell proliferation to inhibit the progression of triple-negative breast cancer[10]. A study has confirmed the role of the KIFs in GC, such as KIF15 promotes GC progression by promoting proliferation and inhibiting apoptosis[11], and KIF2A downregulation inhibits GC cell invasion by inhibiting MT1-MMP[12]. Little research has focused on the regulation of GC progression by KIF26B. One study from 2017 suggests that KIF26B promotes GC cell proliferation and metastasis through VEGF pathway activation[13].
In recent years, many literatures have verified that tumor microenvironment (TME) promotes tumor occurrence, progression, and metastasis[14-16]. One of the main functions of TME is to stimulate the immunosuppressive envi-ronment around tumors through various mechanisms[17,18]. Cancer-associated fibroblasts (CAFs) are the key cells in TME[18,19]. According to pan-cancer analysis, KIF26B expression is highly associated with CAF immune infiltration[20].
Macrophages interact with tumor cells in the TME and cause tumor progression[21]. M1 polarization of macrophage is characterized by increased pro-inflammatory activity, enhanced antigen presentation, and tumor growth inhibition[22,23]. Macrophages’ M2 polarization contributes to malignant angiogenesis, tumor cell proliferation, and growth[24,25]. Cancer cells regulate macrophages to aggravate tumor metastasis[26]. A study has illustrated the relationship axis between CAFs/macrophages/cancer cells, indicating that CAFs can promote macrophage M2 polarization by secreting CXCR12, thereby affecting cancer cell behavior and worsening cancer prognosis[27].
We hypothesized that KIF26B might regulate immune suppression and metastasis of GC through influencing CAF immune infiltration in this study.
We obtained tumor and adjacent normal tissues in pairs (n = 50) from GC patients from July 2019 to March 2023. This study was approved by the Medical Ethics Committee of The 901st Hospital of PLA (No. 202311006). The written consent from each participant has been acquired.
To verify GC patients in the high-KIF26B or low-KIF26B expression subgroups, the Kaplan-Meier analysis with the “survminer” R package was performed. The log-rank test was utilized to determine the significance of differences.
We extracted total RNAs from frozen tissues by the Trizol reagent (Invitrogen, Carlsbad, CA, United States). Complementary DNA from RNAs was synthesized through the reverse transcription kit (Invitrogen, Carlsbad, CA, United States). Quantitative real-time polymerase chain reaction (qRT-PCR) was made through a SYBR® Green qRT-PCR Kit (Promega, Madison, WI, United States). We calculated relative mRNA expression levels by means of the 2-ΔΔct method (GAPDH as the internal reference).
Primer sequences: KIF26B, forward: 5’-CCACCUCUUU GAGAAGGATT-3’, reverse: 5’-UUCCUUCUCAAAGAGGUGGTT-3’; α-SMA, forward: 5’-CGCCCTCGCCACCAGATCTG-3’, reverse: 5’-TAGCCTTCATAGATG GGGAC-3’; ACTA2, forward: 5’-GAGGGAAGGTCCTAACAGCC-3’, reverse: 5’-GCTTCACAGGATTCCCGTCT-3’; CXCL12, forward, 5’-CCGCGCTCTGCCT CAGCGACGGGAAG-3’, reverse, 5’-CTTGTTTAAAGCTTTCTCCAGGTACT-3’; FAP, forward: 5’-TGGGTGTCCAGT-GAACGAGTATG-3’, reverse: 5’-TGTATTT CTTGGTCTGTGCGGC-3’; ITGB1, forward: 5’-CCTCTCAGCCTCCAGCGTTG-3’, reverse: 5’-TGCTCTTGCTCACTCACACTCC-3’; PDPN, forward: 5’-CGAAGATG ATGTGGTGACTC-3’, reverse: 5’-CGATGCGAATGCCTGTTAC-3’; THY1, forward: 5’-GAAGGTCCTCTACTT ATCCGCC-3’, reverse: 5’-TGATGCCCTCACA CTTGACCAG-3’; GAPDH, forward: 5’-GGTGAAGGTCGGAGTCAACG-3’, reverse: 5’-CAAAGTTGTCATGGATGHACC-3’.
We collected paraffin sections of GC tumor tissues and normal adjacent tissues (n = 3) from GC patients as well as tumor tissues from mice. Tumor tissues were fixed, embedded (paraffin), and sliced into sections (4 μm thick). We deparaffinized paraffin sections and then rehydrated them, and restoring the antigen was carried out by means of citrate buffer at a high temperature and pH of 6.0. We incubated these sections mixed with primary antibody anti-KIF26B (ab121952, 1/200, Abcam), anti-Ki67 (ab15580, 0.1 µg/mL, Abcam), anti-α-SMA (ab5694, 1/200, Abcam) and anti-CD163 (ab182422, 1/500, Abcam) at 4 °C overnight. These sections were washed by incubation with secondary antibodies Goat F (ab')2 Anti-Rabbit IgG F (ab')2 (HRP) (ab6112, 1: 500, Abcam) for 30 min. The sections were stained with hematoxylin and diaminobenzidine and observed under a microscope. The immunohistochemistry (IHC) images were analyzed through Image J software.
We purchased HGC-27/AGS and GES-1 cells from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured them in RPMI-1640 medium with 10% foetal bovine serum (FBS) and 1% penicillin at 37 °C in an incubator with 5% CO2.
The plasmids (RiboBio, Beijing, China), including sh-KIF26B, pcDNA-KIF26B, and negative controls (sh-NC, pcDNA-NC) were utilized. We transfected above mentioned plasmids into HGC-27 and AGS cells with Lipofectamine 3000 (Invitrogen, United States) and cultured them for 24 h. The transfection efficiency was detected utilizing western blotting.
We bought mice (5-wk-old; BALB/c nude mice; males) from Vital River Laboratories (Beijing, China). All mice were maintained under controlled conditions (12 h light/dark, temperature: 22–24 °C, humidity: 40%–60%) and were offered free food and sterilized water. AGS cells (5 × 106) transfected with sh-KIF26B/sh-NC were injected into the mice’s inguinal skin. After four weeks, we killed nude mice with an overdose of pentobarbital. All animal experiments were approved by the Animal Ethics Committee of Beijing Viewsolid Biotechnology Co. LTD (No. VS2126A00168). The authors read the ARRIVE Guidelines, and the manuscript was prepared and revised according to the ARRIVE Guidelines. After 28 d, mice were euthanized (the cervical dislocation method), and tumors were removed and weighed. The nodules of lung metastasis and intraperitoneal metastasis were counted. The experimental methods of mice were shown in Supplementary Figure 1.
