Published online Jan 28, 2021. doi: 10.3748/wjg.v27.i4.305
Peer-review started: November 9, 2020
First decision: December 8, 2020
Revised: December 19, 2020
Accepted: December 27, 2020
Article in press: December 27, 2020
Published online: January 28, 2021
Processing time: 76 Days and 9.3 Hours
Genome-wide association studies of complex diseases, including nonalcoholic fatty liver disease (NAFLD), have demonstrated that a large number of variants are implicated in the susceptibility of multiple traits — a phenomenon known as pleiotropy that is increasingly being explored through phenome-wide association studies. We focused on the analysis of pleiotropy within variants associated with hematologic traits and NAFLD. We used information retrieved from large public National Health and Nutrition Examination Surveys, Genome-wide association studies, and phenome-wide association studies based on the general population and explored whether variants associated with NAFLD also present associations with blood cell-related traits. Next, we applied systems biology approaches to assess the potential biological connection/s between genes that predispose affected individuals to NAFLD and nonalcoholic steatohepatitis, and genes that modulate hematological-related traits—specifically platelet count. We reasoned that this analysis would allow the identification of potential molecular mediators that link NAFLD with platelets. Genes associated with platelet count are most highly expressed in the liver, followed by the pancreas, heart, and muscle. Conversely, genes associated with NAFLD presented high expression levels in the brain, lung, spleen, and colon. Functional mapping, gene prioritization, and functional analysis of the most significant loci (P < 1 × 10-8) revealed that loci involved in the genetic modulation of platelet count presented significant enrichment in metabolic and energy balance pathways. In conclusion, variants in genes influencing NAFLD exhibit pleiotropic associations with hematologic traits, particularly platelet count. Likewise, significant enrichment of related genes with variants influencing platelet traits was noted in metabolic-related pathways. Hence, this approach yields novel mechanistic insights into NAFLD pathogenesis.
Core Tip: Pleiotropy within variants associated with hematologic traits and nonalcoholic fatty liver disease.
- Citation: Pirola CJ, Salatino A, Sookoian S. Pleiotropy within gene variants associated with nonalcoholic fatty liver disease and traits of the hematopoietic system. World J Gastroenterol 2021; 27(4): 305-320
- URL: https://www.wjgnet.com/1007-9327/full/v27/i4/305.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i4.305
Nonalcoholic fatty liver disease (NAFLD) is regarded as the most prevalent chronic liver disease[1]. A worldwide increase in NAFLD prevalence is acknowledged not only in the adult but also the pediatric population[1]. Consequently, NAFLD has become a serious health issue on a global scale.
Like many other complex diseases, NAFLD develops due to the combined effect of environmental and genetic factors[2-4]. NAFLD presents phenotypic complexity and inter-individual variability, implying that its natural course is characterized by different histological stages-from simple fat accumulation to steatohepatitis (NASH), cirrhosis, and eventually to hepatocellular carcinoma[1]—and considerable variability in disease progression exists among affected patients. One of the proposed factors contributing to the observed inter-patient differences in the disease prognosis and severity is genetic susceptibility[2-4], which might explain up to approximately 20% of the disease variance[5].
Furthermore, NAFLD has not only a high degree of comorbidity with disorders of the metabolic syndrome, including type 2 diabetes, obesity, and cardiovascular disease, but also shared disease mechanisms and disease pathways[6]. These comorbidities have a strong negative impact on the course of NAFLD and vice versa, whereby the presence of NAFLD substantially modifies the course and prognosis of metabolic syndrome-associated diseases[6]. More importantly, the impact of long-term consequences of these comorbidities on cardiovascular health is, at least in part, independent of the presence of general obesity. In fact, it was shown that lean patients with NAFLD have an altered metabolic profile mostly related to increased visceral adiposity that predispose them to cardiovascular risks[7].
Notably, knowledge gained from genome-wide and exon-wide association studies [Genome-wide association studies (GWAS) and EWAS, respectively] of complex diseases, including NAFLD, shows that a large number of single nucleotide polymorphisms are implicated in the susceptibility of multiple traits, which is known as pleiotropy. Phenome-wide association studies (PheWAS) that exploit a significant amount of clinical characteristics gathered mostly from electronic clinical records have thus become a powerful strategy for uncovering pleiotropy.
Among the many traits explored to date in large GWAS/EWAS and PheWAS, including disease traits and biochemical parameters, pleiotropy within gene variants associated with NAFLD and biochemical traits of the hematopoietic system is remarkably consistent across different datasets, specifically those concerning platelet count and platelet volume[8-10]. However, the mechanisms behind the biological connection between NAFLD and platelet-related traits remain poorly understood.
Therefore, in this review, we focused on the analysis of pleiotropy within variants associated with biochemical hematologic traits and NAFLD. We took advantage of information retrieved from large public GWAS and PheWAS based on the general population and examined whether variants known to be associated with NAFLD also exhibit associations with blood cell-related traits, specifically platelet-related phenotypes. After collecting that information, we adopted systems biology approaches to assess the potential biological connection/s between genes that predispose to NAFLD and NASH, and genes that modulate related hematological traits, including platelet count and platelet crit. We reasoned that focusing on clinically meaningful traits for which associations with liver disorders would be plausible from the biological perspective may help elucidate NAFLD biology. Likewise, these analyses would conceptually allow the identification of potential molecular mediators that link NAFLD with platelets.
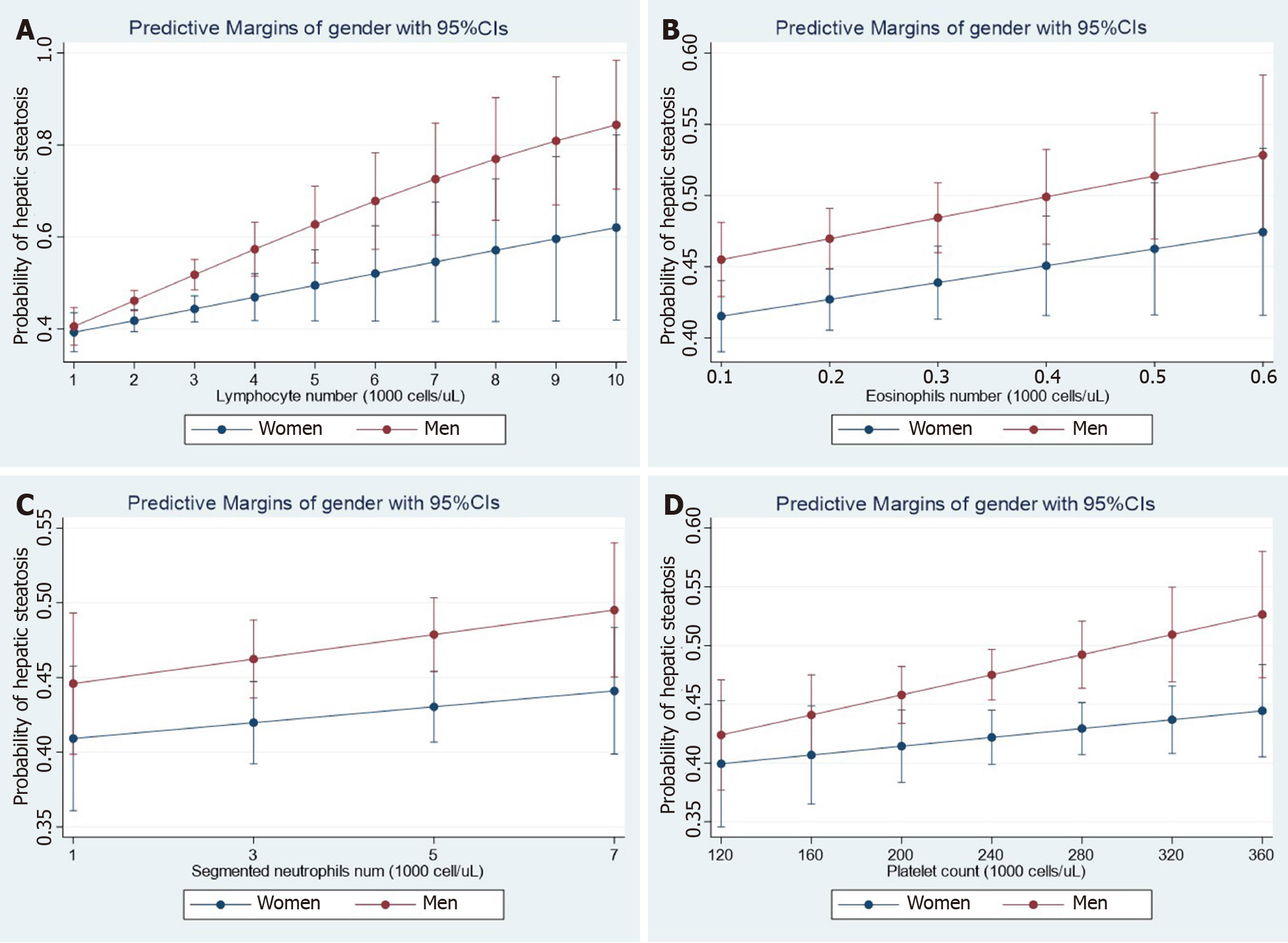
Recent observations shed light on the concept that NAFLD not only co-exists with cardiovascular disease, including functional and structural myocardial abnormalities[11], but is also associated with other diseases, including cancer, kidney disease, and hematological disorders, among many others[5]. To illustrate the relevance of the association between NAFLD and hematological traits, we analyzed data from the National Health and Nutrition Examination Surveys (NHANES) 2017-2018 database. The dataset and further information are freely available online https://www.cdc.gov/nchs/nhanes/index.htm). The NHANES are population-based surveys conducted by the National Center for Health Statistics of the Centers for Disease Control and Prevention of the United States. They are frequently used to study liver disease. The National Center for Health Statistics Research Ethics Review Board approved the NHANES protocol, and informed consent was obtained from all participants. Liver steatosis was defined by the controlled attenuation parameter (CAP) obtained via transient elastography (FibroScan®). Liver steatosis was diagnosed when CAP >268 dB/m was obtained, which is the threshold for significant steatosis based on a large study[12]. We modeled the relationship between NAFLD and relevant factors by linear logistic regression with an interaction term for the specific hematologic trait and gender while adjusting for subjects' demographic and clinical characteristics. Figure 1 shows the effect of white blood cells (WBC), including lymphocytes (Figure 1A), eosinophils (Figure 1B), neutrophils (Figure 1C), and platelet (Figure 1D) count number by gender. Myeloid and lymphoid lineages, as well as platelets show interaction effects with gender even after adjusting for log-transformed confounding factors such as age, waist circumference, diabetes, glycohemoglobin, total cholesterol, and systolic blood pressure. More importantly, NAFLD risk increased steadily with the number of different blood cell types, particularly in men.
