Published online Oct 14, 2020. doi: 10.3748/wjg.v26.i38.5759
Peer-review started: August 14, 2020
First decision: August 22, 2020
Revised: September 3, 2020
Accepted: September 17, 2020
Article in press: September 17, 2020
Published online: October 14, 2020
Processing time: 60 Days and 13 Hours
Chronic infection with viral hepatitis affects half a billion individuals worldwide and can lead to cirrhosis, cancer, and liver failure. Liver cancer is the third leading cause of cancer-associated mortality, of which hepatocellular carcinoma (HCC) represents 90% of all primary liver cancers. Solid tumors like HCC are complex and have heterogeneous tumor genomic profiles contributing to complexity in diagnosis and management. Chronic infection with hepatitis B virus (HBV), hepatitis delta virus (HDV), and hepatitis C virus (HCV) are the greatest etiological risk factors for HCC. Due to the significant role of chronic viral infection in HCC development, it is important to investigate direct (viral associated) and indirect (immune-associated) mechanisms involved in the pathogenesis of HCC. Common mechanisms used by HBV, HCV, and HDV that drive hepatocarcinogenesis include persistent liver inflammation with an impaired antiviral immune response, immune and viral protein-mediated oxidative stress, and deregulation of cellular signaling pathways by viral proteins. DNA integration to promote genome instability is a feature of HBV infection, and metabolic reprogramming leading to steatosis is driven by HCV infection. The current review aims to provide a brief overview of HBV, HCV and HDV molecular biology, and highlight specific viral-associated oncogenic mechanisms and common molecular pathways deregulated in HCC, and current as well as emerging treatments for HCC.
Core Tip: Hepatocellular carcinoma (HCC) is a dreaded complication of viral infection with hepatitis B virus and/or hepatitis delta virus and hepatitis C virus. Many of the direct and indirect molecular mechanisms used by these viruses to co-opt the liver microenvironment for persistence also disrupt cell cycle pathways. Indirectly, the immune system has a major role in the contribution of HCC in the context of viral hepatitis, but direct viral mechanisms also create a pro-tumorigenic environment.
- Citation: D'souza S, Lau KCK, Coffin CS, Patel TR. Molecular mechanisms of viral hepatitis induced hepatocellular carcinoma. World J Gastroenterol 2020; 26(38): 5759-5783
- URL: https://www.wjgnet.com/1007-9327/full/v26/i38/5759.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i38.5759
Liver cancer is the third leading cause of cancer-associated mortality (781631 people/year), despite being ranked seventh on global incidence (841080 people/year)[1]. Approximately 12% of all cancer cases globally arise from chronic infections with bloodborne oncogenic viral pathogens including hepatitis B virus (HBV), hepatitis C virus (HCV), and hepatitis delta virus (HDV)[2]. Although incidence in the majority of cancers has decreased, primary liver cancer incidence is the fastest-growing cancer with regards to incidence and mortality[3]. Hepatocellular carcinoma (HCC) represents 90% of all liver cancer cases and the risk factors are well defined: Viral infection with HBV, (54% of all HCCs) and/or HCV (31% of all HCCs), cirrhosis (80% of all HCCs), high alcohol consumption, obesity, genetic disorders such as hemochromatosis, exposure to aflatoxins, sex (male) and older age (50+)[4-7].