We dissolved the HGC-27 cells with RIPA buffer (Beyotime, Shanghai, China), and the total protein was purified and quantified through bicinchoninic acid protein kits (ThermoFisher, United States). We detached proteins by 10% SDS-PAGE and moved them to PVDF membranes. After blocking with skimmed milk (5%), we incubated proteins with the primary antibodies anti-KIF26B (1:1000, FNab04559, FineTest, Wuhan, China), anti-E-cadherin (1:2000, ab40772, Abcam), anti-MMP-2 (1:5000, ab92536, Abcam), anti-N-cadherin (1:2000, ab76011, Abcam), anti-MMP-9 (1:5000, ab76003, Abcam) and anti-β-actin (1:2000, ab8227, Abcam) overnight at 4 °C. Proteins were incubated with the anti-rabbit secondary antibody (1:5,000; SA00001-2, SanYing, China) for one hour. We examined protein blots by the ECL chemiluminescent system. Image J was applied for the quantification of protein blots.
We resuspended the HGC-27 cells (1 × 105) with 300 µL of complete medium, plated them in 96-well plates, and collected the supernatant after 3 d. ELISA was performed according to ELISA kits’ instructions (Invitrogen, United States). We detected and recorded the absorbance value at 450 nm.
In order to evaluate the ability of cell invasion, the transwell chamber was used. The supernatant (500 μL) collected from HGC-27 or CAFs was added into the upper chamber. Into the bottom chamber, we appended the serum-free medium (about 200 μL). Then, the cell plate was cultured for 2 d at 37 °C with 5% CO2. We removed the bottom chamber’s cells with cotton swabs, and the upper chamber’s cells were exposed to crystal violet (0.2%) to stain for 5 min. We used the inverted microscope for counting the number of invasion cells.
We cultured THP-1 monocytes in 6-well plates (106 cells/well) in RPMI medium together with 5% FBS and the supernatant of CAFs transfected with sh-KIF26B, pcDNA-KIF26B, and negative controls for 72 h. The macrophages were separated and incubated in a complete medium with 0.02% NaN3 for 30 min on ice. The incubation of macrophages with blocking antibodies anti-CD14/anti-CD206 and anti-F4/80/anti-CD206 (BD Biosciences Pharmingen, United States) was done for 20 min in Brilliant stain buffer on ice. We fixed the cells with 1% paraformaldehyde and performed the flow cytometric acquisition by BD LSR Fortessa (Flow cytometry, BD Biosciences Pharmingen, United States). The data analysis was made through FlowJoTM 10.8.1 software.
We used an immunofluorescence assay for the detection of CD206 expression. CD206 is the marker of M2 macrophages. THP-1 cells were cultured to 70% confluence on the glass side. After washing twice with 1 × PBS, we fixed the cells with 4% paraformaldehyde for 1 h at 25 °C. We permeabilized cells with 0.2% Triton X-100 (X100, Sigma) in 1 × PBS at 37 °C for 15 min and closed by 10% normal goat serum at room temperature for 1 h. We incubated the cells with an anti-Mannose Receptor antibody (Abcam, ab64693, dilution 1 µg/mL) overnight at 4 °C. After PBS-washing, we incubated the cells with the goat anti-mouse IgG Alexa 488 conjugated fluorescence secondary antibody (R37120, dilution 1/1000) for 1 h at room temperature. We stained the cells with DAPI and observed them under a microscope.
We incubated AGS and HGC-27 cells (3 × 105 cells/well) in 6-well plates with a complete medium with extracellular matrix molecule (10 μg/mL) until the cell monolayer was formed. We used a pipette tip (200 μL) to scratch a straight line on the plate bottom. Next, we cultured AGS and HGC-27 cells with a medium (serum-free) for 1 d. We observed the wound-healing distance of both types of cells and gauged them by an inverted microscope (Carl Zeiss, Oberkochen, Germany).
HUVECs (2 × 104) were inoculated onto the 96-well plate coated with Matrigel, and the supernatant of CAFs was added. After culturing for 8 h, the tubular structure of HUVECs was observed under the inverted microscope.
Data were represented by the mean ± SD of three independent experiments. GraphPad Prism 7.0 (GraphPad Software, United States) was used for statistical analysis. Multiple group differences were analyzed using a one-way analysis of variance. Two group comparisons were performed using a Student’s t-test. P < 0.05 indicates a statistical significance value.