These results are reproducibly observed in the Asian population. Wang et al[13]reported that WBC count is a significant factor associated with incident NAFLD in Han Chinese[13]. The authors observed that the association between WBC count and NAFLD remained valid after adjusting for confounding factors including age, gender, smoking, regular exercise, BMI, hypertension, hyperglycemia, and lipid traits[13]. Similarly, WBC count was found to be independently associated with NAFLD regardless of components of the metabolic syndrome in the Korean population[14,15].
For many biological reasons, the liver and platelets are biologically connected. For example: (1) The fetal liver is a privileged organ of megakaryocyte progenitor differentiation[16]; (2) Thrombocytopenia is a major and debilitating complication of liver cirrhosis[17]; (3) Platelets are involved in the process of fibrogenesis by remodeling the extracellular matrix[18] and secretion of cytokines, including platelet-derived growth factors[19]; and (4) Platelets functioning in the liver exhibit a collaborative effect with endothelial and Kupffer cells in liver regeneration[20]. A more comprehensive update on the role of platelets in liver disorders has been recently published[21].
Earlier evidence also demonstrates that platelets are involved in NASH-related complications. For example, we have observed that patients with NASH have high expression of TGFB1 (transforming grown factor β1)-mRNA in circulating platelets[22]. Platelets are also involved in the process of atherogenesis by upregulating many molecules[23], including TGFB1[24], which is also related to NAFLD and the risk of cardiovascular disease. Results yielded by a recent study suggest that platelet-mediated inflammation in NAFLD drives hepatocellular carcinogenesis[25].
The nonsynonymous rs738409 C/G variant in PNPLA3 (patatin-like phospholipase domain containing protein 3, also known as adiponutrin or calcium-independent phospholipase A2-epsilon), which encodes the amino acid substitution I148M, is regarded as the major genetic variant associated with the susceptibility to NAFLD and NASH[26,27].
The rs738409 has also been associated with alcoholic liver disease[28-30], hepatitis C[31], hepatitis B[32], and hepatocellular carcinoma[33],[34].
Results of a large PheWAS performed in subjects of European ancestry (816903 participants) with genome-wide genotyped data linked to phenotypic information, including the United Kingdom Biobank cohort, 23andMe cohort, FINRISK (working-age population of Finland), and Children’s Hospital of Philadelphia, confirmed that rs738409 presents pleiotropic effects beyond the liver. For instance, rs738409-G was associated with increased risk of type 2 diabetes and decreased risk of high total cholesterol, acne, gout, and gallstones; all these associations remained significant after adjusting for elevated transaminases[35].
In addition, it was suggested that rs738409 might be used to predict race-related hepatotoxicity in pediatric patients with acute lymphoblastic leukemia[9].
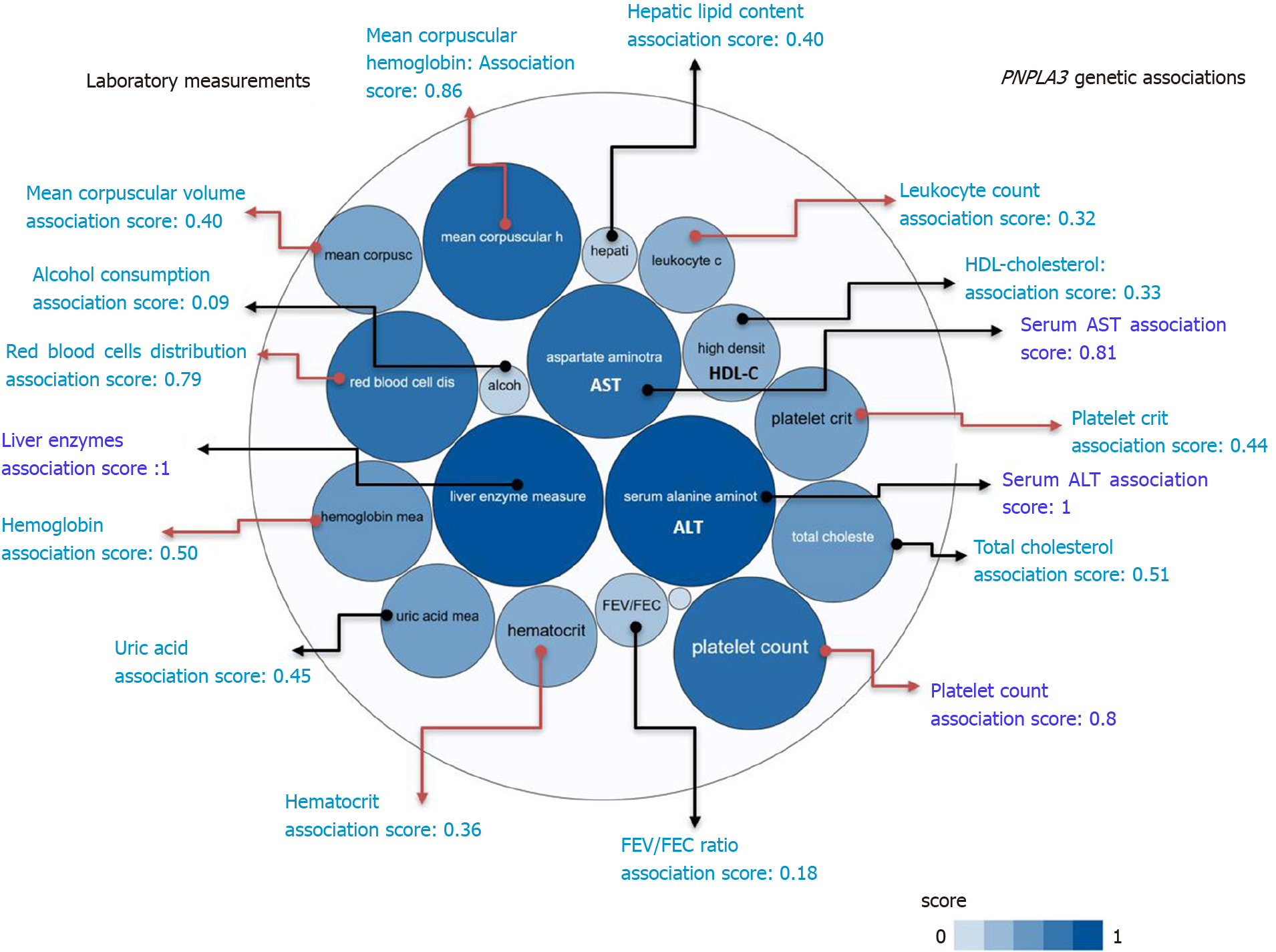
Another study revealed the association of rs738409 with mean corpuscular hemoglobin (P = 6 × 10-9) based on the analysis of individuals of 116666 British ancestry[36]. Consistently, Kichaev et al[37] established the association of rs738409 with mean corpuscular hemoglobin (P = 4 × 10-25) by using genome-wide genotyping array in a sample of 443000 individuals of European ancestry[37]. Variants in or near PNPLA3 have also been associated with the aspartate transaminase-to-platelet ratio index[38].