Virus-induced HCC is present worldwide, however, there are considerable differences in populations that develop HBV or HDV induced HCC vs HCV induced HCC. HBV and HDV associated HCC is more common in low and middle-human development index countries, while HCV induced HCC is more common in high and very high-human development index[2]. Chronic hepatitis B (CHB) infection affects around 257 million people worldwide, of which 48-60 million people are co-infected with HDV and an estimated 2.6 million are co-infected with HCV[8-10]. Exposure to infected blood/bodily fluids is the primary mode of transmission for HBV and HBV/HDV, with majority of exposures occurring from mother to child during birth or early years of life. Unvaccinated neonates and children who have been exposed to HBV have > 95% risk of developing chronic disease, while infection during adulthood results in < 2% chance of developing chronic disease[11]. HBV/HDV co-infection have the highest mortality rate (20%) associated with any viral hepatitis infection and most severe liver disease (i.e. acute liver failure, cirrhosis within 5 years, and HCC within 10 years)[10,12,13]. HCV has established chronic infection in 70 million people primarily through horizontal blood-borne transmission routes such as intravenous drug use, needle pricks, unscreened blood transfusions, and high-risk sexual practices[11]. In comparison to HBV or HCV mono-infection, individuals who are co-infected with HBV/HCV have increased rates of HCC development. Overall, viral etiologies represent approximately 80% of all HCC related cases, highlighting the importance of investigating the role of these viruses in the development of liver cancer.
Preventative measures against HBV and HDV induced liver cancer include birth-dose vaccinations, hepatitis B immunoglobulin treatment for children born to infected mothers as well as treatment of mothers with high HBV viral load with nucleos(t)ide inhibitors in the third trimester[14]. For those individuals who are already chronic carriers of HBV/HDV, there is no virological cure; however, treatment with nucleos(t)ide analogs can lower the risk of HCC development[15]. There is no protective vaccine available for HCV, but there are effective direct-acting antivirals that can cure > 90% of chronic carriers. Those who have a sustained virological response from direct-acting antiviral treatment have a significantly lower risk of HCC development if cirrhosis is absent[16]. Although there are treatment options to lower the risk of HCC in those who have chronic viral hepatitis infection, globally many individuals are unaware of their status, lack access to testing, and effective treatment.