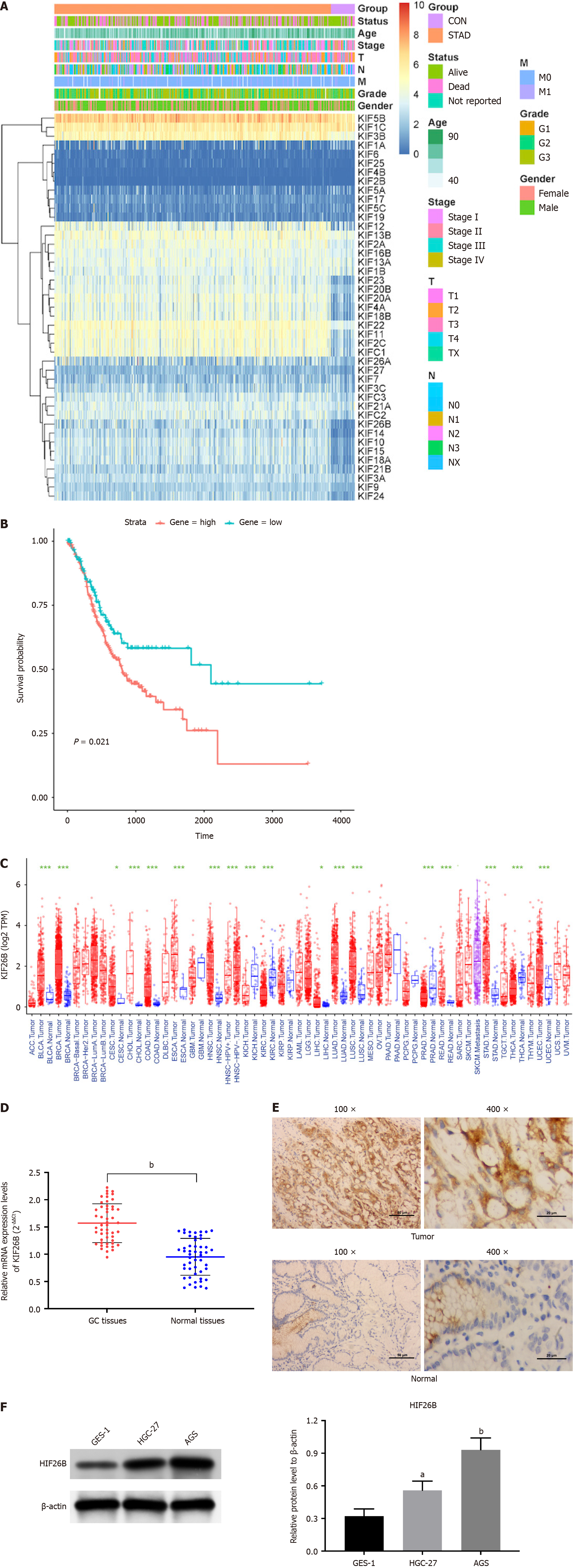
We examined kinesin superfamily expression levels in normal and tumor tissues and identified 18 DEGs (differentially expressed genes), of which 17 showed up-regulation, and 1 showed down-regulation (Figure 1A). We listed the DEGs of KIFs and quantified them, and found that the most significant difference in expression of KIF26B was observed in GC tissues (Supplementary Table 1). According to survival analysis, higher KIF26B expression resulted in poor prognosis (Figure 1B). We chose KIF26B as the follow-up study gene. We conducted a pan-cancer analysis on KIF26B and found that KIF26B was up-regulated in most cancers, with markedly significant differences in stomach adenocarcinoma (Figure 1C). qRT-PCR demonstrated that KIF26B expression was higher in GC tissues than in normal tissues (Figure 1D). IHC results presented high KIF26B (located in the cytoplasm) protein expression in GC tissues from patients (Figure 1E). Western blotting indicated that KIF26B protein expression in HGC-27/AGS cells was markedly higher compared with that in GES-1 cells (Figure 1F). A significant relation between high levels of KIF26B and infiltration depth, distant metastasis, lymph node metastasis, and tumor-node-metastasis (TNM) staging (T1 + T2 vs T3 + T4) was observed (Table 1).
| Variable | Total | KIF26B expression (n = 50) | P value | |
| Low | High | |||
| Age | 0.5215 | |||
| < 60 yr | 18 | 9 | 9 | |
| 60 yr | 32 | 19 | 13 | |
| Sex | 0.7412 | |||
| Female | 23 | 13 | 10 | |
| Male | 27 | 14 | 13 | |
| Tumor size, cm | 0.0336a | |||
| < 2 | 32 | 17 | 15 | |
| 2 | 18 | 4 | 14 | |
| TNM stage | 0.0016b | |||
| I/II | 30 | 18 | 12 | |
| III/IV | 20 | 3 | 17 | |
| Lymph node metastasis | 0.0227a | |||
| Negative | 25 | 15 | 10 | |
| Positive | 25 | 7 | 18 | |
| Peritoneal metastasis | 0.0009c | |||
| Negative | 29 | 16 | 13 | |
| Positive | 21 | 2 | 19 | |
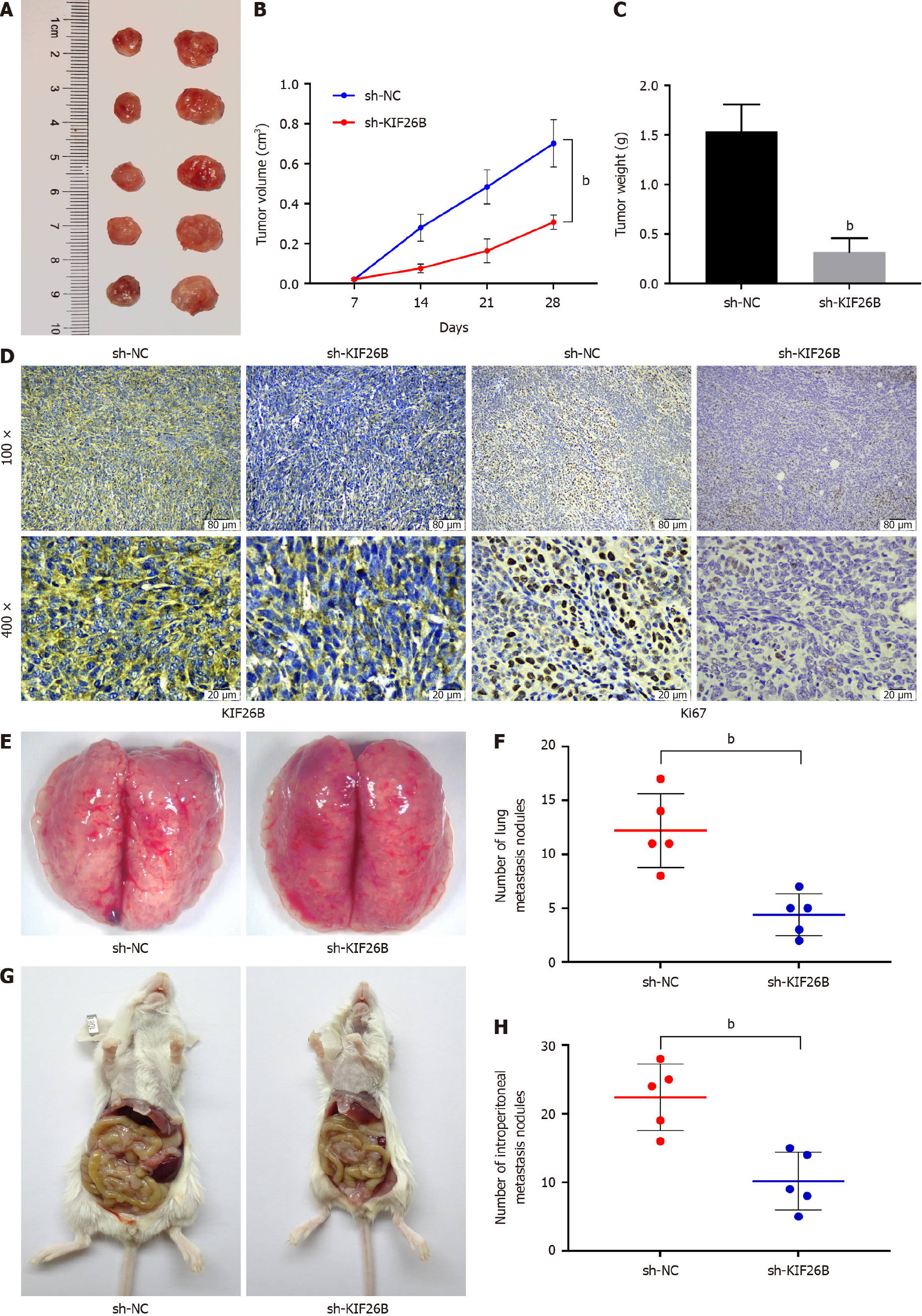
We determined a xenograft tumor nude mouse model. We learned from the xenograft tumor experiment that knockdown of KIF26B could inhibit tumor growth of GC (Figure 2A-C). Immunohistochemical results demonstrated that KIF26B and Ki67 (located in the nucleus) expression levels were decreased after KIF26B suppression in tumor tissues from mice, indicating that there was an inhibition on the proliferative activity of GC cells after knocking down KIF26B (Figure 2D). The GC cells in the sh-KIF26B group decreased their metastasis to the pulmonary lymph node compared with the sh-NC group (Figure 2E and F). Knockdown of KIF26B inhibited the metastasis of GC cells to the peritoneum (Figure 2G and H). Knockdown of KIF26B could inhibit the occurrence of tumors, lung metastasis, and abdominal metastasis of GC in vivo.