A summary picture illustrating the genetic associations with PNPLA3 locus, including its pleiotropic effects on diverse laboratory measurements, is shown in Figure 2. It reveals that the strength of the association scores of rs738409 and levels of liver enzymes, particularly ALT levels (association score = 1), is shared with the effect of this variant on many blood-related traits, for instance platelet count (association score = 0.8) (Figure 2).
We next used information sourced from electronic health records and GWAS data computed from United Kingdom Biobank entries pertaining to 452264 individuals, which was retrieved from the Gene ATLAS (http://geneatlas.roslin.ed.ac.uk) and Neale's database (http://www.nealelab.is/uk-biobank/).
We specifically searched for PheWAS associations of variants influencing NAFLD, including PNPLA3-rs738409, TM6SF2-rs58542916, MBOAT7-TMC4 rs641738, GCKR-rs780094, and HSD17B13-rs72613567. As expected, rs738409, rs58542916, and rs72613567 were associated with liver-related traits in the United Kingdom-Biobank GWAS (Table 1).
| Variant | Trait1 | Effect (beta) | P value |
| PNPLA3-rs738409. Chromosomal position: 44324727. Allele C MAF: 0.21 | |||
| Blood-related trait | Platelet count | 1.5369 | 2.8659e-45 |
| Hemoglobin concentration | -0.025607 | 9.0819e-35 | |
| Platelet crit | 0.0010141 | 3.6242e-29 | |
| Mean corpuscular volume | -0.069237 | 9.4452e-16 | |
| Hematocrit percentage | -0.063676 | 2.9494e-25 | |
| Mean corpuscular hemoglobin | -0.031471 | 2.2846e-19 | |
| Monocyte percentage | -0.035411 | 4.7043e-15 | |
| Mean platelet (thrombocyte) volume | -0.016089 | 9.5385e-18 | |
| Liver diseases | K76 Other diseases of liver | -0.0022213 | 4.5111e-22 |
| K70 Alcoholic liver disease | -0.00087844 | 1.7073e-14 | |
| K70-K77 Diseases of liver | -0.0028804 | 3.3855e-25 | |
| K74 Fibrosis and cirrhosis of liver | -0.00075775 | 2.3186e-11 | |
| TM6SF2-rs58542926. Chromosomal position: 19379549. Allele C MAF: 0.975 | |||
| Blood-related trait | Lymphocyte percentage | 0.22273 | 1.8966e-19 |
| Neutrophil percentage | -0.20618 | 8.0938e-13 | |
| Platelet crit | -0.001243 | 2.6606e-18 | |
| Platelet count | -0.96935 | 1.5736e-08 | |
| Red blood cell (erythrocyte) count | -0.008401 | 4.1569e-14 | |
| Liver diseases | K76 Other diseases of liver | -0.0020969 | 5.9013e-09 |
| K74 Fibrosis and cirrhosis of liver | -0.00078964 | 8.6924e-06 | |
| K70-K77 Diseases of liver | -0.0023442 | 7.1296e-08 | |
| HSD17B13-rs72613567. Chromosomal position: 88231392. Allele T MAF: 0.27 | |||
| Blood-related trait | Platelet count | -1.0628 | 6.675e-26 |
| Platelet crit | -0.00068946 | 2.0658e-16 | |
| Mean platelet (thrombocyte) volume | 0.012228 | 2.4506e-12 | |
| Mean corpuscular hemoglobin | 0.020281 | 2.7946e-10 | |
| Liver diseases | K70-K77 Diseases of liver | 0.00095351 | 0.0002012 |
| K74 Fibrosis and cirrhosis of liver | 0.00032971 | 0.0016351 | |
| K70 Alcoholic liver disease | 0.00030904 | 0.0034794 | |
| MBOAT7-TMC4 rs641738. Chromosomal position: 54676763. Allele T MAF: 0.44 | |||
| Blood-related trait | Platelet crit | 0.00079482 | 1.0509e-25 |
| Platelet count | 0.84392 | 2.3213e-20 | |
| White blood cell (leukocyte) count | -0.012957 | 3.4321e-05 | |
| Mean platelet (thrombocyte) volume | 0.0013881 | 0.37852 | |
| Liver diseases | K76 Other diseases of liver | 0.0005036 | 0.0086505 |
| K70-K77 Diseases of liver | 0.00058356 | 0.011748 | |
| K74 Fibrosis and cirrhosis of liver | 0.00019063 | 0.043702 | |
| K70 Alcoholic liver disease | 0.00017298 | 0.069988 | |
| GCKR-rs780094. Chromosomal position: 27741237. Allele C MAF: 0.38 | |||
| Blood-related trait | Mean reticulocyte volume | 0.16064 | 5.9197e-33 |
| Neutrophil percentage | -0.14035 | 2.7743e-19 | |
| Neutrophil count | -0.037139 | 6.3804e-47 | |
| Monocyte percentage | 0.073076 | 7.914e-79 | |
| Mean platelet (thrombocyte) volume | 0.014342 | 1.3297e-18 | |
| Platelet crit | -0.0013887 | 1.3581e-71 | |
| Platelet count | -1.9052 | 6.2404e-92 | |
| Hematocrit percentage | 0.049511 | 4.3172e-21 | |
| Liver diseases | K76 Other diseases of liver | -0.00058708 | 0.0026207 |
| K70-K77 Diseases of liver | -0.0004415 | 0.060918 | |
| K74 Fibrosis and cirrhosis of liver | -0.00011872 | 0.2169 | |
| K75 Other inflammatory liver diseases | -0.00012854 | 0.13954 | |
| K70 Alcoholic liver disease | 9.7922e-05 | 0.31331 | |
Of note, all aforementioned variants showed GWAS-significant associations (P < 5 × 10−8) with blood-related traits. The strongest associations pertained to platelet traits, including platelet crit (which represents the proportion of blood volume that is occupied by platelets, expressed as a percentage), platelet volume, and platelet count (Table 1). Specifically, PNPLA3-rs738409 was associated with platelet count (P = 2.9 × 10-45) and platelet crit (P = 3.6 × 10-29) with the P values for association exceeding those for association with liver diseases (Table 1). A similar pattern was obtained for TM6SF2-rs58542926 and HSD17B13-rs72613567 (Table 1) variants and their associations with platelet traits.
To explore shared pathways between NAFLD and platelet-related traits, we retrieved from the Open Target Genetics platform (https://genetics.opentargets.org) the list of genes (human protein-coding genes) associated with phenotypes of interest. Specifically, we focused on platelet count, which expresses the number of platelets per unit volume in a sample of venous blood.
The genetic associations in the Open Target platform are derived from GWAS Catalog (https://www.ebi.ac.uk/gwas) and PheWAS (https://phewascatalog.org/), whereby the former contains publications indexed in PubMed and the latter is a repository of electronic medical records with links to the Vanderbilt DNA biobank. The list of genes associated with platelet count (n = 305) and NAFLD (n = 161) is shown in Supplementary Table 1 and Supplementary Table 2, respectively.