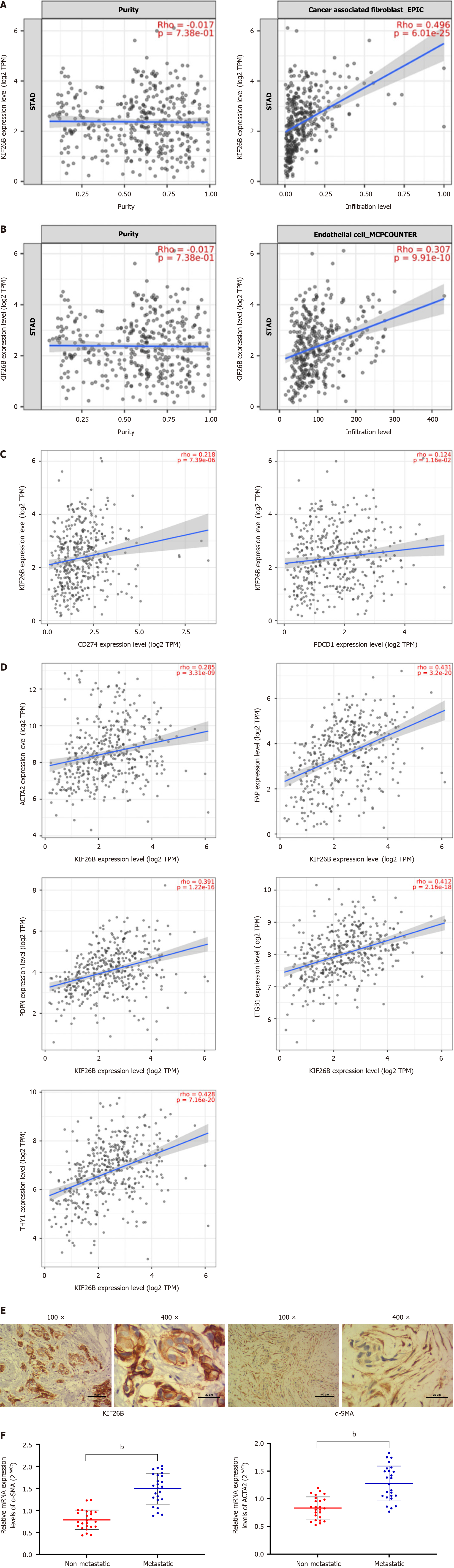
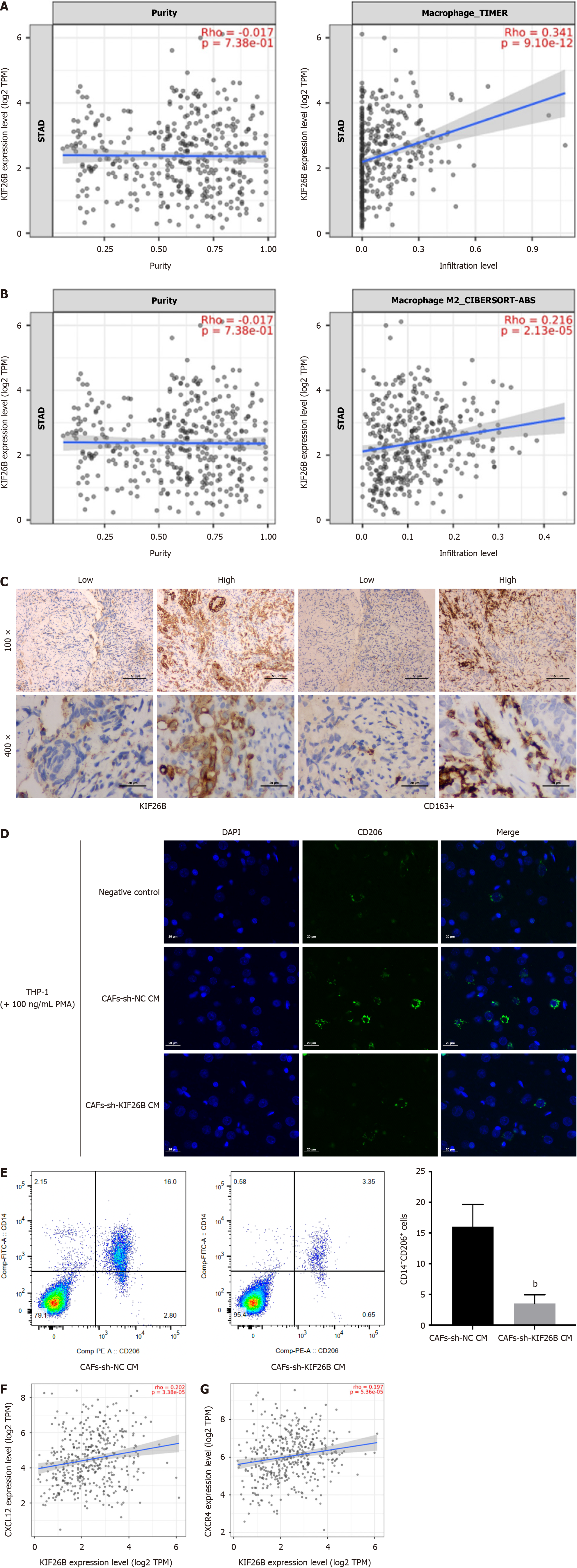
KIF26B is highly correlated with the infiltration degree of CAFs, followed by endothelial cells (Figure 3A and B). We found a positive correlation between expression levels of KIF26B and PD-1 (PDCD1) and PD-L1 (CD274) (Figure 3C). To verify the correlation between KIF26B and the CAFs, we utilized the Timer 2.0 database. We confirmed that KIF26B was significantly correlated with CAFs biomarkers, such as ACTA2, FAP, ITGB1, PDPN, and THY1 (Figure 3D). In the meanwhile, IHC experiments have found that both KIF26B and α-SMA (activation biomarker of CAFs) can be detected simultaneously in GC tissue from patients (Figure 3E). The results of qRT-PCR confirmed that α-SMA/ACTA2 mRNA in metastatic GC tissues were markedly higher while comparing with non-metastatic GC tissues (Figure 3F). High expression of KIF26B may affect the activation and infiltration of CAFs, influencing the metastasis of GC.