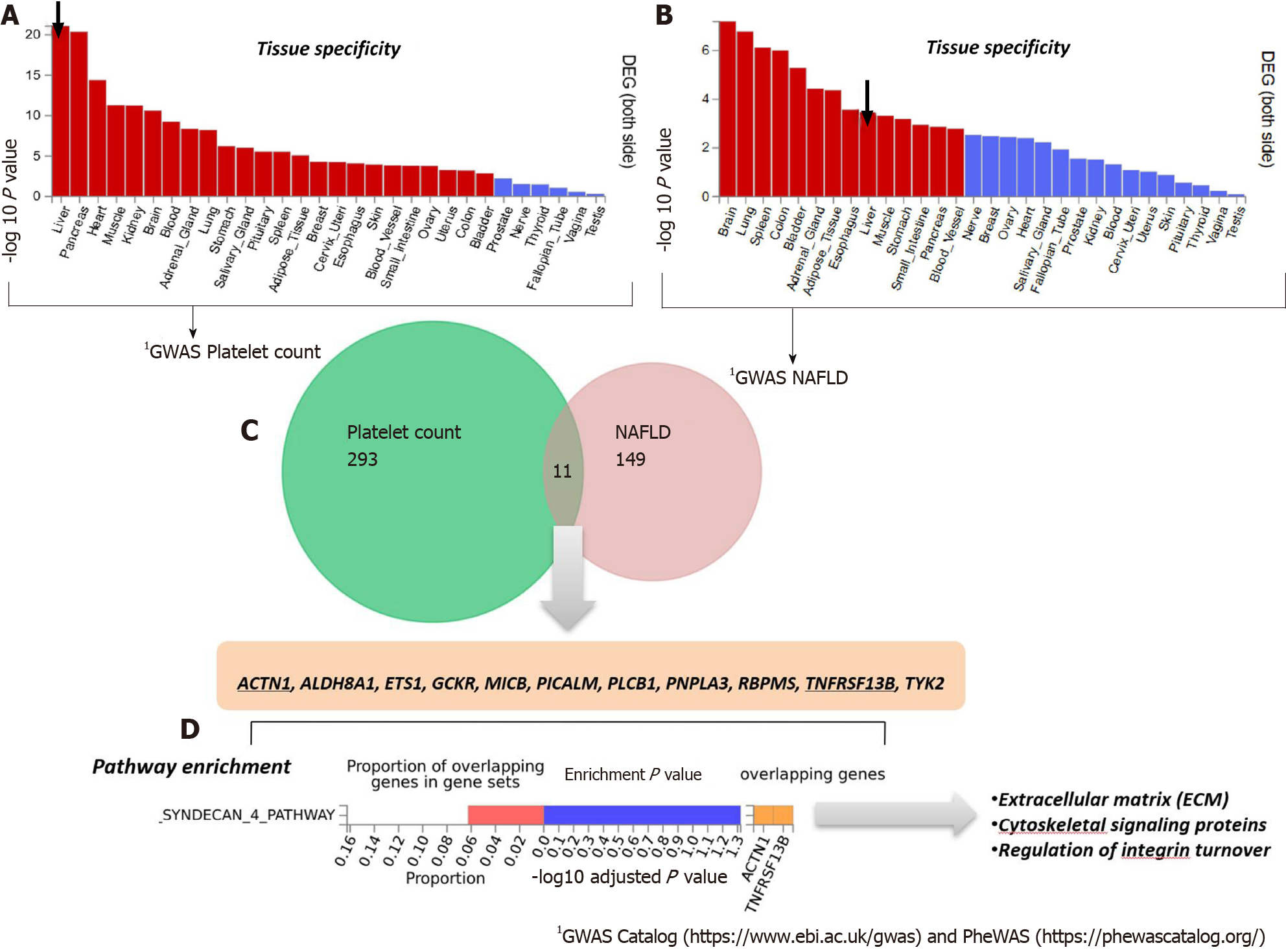
To analyze and interpret the pathways shared between genes associated with NAFLD and those associated with platelet count, we used the FUMA platform available at https://fuma.ctglab.nl/. FUMA utilizes positional expression quantitative trait loci and chromatin interaction mappings to build gene-based pathways and tissue enrichment heatmaps[39]. Hence, we first tested the tissue specificity of the list of genes/proteins associated with each phenotype, namely NAFLD and platelet count. Specifically, we explored tissues in which those genes/proteins present higher expression levels (differentially expressed genes are defined for each label of each expression dataset). Genes showing a P ≤ 0.05 after Bonferroni correction and absolute log fold change ≥ 0.58 were defined as differentially expressed. Interestingly, genes associated with platelet count were most highly expressed in the liver, followed by the pancreas, heart, and muscle (Figure 3A). Conversely, genes associated with NAFLD presented high expression levels in the brain, lung, spleen, and colon (Figure 3B).
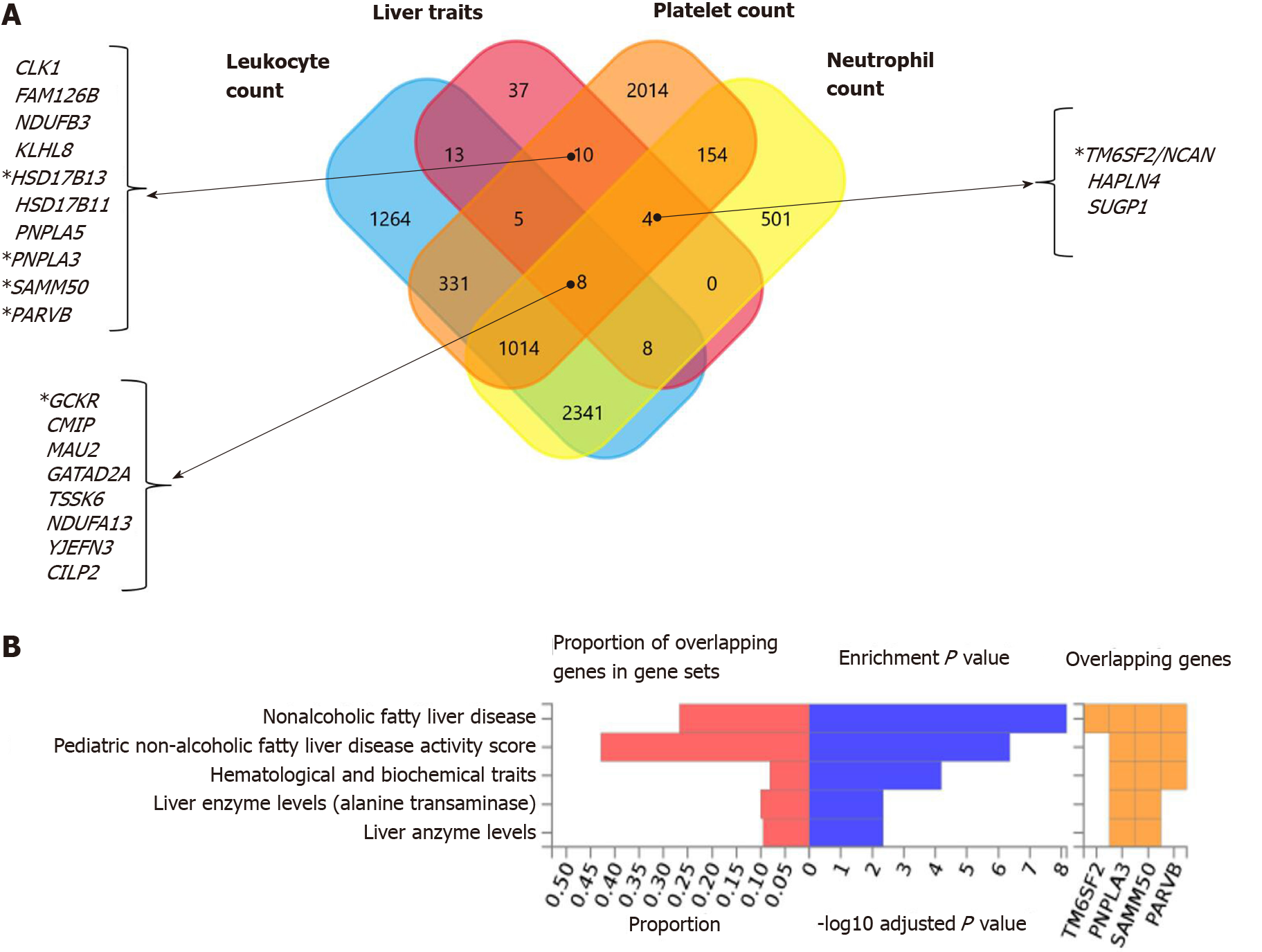
The Venn diagram provided in Figure 3C shows the genes shared between NAFLD and platelet count according to the information retrieved from GWAS and PheWAS catalogs, as explained earlier, among which we found eleven shared loci that included PNPLA3. Next, we performed pathway analysis on the list of shared genes, which revealed an enrichment of genes (ACTN1 and TNFRSF13B) belonging to the predicted pathway “Syndecan 4 pathway” (PID_SYNDECAN_4_PATHWAY) (Figure 3D). The Syndecan 4 pathway is involved in cell growth, differentiation, and adhesion, and in the modulation of extracellular matrix proteins[40]. Syndecans are type I transmembrane proteins with an N-terminal ectodomain that contains several consensus sequences for attachment to glycosaminoglycan, heparan sulfate, and to a lesser extent chondroitin sulfate chains, and a short C-terminal cytoplasmic domain. Syndecans may act as integrin co-receptors. Interactions between fibronectin and syndecans are modulated by tenascin-C. Syndecans bind a wide variety of soluble and insoluble ligands, including extracellular matrix components, cell adhesion molecules, and growth factors, including VEGFs, cytokines, and proteinases[41]. It is worth noting that parvin beta (PARVB), which has been significantly associated with NAFLD[42,43] and hematological traits[44], encodes a member of the parvin family of actin-binding proteins that play a role in cytoskeleton organization and cell adhesion. This family member binds to alphaPIX and alpha-actinin, and can inhibit the activity of integrin-linked kinase. This protein also functions as a tumor suppressor. As the PARVB locus is located near PNPLA3, further studies are needed to establish whether the association of the locus with NAFLD and hematological traits merely reflects a linkage between the two genetic loci.