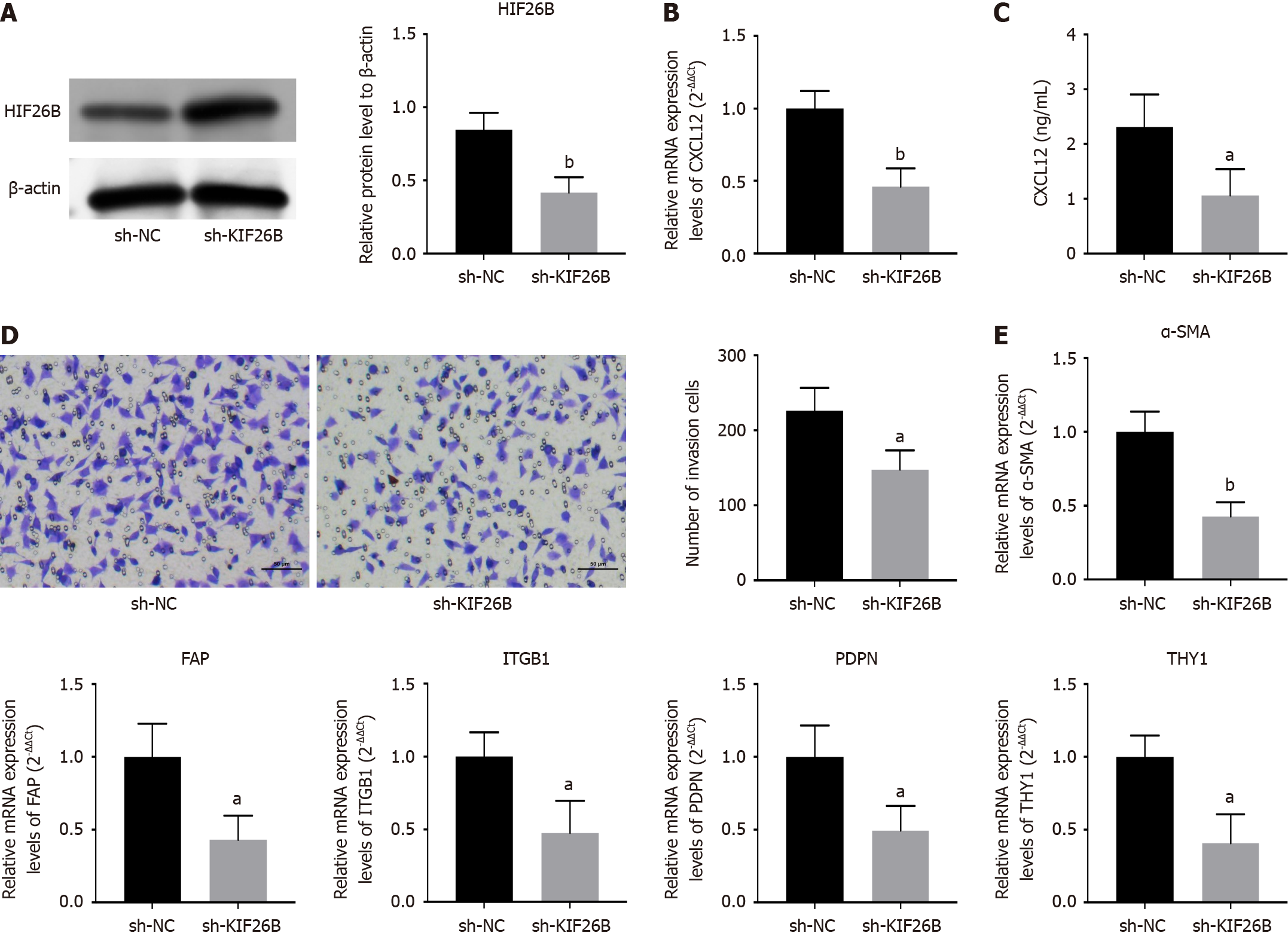
We incubated primary human foreskin fibroblasts (HFF) with the supernatant of sh-KIF26B and sh-NC-transfected HGC-27 cells. The transfection’s efficacy was confirmed by western blotting (Figure 4A). qRT-PCR and ELISA assays were used for detecting chemokines (CXCL12) in HFF cells, and KIF26B knockdown decreased CXCL12 expression, indicating reducing the chemotaxis of HFF cells (Figure 4B and C). The transwell assay also demonstrated that suppression of KIF26B could decline HFF’s invasive ability (Figure 4D). We detected the biomarkers of CAFs, including α-SMA, FAP, ITGB1, PDPN, and THY1, and inhibition of KIF26B decreased the expression levels of CAFs biomarkers (Figure 4E). It indicates that KIF26B can promote the activation of fibroblasts to form the CAF phenotype.
A positive relation was found between KIF26B expression and the number of M2 macrophages (Figure 5A and B). Immunohistochemical results reported that KIF26B and CD163+ expressions were increased in GC tissues (high infiltration) from patients, indicating that KIF26B expression was positively related with the CD163+ expression (Biomarker of M2 macrophage) (Figure 5C). We transfected primary CAFs with sh-KIF26B, collected the supernatant and incubated THP-1 cells (induced by 100 ng/mL). Immunofluorescence assay detected CD206 (Surface biomarker of M2 macrophage) in THP-1 cells and indicated that the fluorescence intensity in the sh-KIF26B group was decreased markedly compared with sh-NC group (Figure 5D). We counted macrophages using flow cytometry and found the number of M2 macrophages was a decreased in the sh-KIF26B group comparing with sh-NC group (Figure 5E). Data analysis found a significant positive correlation between KIF26B and chemokine CXCL12 expression, and its corresponding receptor CXCR4 (Figure 5F and G). KIF26B in GC cells may affect the M2 polarization of macrophage through CXCL12 secreted by CAFs.
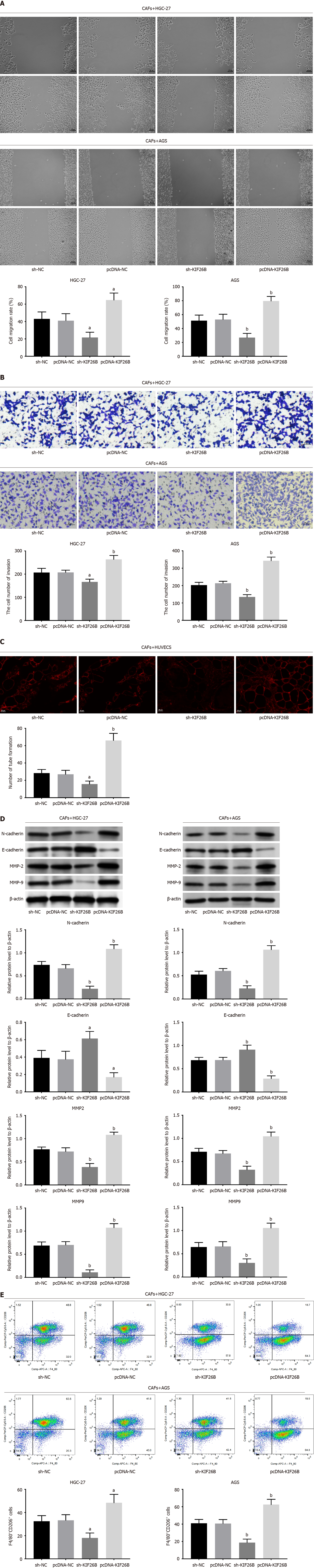
We collected the supernatant of CAFs that transfected with sh-NC, sh-KIF26B, pcDNA-NC, and pcDNA-KIF26B as a conditioned medium to incubate GC cells (AGS and HGC-27 cells). Overexpression of KIF26B enhanced the migration and invasiveness of HGC-27 and AGS cells when compared with the negative control group, while knockdown of KIF26B could reduce the migration and invasiveness of AGS and HGC-27 cells (Figure 6A and B). Tube formation assay reported that the tube forming ability of cells was enhanced when KIF26B was overexpressed, and it was inhibited after KIF26B knockdown (Figure 6C). We detected epithelial-mesenchymal transition (EMT)-related protein expressions in HGC-27 and AGS cells, such as N-cadherin, E-cadherin, MMP2, and MMP9 (Figure 6D). While comparing with the control group, the sh-KIF26B group had lower N-cadherin, MMP2, and MMP9 protein expression levels, while the pcDNA-KIF26B group had higher N-cadherin, MMP2, and MMP9 protein expression levels, while the expression level of E-cadherin was opposite to other proteins (Figure 6D). M2 polarized macrophages’ infiltration quantity in the pcDNA-KIF26B group was higher compared to the control group, while the infiltration quantity of M2 polarized macrophages in the sh-KIF26B group was lower (Figure 6E). Based on the above experimental results, it is revealed that KIF26B can regulate M2 polarization of macrophages through fibroblast activation, regulating invasiveness, angiogenesis, and EMT processes of GC cells.
GC is one of the most severe cancers globally[28,29]. Exploring GC progression mechanisms and developing new treatments have become necessary.
By analyzing the TCGA database, we discovered overexpression of KIF26B in most tumors. Highly expressed KIF26B was found in GC tissues and cells. Through survival analysis, we found that higher KIF26B expression was associated with poor GC prognosis. The clinical data analysis showed a high level of KIF26B significant association with invasion depth, lymph node metastasis, and TNM stage of tumor. Like our results, Teng et al[30] reported that the level of KIF26B is significantly increased in breast cancer cells and tissues, and KIF26B level was positively related to the tumor size, TNM grading, and differentiation of breast cancer patients[30]. Data from Wang et al[31] suggest that KIF26B causes the occurrence of colorectal cancer (CC) and acts as a possible therapeutic target for CC[31]. Higher expression of KIF26B is observed in hepatocellular carcinoma tissues and cell lines[32]. Also, increased KIF26B expression is related to poor survival and differentiation and advanced TNM[32].
GC metastasis is the leading cause of death for patients[33], and the 5-year survival rate of patients in China with metastatic GC is less than 10%[34,35]. We found that knockdown of KIF26B decreased the occurrence, lung metastasis, and abdominal metastasis of GC. Previous research showed that KIF26B had an impact on the metastasis of breast cancer[30] and GC[13]. These are good supports for our results.
CAFs are essential to various tumors, such as GC[36,37]. CAFs aid in the invasion and metastasis of tumors during their occurrence and development[38]. The interaction mechanism between GC cells and CAFs is still unclear. A previous study found that knockdown of KIF26B inhibited the activation of renal fibroblasts[39]. We indicated that the high expression level of KIF26B promoted the activation and infiltration of CAFs through data analysis and experiments on GC tissues and cell lines. We found a positive relation between KIF26B expression and M2 macrophage activation. Cai et al[40] show that the fibroblast activating protein is associated with the invasion of M2 macrophages in gastrointestinal cancer[40]. Our subsequent experimental findings are similar to those of Cai et al[40] where KIF26B enhances crosstalk between tumor fibroblasts and macrophages, mediating the M2 polarization of macrophages. We found that KIF26B in GC cells might affect macrophage M2 polarization through CXCL12 secreted by CAFs.
The invasion[41], tube formation[42,43], and EMT process[44] of tumor cells are related to the cancer progression. Our research results indicated that KIF26B could regulate M2 polarization of macrophages through fibroblast activation, regulating tumor cell migration, invasion, angiogenesis, and EMT processes and completing the regulation of GC development and metastasis.