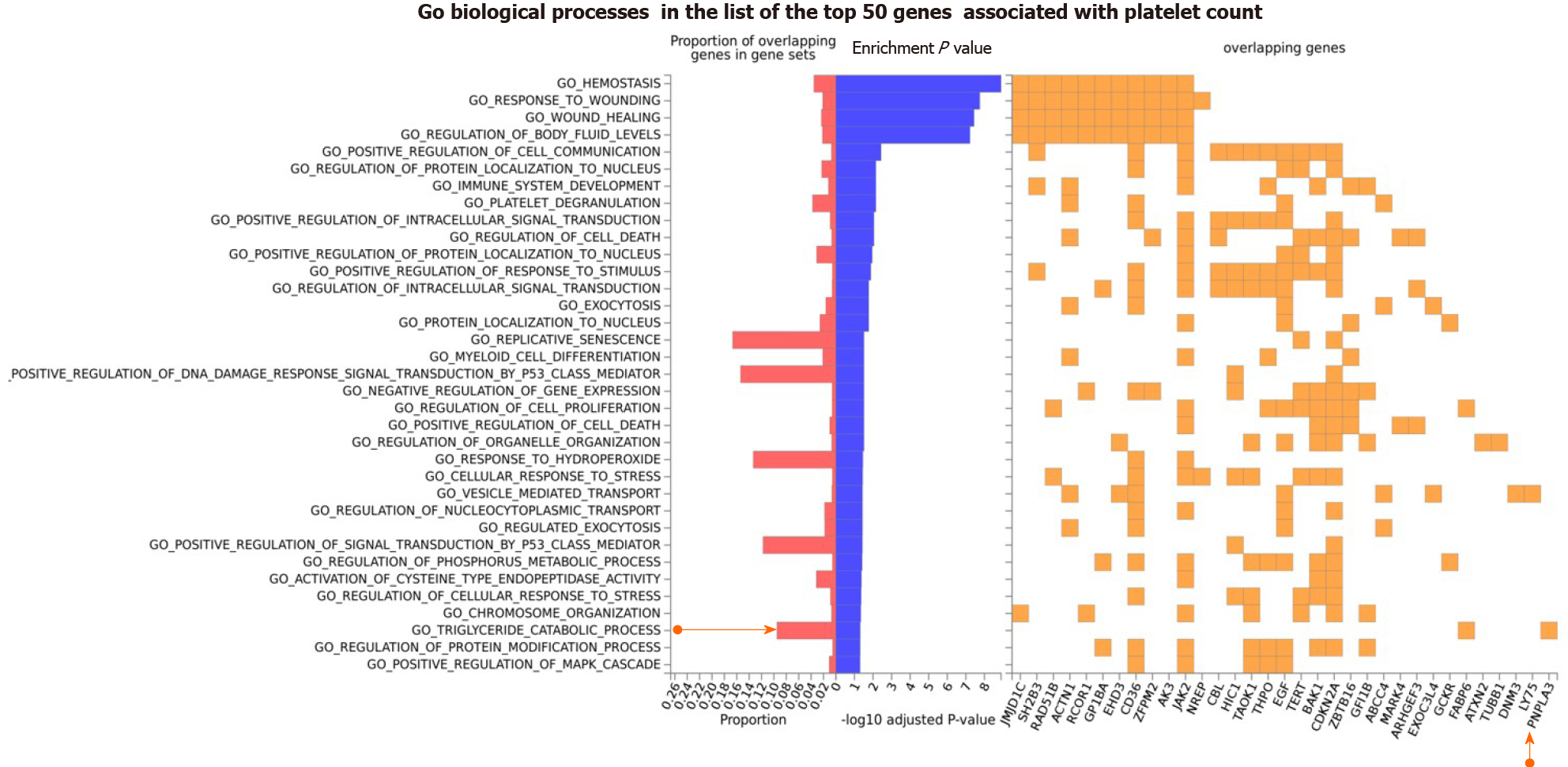
We further explored the Gene Ontology (GO) biological processes in which the list of genes associated with platelet count was specifically enriched. We found that, in addition to expected pathways that included hemostasis, response to wound healing, and platelet degranulation, there were pathways that support the plausibility of sharing genes with NAFLD, for example the triglyceride catalytic process. The GO biological processes associated with the top 50 genes on the list of loci associated with platelet count is shown in Figure 4. Remarkably, PNPLA3 and FABP6 (Fatty Acid Binding Protein 6) are the two overlapping loci that would be responsible for the enrichment of triglyceride metabolism (Figure 4).
It is also noteworthy that, despite the shared genes, there are differences in the biological processes in which the sets of genes associated with platelet count and with NAFLD are involved. To explore these pathways, we performed overrepresentation analysis based on a more comprehensive list of genes associated with each phenotype.
The input list of platelet count-associated variants (P < 1 × 10-6) was generated by searching the United Kingdom Biobank GWAS database as provided by Neale’s lab resource (http://www.nealelab.is/uk-biobank/), which shows variants in about 2424 loci with genome-wide significance (P < 5 × 10-8) for an association with platelet count as a continuous trait (109 cells/L; mean = 252.023 ± Std.dev = 60.0604). We used the United Kingdom Biobank GWAS database because it provides one of the largest publically available sources of genetic associations with laboratory traits in the general population involving 479367 individuals of both sexes (http://biobank.ctsu.ox.ac.uk/crystal/field.cgi?id=30080).
In the case of NAFLD, and to avoid issues arising from the paucity of GWAS/EWAS discovered genes, we included a more comprehensive list of 928 loci obtained by data mining[5] that represents genetic and molecular associations with the disease.
We chose to conduct the overrepresentation analysis on the list of loci obtained from data mining because NAFLD as the disease trait (K76.0 Fatty change of liver: http://biobank.ctsu.ox.ac.uk/crystal/field.cgi?id=41202) is underrepresented in the United Kingdom Biobank GWAS database, affecting only 460 of 410332 individuals whose data are stored in this repository.
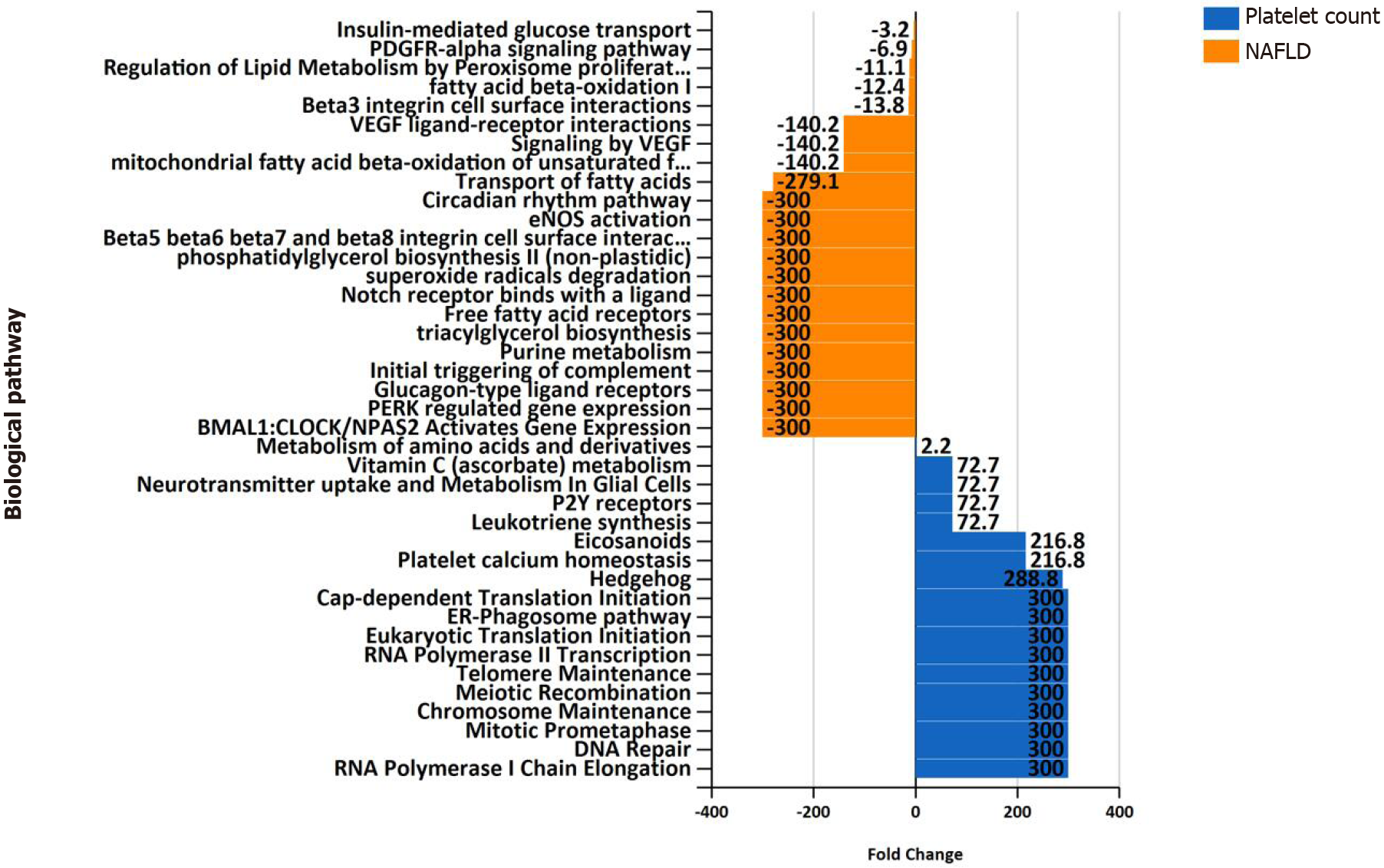
Differences in biological pathway enrichment between the two datasets (NAFLD and platelet count) are summarized in Figure 5, which shows specificity in the function of genes in each of the gene lists. For instance, the NAFLD list of associated genes/proteins is enriched in expected pathways, such as metabolism of lipids and amino acids, purine metabolism, and circadian rhythm, among others (Figure 5). The platelet count list is enriched with genes involved in DNA repair, telomere maintenance, and the Hedgehog pathway, among many others, as shown in
To understand the potential involvement of genes associated with laboratory hematological-related traits in NAFLD biology, we performed functional analysis. Based on the reverse biology premise, we reasoned that genes involved in hematological-related traits might be associated with some metabolic function/s that would presumably affect NAFLD pathogenesis. Thus, the top blood-related associated traits in terms of their statistical significance were further searched for genetic associations in the entire United Kingdom Biobank dataset. Specifically, approximately 30 million variants in the United Kingdom Biobank from the Gene ATLAS (http://geneatlas.roslin.ed.ac.uk) and Neale's database (http://www.nealelab.is/uk-biobank/) resources were comprehensively tested for associations with blood cell-associated traits, including platelet, leukocyte and neutrophil counts. We further explored the pathways in which the lists of genes associated with these traits are involved. For this purpose, we used the FUMA resource that allows using functional and biological information to prioritize genes based on GWAS outcomes.