Although advances in basic and clinical studies have reduced the mortality rate of GC over the past decade, its prognosis remains poor due to incomplete diagnosis, high metastasis rate, and high chemotherapy resistance. We found that higher KIF26B expression could promote CAFs activation, mediating macrophage M2 polarization and affecting the occurrence, lung metastasis, and abdominal metastasis of GC. This study provides useful insights for exploring new mechanisms of GC and delaying its progression.
| 1. | Miki H, Setou M, Kaneshiro K, Hirokawa N. All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci U S A. 2001;98:7004-7011. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 486] [Article Influence: 19.4] [Reference Citation Analysis (2)] |
| 2. | Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88:1089-1118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 369] [Article Influence: 20.5] [Reference Citation Analysis (3)] |
| 3. | Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682-696. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1142] [Cited by in RCA: 1349] [Article Influence: 79.4] [Reference Citation Analysis (3)] |
| 4. | Li X, Tai Y, Liu S, Gao Y, Zhang K, Yin J, Zhang H, Wang X, Li X, Zhang D. Bioinformatics analysis: relationship between adrenocortical carcinoma and KIFs. Biotechnol Genet Eng Rev. 2022;1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (2)] |
| 5. | Li TF, Zeng HJ, Shan Z, Ye RY, Cheang TY, Zhang YJ, Lu SH, Zhang Q, Shao N, Lin Y. Overexpression of kinesin superfamily members as prognostic biomarkers of breast cancer. Cancer Cell Int. 2020;20:123. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 89] [Article Influence: 14.8] [Reference Citation Analysis (2)] |
| 6. | Lucanus AJ, Yip GW. Kinesin superfamily: roles in breast cancer, patient prognosis and therapeutics. Oncogene. 2018;37:833-838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (3)] |
| 7. | Wu Z, Zhang H, Sun Z, Wang C, Chen Y, Luo P, Yan W. Knockdown of Kinesin Family 15 Inhibits Osteosarcoma through Suppressing Cell Proliferation and Promoting Cell Apoptosis. Chemotherapy. 2019;64:187-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (2)] |
| 8. | Zhou J, Chen WR, Yang LC, Wang J, Sun JY, Zhang WW, He ZY, Wu SG. KIF11 Functions as an Oncogene and Is Associated with Poor Outcomes from Breast Cancer. Cancer Res Treat. 2019;51:1207-1221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 52] [Article Influence: 6.5] [Reference Citation Analysis (2)] |
| 9. | Li ZY, Wang ZX, Li CC. Kinesin family member 20B regulates tongue cancer progression by promoting cell proliferation. Mol Med Rep. 2019;19:2202-2210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (2)] |
| 10. | Sheng J, Xue X, Jiang K. Knockdown of Kinase Family 15 Inhibits Cancer Cell Proliferation In vitro and its Clinical Relevance in Triple-Negative Breast Cancer. Curr Mol Med. 2019;19:147-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (2)] |
| 11. | Ding L, Li B, Yu X, Li Z, Li X, Dang S, Lv Q, Wei J, Sun H, Chen H, Liu M, Li G. KIF15 facilitates gastric cancer via enhancing proliferation, inhibiting apoptosis, and predict poor prognosis. Cancer Cell Int. 2020;20:125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (2)] |
| 12. | Zhao P, Lan F, Zhang H, Zeng G, Liu D. Down-regulation of KIF2A inhibits gastric cancer cell invasion via suppressing MT1-MMP. Clin Exp Pharmacol Physiol. 2018;45:1010-1018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (3)] |
| 13. | Zhang H, Ma RR, Wang XJ, Su ZX, Chen X, Shi DB, Guo XY, Liu HT, Gao P. KIF26B, a novel oncogene, promotes proliferation and metastasis by activating the VEGF pathway in gastric cancer. Oncogene. 2017;36:5609-5619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 54] [Article Influence: 6.0] [Reference Citation Analysis (2)] |
| 14. | Xiao Y, Yu D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther. 2021;221:107753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 1364] [Article Influence: 227.3] [Reference Citation Analysis (2)] |
| 15. | Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019;79:4557-4566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 750] [Cited by in RCA: 2329] [Article Influence: 332.7] [Reference Citation Analysis (2)] |
| 16. | Deepak KGK, Vempati R, Nagaraju GP, Dasari VR, S N, Rao DN, Malla RR. Tumor microenvironment: Challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol Res. 2020;153:104683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 434] [Article Influence: 72.3] [Reference Citation Analysis (2)] |
| 17. | Bader JE, Voss K, Rathmell JC. Targeting Metabolism to Improve the Tumor Microenvironment for Cancer Immunotherapy. Mol Cell. 2020;78:1019-1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 566] [Cited by in RCA: 725] [Article Influence: 120.8] [Reference Citation Analysis (2)] |
| 18. | Pitt JM, Marabelle A, Eggermont A, Soria JC, Kroemer G, Zitvogel L. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann Oncol. 2016;27:1482-1492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 1001] [Article Influence: 100.1] [Reference Citation Analysis (2)] |
| 19. | Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3347] [Cited by in RCA: 3595] [Article Influence: 179.8] [Reference Citation Analysis (1)] |
| 20. | Sun F, Lian Y, Wang J, Hu L, Luo J, Yu J. KIF26B in the Prognosis and Immune Biomarking of Various Cancers: A Pan-Cancer Study. J Oncol. 2022;2022:4829697. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (1)] |
| 21. | Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1523] [Cited by in RCA: 1691] [Article Influence: 84.6] [Reference Citation Analysis (2)] |
| 22. | Ma J, Liu L, Che G, Yu N, Dai F, You Z. The M1 form of tumor-associated macrophages in non-small cell lung cancer is positively associated with survival time. BMC Cancer. 2010;10:112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 339] [Cited by in RCA: 358] [Article Influence: 22.4] [Reference Citation Analysis (1)] |
| 23. | Ong SM, Tan YC, Beretta O, Jiang D, Yeap WH, Tai JJ, Wong WC, Yang H, Schwarz H, Lim KH, Koh PK, Ling KL, Wong SC. Macrophages in human colorectal cancer are pro-inflammatory and prime T cells towards an anti-tumour type-1 inflammatory response. Eur J Immunol. 2012;42:89-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 114] [Article Influence: 7.6] [Reference Citation Analysis (1)] |