Interestingly, functional mapping, gene prioritization, and functional analysis using FUMA of the most significant genetic variants (P < 1 × 10-6) revealed 85 mapped genes in the full list of loci associated with hematological traits that are also associated with liver traits, including chronic liver diseases, fibrosis and liver cirrhosis, NAFLD, and other inflammatory liver diseases (Figure 6A). PNPLA3, SAMM50, PARVB and HSD17B13 are among the ten genes shared by liver traits and platelet count. TM6SF2 is shared by liver traits, platelet count, and neutrophil count, and GCKR is shared by liver and all hematological traits (Figure 6A).
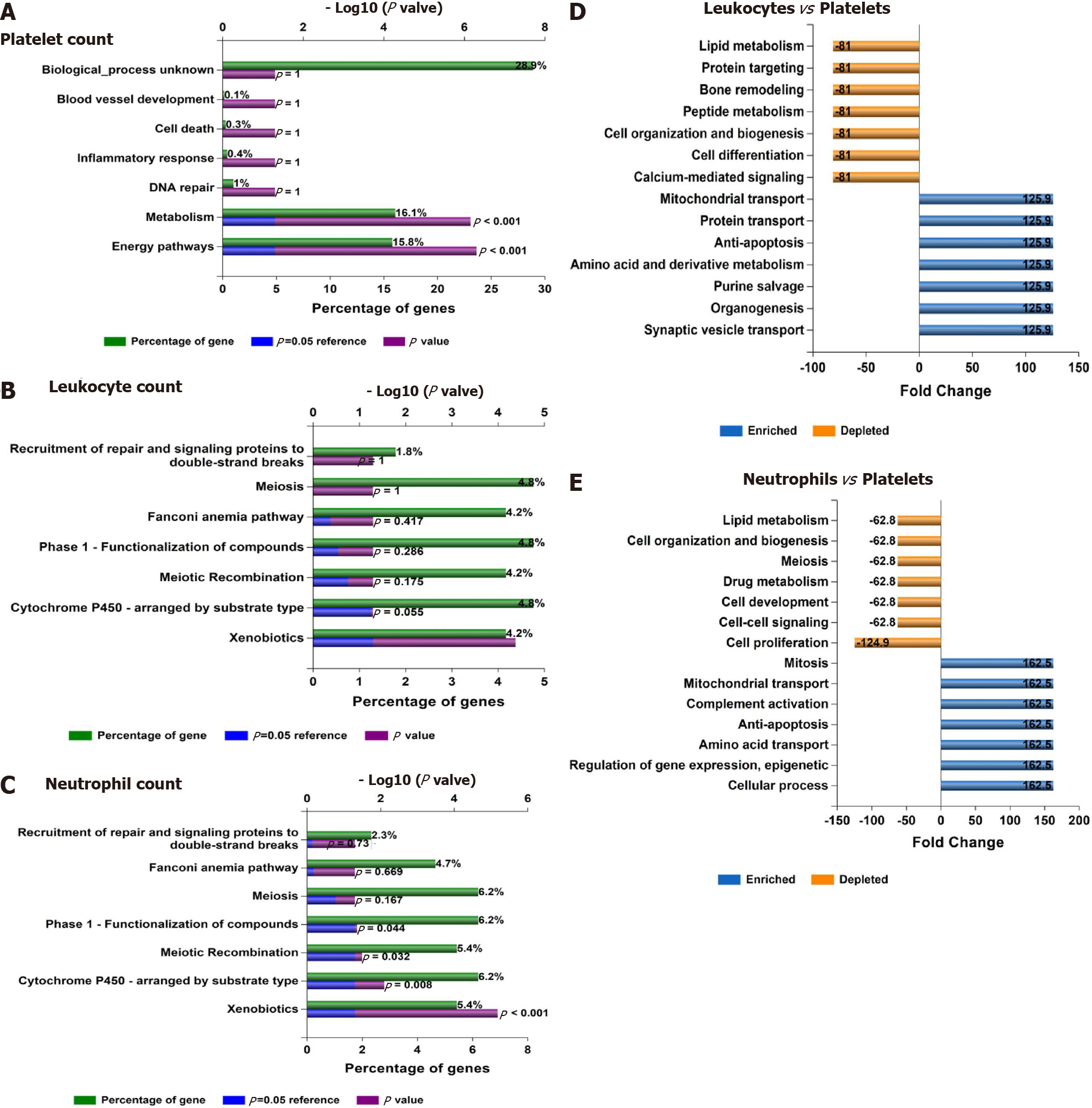
In addition, Figure 6B shows the consistent association of PNPLA3, TM6SF2, SAMM50, and PARVB with liver and hematological traits identified via FUMA analysis in reported GWAS. Finally, we performed functional enrichment analysis on genes significantly associated with platelet count, leukocyte count, and neutrophil count by applying the FunRich tool. Notably, loci involved in the genetic modulation of platelet, leukocyte, and neutrophil counts presented significant enrichment in metabolic, energy balance, xenobiotics, and CYP-450-related pathways (Figure 7A−C). However, platelet-related loci are particularly involved in regulating key aspects of metabolism, while leukocyte and neutrophil counts are related to more general homeostasis regulation processes (Figure 7D−F).
PheWas revealed that variants in genes influencing NAFLD present pleiotropic associations with laboratory-related hematologic traits and are relevant to the hematopoietic liver function. Similarly, related genes with variants influencing hematological traits, platelet count in particular, presented significant enrichment in metabolic and energy balance-related pathways.
By using different resources and datasets of variants associated with the genetics of platelet count and NAFLD, we found consistency in the results, suggesting that there are shared mechanisms and pathways between the two phenotypes. In particular, we found metabolic and lipid pathways shared by NAFLD and platelet traits. It is anticipated that potential therapeutic targets, including novel ligands of peroxisome proliferator-activated receptors may also play a role in modulating platelet-related phenotypes such as platelet activation and the cascade of events associated with inflammation and cardiovascular risk.