| 24. | Rhee I. Diverse macrophages polarization in tumor microenvironment. Arch Pharm Res. 2016;39:1588-1596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 222] [Article Influence: 22.2] [Reference Citation Analysis (1)] |
| 25. | Zhang M, He Y, Sun X, Li Q, Wang W, Zhao A, Di W. A high M1/M2 ratio of tumor-associated macrophages is associated with extended survival in ovarian cancer patients. J Ovarian Res. 2014;7:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 359] [Cited by in RCA: 421] [Article Influence: 35.1] [Reference Citation Analysis (1)] |
| 26. | Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X, Liu Q, Dou R, Xiong B. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer. 2019;18:64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 238] [Cited by in RCA: 636] [Article Influence: 90.9] [Reference Citation Analysis (1)] |
| 27. | Li X, Sun Z, Peng G, Xiao Y, Guo J, Wu B, Li X, Zhou W, Li J, Li Z, Bai C, Zhao L, Han Q, Zhao RC, Wang X. Single-cell RNA sequencing reveals a pro-invasive cancer-associated fibroblast subgroup associated with poor clinical outcomes in patients with gastric cancer. Theranostics. 2022;12:620-638. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 237] [Article Influence: 59.3] [Reference Citation Analysis (1)] |
| 28. | Deng H, Gao J, Cao B, Qiu Z, Li T, Zhao R, Li H, Wei B. LncRNA CCAT2 promotes malignant progression of metastatic gastric cancer through regulating CD44 alternative splicing. Cell Oncol (Dordr). 2023;46:1675-1690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (1)] |
| 29. | Chen G, Luo D, Qi X, Li D, Zheng J, Luo Y, Zhang C, Ren Q, Lu Y, Chan YT, Chen B, Wu J, Wang N, Feng Y. Characterization of cuproptosis in gastric cancer and relationship with clinical and drug reactions. Front Cell Dev Biol. 2023;11:1172895. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (1)] |
| 30. | Teng Y, Guo B, Mu X, Liu S. KIF26B promotes cell proliferation and migration through the FGF2/ERK signaling pathway in breast cancer. Biomed Pharmacother. 2018;108:766-773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (2)] |
| 31. | Wang J, Cui F, Wang X, Xue Y, Chen J, Yu Y, Lu H, Zhang M, Tang H, Peng Z. Elevated kinesin family member 26B is a prognostic biomarker and a potential therapeutic target for colorectal cancer. J Exp Clin Cancer Res. 2015;34:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 35] [Article Influence: 3.2] [Reference Citation Analysis (1)] |
| 32. | Li H, Shen S, Chen X, Ren Z, Li Z, Yu Z. miR-450b-5p loss mediated KIF26B activation promoted hepatocellular carcinoma progression by activating PI3K/AKT pathway. Cancer Cell Int. 2019;19:205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 5.3] [Reference Citation Analysis (1)] |
| 33. | Yu B, Zhu N, Fan Z, Li J, Kang Y, Liu B. miR-29c inhibits metastasis of gastric cancer cells by targeting VEGFA. J Cancer. 2022;13:3566-3574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (1)] |
| 34. | Chen W, Sun K, Zheng R, Zeng H, Zhang S, Xia C, Yang Z, Li H, Zou X, He J. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 483] [Cited by in RCA: 714] [Article Influence: 89.3] [Reference Citation Analysis (2)] |
| 35. | Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11444] [Cited by in RCA: 13325] [Article Influence: 1332.5] [Reference Citation Analysis (4)] |
| 36. | Zhao X, He Y, Gao J, Fan L, Li Z, Yang G, Chen H. Caveolin-1 expression level in cancer associated fibroblasts predicts outcome in gastric cancer. PLoS One. 2013;8:e59102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (1)] |
| 37. | Özdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, Laklai H, Sugimoto H, Kahlert C, Novitskiy SV, De Jesus-Acosta A, Sharma P, Heidari P, Mahmood U, Chin L, Moses HL, Weaver VM, Maitra A, Allison JP, LeBleu VS, Kalluri R. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719-734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1955] [Cited by in RCA: 2027] [Article Influence: 168.9] [Reference Citation Analysis (1)] |
| 38. | Chung HW, Lim JB. Role of the tumor microenvironment in the pathogenesis of gastric carcinoma. World J Gastroenterol. 2014;20:1667-1680. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 65] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (1)] |
| 39. | Yamamura Y, Iwata Y, Furuichi K, Kato T, Yamamoto N, Horikoshi K, Ogura H, Sato K, Oshima M, Nakagawa S, Miyagawa T, Kitajima S, Toyama T, Hara A, Sakai N, Shimizu M, Horike S, Daikoku T, Nishinakamura R, Wada T. Kif26b contributes to the progression of interstitial fibrosis via migration and myofibroblast differentiation in renal fibroblast. FASEB J. 2022;36:e22606. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (1)] |
| 40. | Cai J, Yang D, Sun H, Xiao L, Han F, Zhang M, Zhou L, Jiang M, Jiang Q, Li Y, Nie H. A multifactorial analysis of FAP to regulate gastrointestinal cancers progression. Front Immunol. 2023;14:1183440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (1)] |
| 41. | Li S, Cong X, Gao H, Lan X, Li Z, Wang W, Song S, Wang Y, Li C, Zhang H, Zhao Y, Xue Y. Tumor-associated neutrophils induce EMT by IL-17a to promote migration and invasion in gastric cancer cells. J Exp Clin Cancer Res. 2019;38:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 218] [Article Influence: 31.1] [Reference Citation Analysis (1)] |
| 42. | Gianni-Barrera R, Di Maggio N, Melly L, Burger MG, Mujagic E, Gürke L, Schaefer DJ, Banfi A. Therapeutic vascularization in regenerative medicine. Stem Cells Transl Med. 2020;9:433-444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 60] [Article Influence: 10.0] [Reference Citation Analysis (1)] |
| 43. | Yang Y, Cao Y. The impact of VEGF on cancer metastasis and systemic disease. Semin Cancer Biol. 2022;86:251-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 188] [Article Influence: 47.0] [Reference Citation Analysis (1)] |
| 44. | Tian S, Peng P, Li J, Deng H, Zhan N, Zeng Z, Dong W. SERPINH1 regulates EMT and gastric cancer metastasis via the Wnt/β-catenin signaling pathway. Aging (Albany NY). 2020;12:3574-3593. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 157] [Article Influence: 26.2] [Reference Citation Analysis (1)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/