In summary, our approach provides novel mechanistic insights into NAFLD pathogenesis. Further research is nonetheless necessary to ascertain whether genes associated with liver diseases present ample pleiotropy and, therefore, modify functions of diverse organs simultaneously. If, conversely, some phenotypes are found to act as intermediaries between genes and disease, a Mendelian Randomization approach can be used to study the relationship between, in this case, hematological and liver traits or vice versa.
| 1. | Brunt EM, Wong VW, Nobili V, Day CP, Sookoian S, Maher JJ, Bugianesi E, Sirlin CB, Neuschwander-Tetri BA, Rinella ME. Nonalcoholic fatty liver disease. Nat Rev Dis Primers. 2015;1:15080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 466] [Cited by in RCA: 639] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 2. | Sookoian S, Pirola CJ. Precision medicine in nonalcoholic fatty liver disease: New therapeutic insights from genetics and systems biology. Clin Mol Hepatol. 2020;26:461-475. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 3. | Pirola CJ, Sookoian S. Epigenetics factors in nonalcoholic fatty liver disease. Expert Rev Gastroenterol Hepatol. 2020;1-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 4. | Sookoian S, Pirola CJ, Valenti L, Davidson NO. Genetic Pathways in Nonalcoholic Fatty Liver Disease: Insights From Systems Biology. Hepatology. 2020;72:330-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 109] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 5. | Sookoian S, Pirola CJ. Genetics of Nonalcoholic Fatty Liver Disease: From Pathogenesis to Therapeutics. Semin Liver Dis. 2019;39:124-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 55] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 6. | Sookoian S, Pirola CJ. Review article: shared disease mechanisms between non-alcoholic fatty liver disease and metabolic syndrome - translating knowledge from systems biology to the bedside. Aliment Pharmacol Ther. 2019;49:516-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 7. | Sookoian S, Pirola CJ. Systematic review with meta-analysis: risk factors for non-alcoholic fatty liver disease suggest a shared altered metabolic and cardiovascular profile between lean and obese patients. Aliment Pharmacol Ther. 2017;46:85-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 175] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 8. | Eicher JD, Chami N, Kacprowski T, Nomura A, Chen MH, Yanek LR, Tajuddin SM, Schick UM, Slater AJ, Pankratz N, Polfus L, Schurmann C, Giri A, Brody JA, Lange LA, Manichaikul A, Hill WD, Pazoki R, Elliot P, Evangelou E, Tzoulaki I, Gao H, Vergnaud AC, Mathias RA, Becker DM, Becker LC, Burt A, Crosslin DR, Lyytikäinen LP, Nikus K, Hernesniemi J, Kähönen M, Raitoharju E, Mononen N, Raitakari OT, Lehtimäki T, Cushman M, Zakai NA, Nickerson DA, Raffield LM, Quarells R, Willer CJ, Peloso GM, Abecasis GR, Liu DJ; Global Lipids Genetics Consortium; Deloukas P; Samani NJ; Schunkert H; Erdmann J; CARDIoGRAM Exome Consortium; Myocardial Infarction Genetics Consortium; Fornage M; Richard M; Tardif JC; Rioux JD; Dube MP; de Denus S; Lu Y; Bottinger EP; Loos RJ; Smith AV; Harris TB; Launer LJ; Gudnason V; Velez Edwards DR; Torstenson ES; Liu Y; Tracy RP; Rotter JI; Rich SS; Highland HM; Boerwinkle E; Li J; Lange E; Wilson JG; Mihailov E; Mägi R; Hirschhorn J; Metspalu A; Esko T; Vacchi-Suzzi C; Nalls MA; Zonderman AB; Evans MK; Engström G; Orho-Melander M; Melander O; O'Donoghue ML; Waterworth DM; Wallentin L; White HD; Floyd JS; Bartz TM; Rice KM; Psaty BM; Starr JM; Liewald DC; Hayward C; Deary IJ; Greinacher A; Völker U; Thiele T; Völzke H; van Rooij FJ; Uitterlinden AG; Franco OH; Dehghan A; Edwards TL; Ganesh SK; Kathiresan S; Faraday N; Auer PL; Reiner AP; Lettre G; Johnson AD. Platelet-Related Variants Identified by Exomechip Meta-analysis in 157,293 Individuals. Am J Hum Genet. 2016;99:40-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 9. | Liu Y, Fernandez CA, Smith C, Yang W, Cheng C, Panetta JC, Kornegay N, Liu C, Ramsey LB, Karol SE, Janke LJ, Larsen EC, Winick N, Carroll WL, Loh ML, Raetz EA, Hunger SP, Devidas M, Yang JJ, Mullighan CG, Zhang J, Evans WE, Jeha S, Pui CH, Relling MV. Genome-Wide Study Links PNPLA3 Variant With Elevated Hepatic Transaminase After Acute Lymphoblastic Leukemia Therapy. Clin Pharmacol Ther. 2017;102:131-140. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 42] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 10. | Verma A, Bang L, Miller JE, Zhang Y, Lee MTM, Zhang Y, Byrska-Bishop M, Carey DJ, Ritchie MD, Pendergrass SA, Kim D; DiscovEHR Collaboration. Human-Disease Phenotype Map Derived from PheWAS across 38,682 Individuals. Am J Hum Genet. 2019;104:55-64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 11. | Lonardo A, Sookoian S, Pirola CJ, Targher G. Non-alcoholic fatty liver disease and risk of cardiovascular disease. Metabolism. 2016;65:1136-1150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 178] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 12. | Serra-Burriel M, Graupera I, Torán P, Thiele M, Roulot D, Wai-Sun Wong V, Neil Guha I, Fabrellas N, Arslanow A, Expósito C, Hernández R, Lai-Hung Wong G, Harman D, Darwish Murad S, Krag A, Pera G, Angeli P, Galle P, Aithal GP, Caballeria L, Castera L, Ginès P, Lammert F; investigators of the LiverScreen Consortium. Transient elastography for screening of liver fibrosis: Cost-effectiveness analysis from six prospective cohorts in Europe and Asia. J Hepatol. 2019;71:1141-1151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 116] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 13. | Wang S, Zhang C, Zhang G, Yuan Z, Liu Y, Ding L, Sun X, Jia H, Xue F. Association between white blood cell count and non-alcoholic fatty liver disease in urban Han Chinese: a prospective cohort study. BMJ Open. 2016;6:e010342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 32] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 14. | Lee YJ, Lee HR, Shim JY, Moon BS, Lee JH, Kim JK. Relationship between white blood cell count and nonalcoholic fatty liver disease. Dig Liver Dis. 2010;42:888-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 60] [Article Influence: 3.8] [Reference Citation Analysis (1)] |
| 15. | Chung GE, Yim JY, Kim D, Kwak MS, Yang JI, Chung SJ, Yang SY, Kim JS. Associations between White Blood Cell Count and the Development of Incidental Nonalcoholic Fatty Liver Disease. Gastroenterol Res Pract. 2016;2016:7653689. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 16. | Brouard N, Jost C, Matthias N, Albrecht C, Egard S, Gandhi P, Strassel C, Inoue T, Sugiyama D, Simmons PJ, Gachet C, Lanza F. A unique microenvironment in the developing liver supports the expansion of megakaryocyte progenitors. Blood Adv. 2017;1:1854-1866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 17. | Afdhal N, McHutchison J, Brown R, Jacobson I, Manns M, Poordad F, Weksler B, Esteban R. Thrombocytopenia associated with chronic liver disease. J Hepatol. 2008;48:1000-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 408] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 18. | Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3381] [Cited by in RCA: 4218] [Article Influence: 200.9] [Reference Citation Analysis (11)] |
| 19. | Pinzani M, Marra F. Cytokine receptors and signaling in hepatic stellate cells. Semin Liver Dis. 2001;21:397-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 330] [Cited by in RCA: 337] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 20. | Kurokawa T, Ohkohchi N. Platelets in liver disease, cancer and regeneration. World J Gastroenterol. 2017;23:3228-3239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 62] [Cited by in RCA: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 21. | Chauhan A, Adams DH, Watson SP, Lalor PF. Platelets: No longer bystanders in liver disease. Hepatology. 2016;64:1774-1784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 111] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 22. | Sookoian S, Gianotti TF, Rosselli MS, Burgueño AL, Castaño GO, Pirola CJ. Liver transcriptional profile of atherosclerosis-related genes in human nonalcoholic fatty liver disease. Atherosclerosis. 2011;218:378-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 79] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 23. | Coppinger JA, Cagney G, Toomey S, Kislinger T, Belton O, McRedmond JP, Cahill DJ, Emili A, Fitzgerald DJ, Maguire PB. Characterization of the proteins released from activated platelets leads to localization of novel platelet proteins in human atherosclerotic lesions. Blood. 2004;103:2096-2104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 570] [Cited by in RCA: 606] [Article Influence: 26.3] [Reference Citation Analysis (0)] |
| 24. | Assoian RK, Sporn MB. Type beta transforming growth factor in human platelets: release during platelet degranulation and action on vascular smooth muscle cells. J Cell Biol. 1986;102:1217-1223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 388] [Cited by in RCA: 427] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 25. | Malehmir M, Pfister D, Gallage S, Szydlowska M, Inverso D, Kotsiliti E, Leone V, Peiseler M, Surewaard BGJ, Rath D, Ali A, Wolf MJ, Drescher H, Healy ME, Dauch D, Kroy D, Krenkel O, Kohlhepp M, Engleitner T, Olkus A, Sijmonsma T, Volz J, Deppermann C, Stegner D, Helbling P, Nombela-Arrieta C, Rafiei A, Hinterleitner M, Rall M, Baku F, Borst O, Wilson CL, Leslie J, O'Connor T, Weston CJ, Adams DH, Sheriff L, Teijeiro A, Prinz M, Bogeska R, Anstee N, Bongers MN, Notohamiprodjo M, Geisler T, Withers DJ, Ware J, Mann DA, Augustin HG, Vegiopoulos A, Milsom MD, Rose AJ, Lalor PF, Llovet JM, Pinyol R, Tacke F, Rad R, Matter M, Djouder N, Kubes P, Knolle PA, Unger K, Zender L, Nieswandt B, Gawaz M, Weber A, Heikenwalder M. Platelet GPIbα is a mediator and potential interventional target for NASH and subsequent liver cancer. Nat Med. 2019;25:641-655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 310] [Cited by in RCA: 314] [Article Influence: 44.9] [Reference Citation Analysis (0)] |
| 26. | Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461-1465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2701] [Cited by in RCA: 2701] [Article Influence: 150.1] [Reference Citation Analysis (2)] |
| 27. | Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883-1894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 756] [Cited by in RCA: 757] [Article Influence: 50.5] [Reference Citation Analysis (1)] |
| 28. | Chamorro AJ, Torres JL, Mirón-Canelo JA, González-Sarmiento R, Laso FJ, Marcos M. Systematic review with meta-analysis: the I148M variant of patatin-like phospholipase domain-containing 3 gene (PNPLA3) is significantly associated with alcoholic liver cirrhosis. Aliment Pharmacol Ther. 2014;40:571-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 29. | Stickel F, Buch S, Lau K, Meyer zu Schwabedissen H, Berg T, Ridinger M, Rietschel M, Schafmayer C, Braun F, Hinrichsen H, Günther R, Arlt A, Seeger M, Müller S, Seitz HK, Soyka M, Lerch M, Lammert F, Sarrazin C, Kubitz R, Häussinger D, Hellerbrand C, Bröring D, Schreiber S, Kiefer F, Spanagel R, Mann K, Datz C, Krawczak M, Wodarz N, Völzke H, Hampe J. Genetic variation in the PNPLA3 gene is associated with alcoholic liver injury in caucasians. Hepatology. 2011;53:86-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 219] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 30. | Tian C, Stokowski RP, Kershenobich D, Ballinger DG, Hinds DA. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet. 2010;42:21-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 349] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 31. | Valenti L, Rumi M, Galmozzi E, Aghemo A, Del Menico B, De Nicola S, Dongiovanni P, Maggioni M, Fracanzani AL, Rametta R, Colombo M, Fargion S. Patatin-like phospholipase domain-containing 3 I148M polymorphism, steatosis, and liver damage in chronic hepatitis C. Hepatology. 2011;53:791-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 205] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 32. | Viganò M, Valenti L, Lampertico P, Facchetti F, Motta BM, D'Ambrosio R, Romagnoli S, Dongiovanni P, Donati B, Fargion S, Colombo M. Patatin-like phospholipase domain-containing 3 I148M affects liver steatosis in patients with chronic hepatitis B. Hepatology. 2013;58:1245-1252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 33. | Falleti E, Fabris C, Cmet S, Cussigh A, Bitetto D, Fontanini E, Fornasiere E, Bignulin S, Fumolo E, Bignulin E, Pirisi M, Toniutto P. PNPLA3 rs738409C/G polymorphism in cirrhosis: relationship with the aetiology of liver disease and hepatocellular carcinoma occurrence. Liver Int. 2011;31:1137-1143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 34. | Trépo E, Nahon P, Bontempi G, Valenti L, Falleti E, Nischalke HD, Hamza S, Corradini SG, Burza MA, Guyot E, Donati B, Spengler U, Hillon P, Toniutto P, Henrion J, Franchimont D, Devière J, Mathurin P, Moreno C, Romeo S, Deltenre P. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: Evidence from a meta-analysis of individual participant data. Hepatology. 2014;59:2170-2177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 196] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 35. | Diogo D, Tian C, Franklin CS, Alanne-Kinnunen M, March M, Spencer CCA, Vangjeli C, Weale ME, Mattsson H, Kilpeläinen E, Sleiman PMA, Reilly DF, McElwee J, Maranville JC, Chatterjee AK, Bhandari A, Nguyen KH, Estrada K, Reeve MP, Hutz J, Bing N, John S, MacArthur DG, Salomaa V, Ripatti S, Hakonarson H, Daly MJ, Palotie A, Hinds DA, Donnelly P, Fox CS, Day-Williams AG, Plenge RM, Runz H. Phenome-wide association studies across large population cohorts support drug target validation. Nat Commun. 2018;9:4285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 141] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 36. | Pilling LC, Atkins JL, Kuchel GA, Ferrucci L, Melzer D. Red cell distribution width and common disease onsets in 240,477 healthy volunteers followed for up to 9 years. PLoS One. 2018;13:e0203504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 37. | Kichaev G, Bhatia G, Loh PR, Gazal S, Burch K, Freund MK, Schoech A, Pasaniuc B, Price AL. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am J Hum Genet. 2019;104:65-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 632] [Cited by in RCA: 695] [Article Influence: 99.3] [Reference Citation Analysis (0)] |
| 38. | Chen VL, Chen Y, Du X, Handelman SK, Speliotes EK. Genetic variants that associate with cirrhosis have pleiotropic effects on human traits. Liver Int. 2020;40:405-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 45] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 39. | Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1354] [Cited by in RCA: 2815] [Article Influence: 312.8] [Reference Citation Analysis (0)] |
| 40. | Elfenbein A, Simons M. Syndecan-4 signaling at a glance. J Cell Sci. 2013;126:3799-3804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 151] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 41. | Morgan MR, Humphries MJ, Bass MD. Synergistic control of cell adhesion by integrins and syndecans. Nat Rev Mol Cell Biol. 2007;8:957-969. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 471] [Cited by in RCA: 459] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 42. | Kitamoto T, Kitamoto A, Ogawa Y, Honda Y, Imajo K, Saito S, Yoneda M, Nakamura T, Nakajima A, Hotta K. Targeted-bisulfite sequence analysis of the methylation of CpG islands in genes encoding PNPLA3, SAMM50, and PARVB of patients with non-alcoholic fatty liver disease. J Hepatol. 2015;63:494-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 71] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 43. | Wu G, Wang K, Xue Y, Song G, Wang Y, Sun X, Zhong L, Zhou C, Shen B, Chen J, Yu Y, Tang H, Peng Z, Sun P, Wang X. Association of rs5764455 and rs6006473 polymorphisms in PARVB with liver damage of nonalcoholic fatty liver disease in Han Chinese population. Gene. 2016;575:270-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 44. | Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, Nakamura Y, Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42:210-215. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 410] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 45. | Min HK, Sookoian S, Pirola CJ, Cheng J, Mirshahi F, Sanyal AJ. Metabolic profiling reveals that PNPLA3 induces widespread effects on metabolism beyond triacylglycerol remodeling in Huh-7 hepatoma cells. Am J Physiol Gastrointest Liver Physiol. 2014;307:G66-G76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 46. | Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, Mouradov D, Sieber OM, Simpson RJ, Salim A, Bacic A, Hill AF, Stroud DA, Ryan MT, Agbinya JI, Mariadason JM, Burgess AW, Mathivanan S. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597-2601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 993] [Article Influence: 90.3] [Reference Citation Analysis (0)] |
| 47. | Sookoian S, Pirola CJ. PNPLA3, the history of an orphan gene of the potato tuber protein family that found an organ: the liver. Hepatology. 2014;59:2068-2071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 48. | Sookoian S, Pirola CJ. PNPLA3, the triacylglycerol synthesis/hydrolysis/storage dilemma, and nonalcoholic fatty liver disease. World J Gastroenterol. 2012;18:6018-6026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 43] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 49. | Mitsche MA, Hobbs HH, Cohen JC. Patatin-like phospholipase domain-containing protein 3 promotes transfer of essential fatty acids from triglycerides to phospholipids in hepatic lipid droplets. J Biol Chem. 2018;293:9232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 2.1] [Reference Citation Analysis (1)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Corresponding Author's Membership in Professional Societies: American Association for the Study of Liver Diseases.
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Argentina
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Elshaarawy O S-Editor: Zhang L L-Editor: Webster JR P-Editor: Liu JH