In this review article, we discuss the molecular biology of HBV, HCV, and HDV, common features associated with virus-induced cancers, viral oncogenic mechanisms leading to HCC relating to the hallmarks of cancer, common molecular pathways deregulated in HCC, and current as well as emerging treatments for HCC.
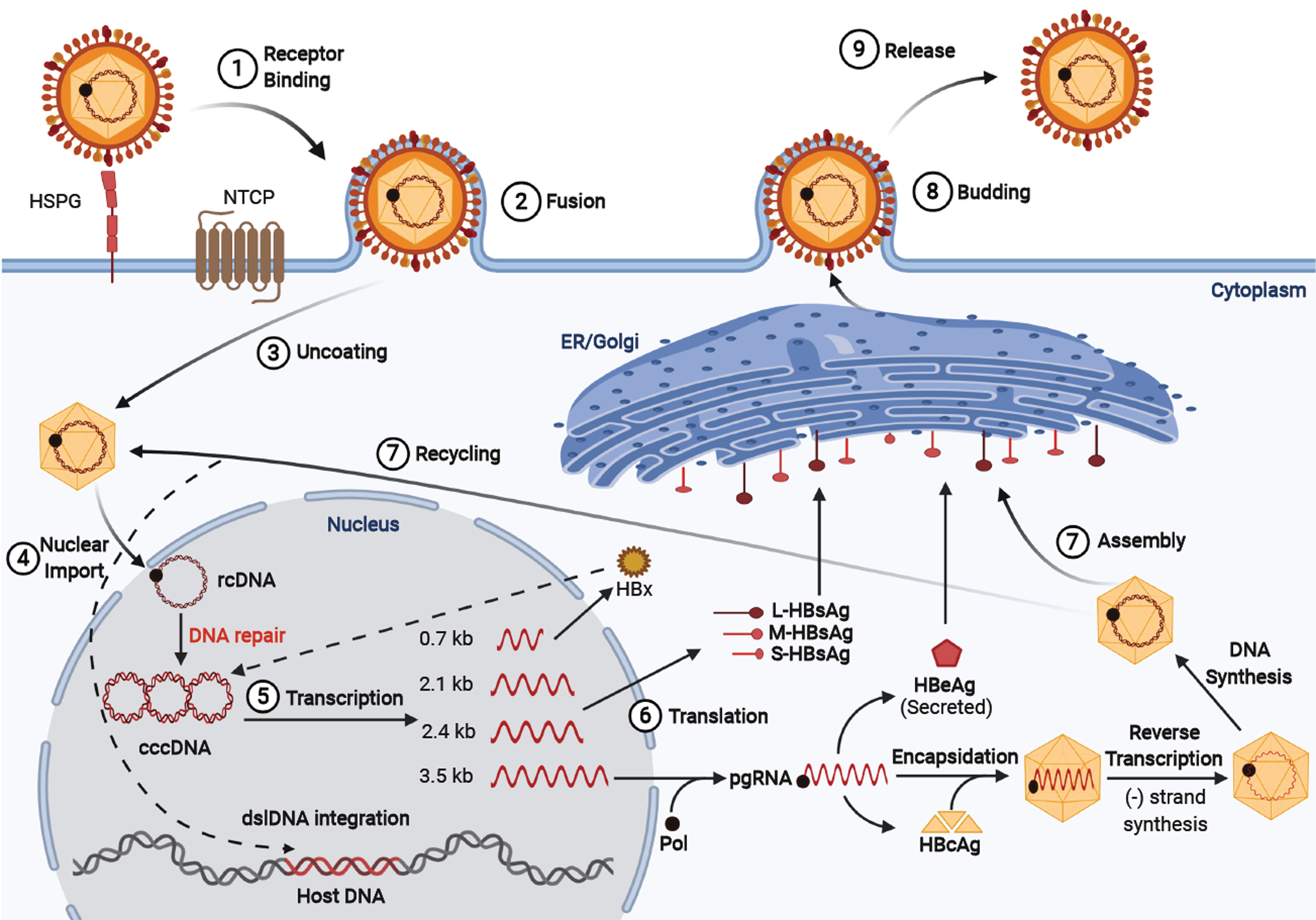
The HBV is a member of the Hepadnaviridae family, which has a cellular tropism for hepatocytes, but has also been detected in extra-hepatic reservoirs such as the lymphoid cells (i.e. peripheral blood mononuclear cells)[17-20]. HBV has a compact 3.2 kb partially double-stranded relaxed circular DNA genome (rcDNA) containing four overlapping open reading frames: Pre-S/S, X, P, and pre-C/C, which are under the transcription control of the pre-S1 promoter, pre-S2/S promoter, enhancer I/X and enhancer II/basal core promoter[21]. The viral protein products include three surface proteins (large/pre-S1, middle/pre-S2, and small/S - also known as HBsAg), the core antigen (HBcAg), the excreted “e” antigen (HBeAg), the viral polymerase (which has reverse transcriptase, DNA polymerase, and RNaseH activity), and the X protein (HBx) that plays an essential role in HBV pathogenesis and viral transcription[21]. Upon viral attachment of the envelope HBV preS1 protein to the sodium taurocholate co-transporting polypeptide receptor, the virus is endocytosed (Figure 1). The nucleocapsid is transported via microtubules from the cytoplasm to the nucleus where the rcDNA is converted to covalently closed circular DNA (cccDNA)[22,23]. The cccDNA associates with histone and non-histone proteins which form a viral minichromosome that persists in the hepatocyte to serve as the template for transcription of pregenomic RNA (pgRNA) and subgenomic RNAs by host RNA polymerase II[24]. The exported pgRNA and subgenomic transcripts are translated to produce the core protein, viral envelope surface proteins, HBeAg, polymerase, and X proteins. In addition, the pgRNA transcript is packaged by the capsid proteins and reverse transcribed by the viral polymerase into rcDNA. The newly packaged rcDNA can either localize back to the nucleus to replenish the cellular cccDNA population or gain their coat through the endoplasmic reticulum (ER)/Golgi and proceed to bud out of the to infect other cells[25]. Current nucleos(t)ide antivirals target the viral reverse transcriptase to produce aberrant transcripts that cannot produce infectious virions. Additional details about the lifecycle and host-transcription factors/proteins required for HBV replication are included in our previous article by Turton et al[26].
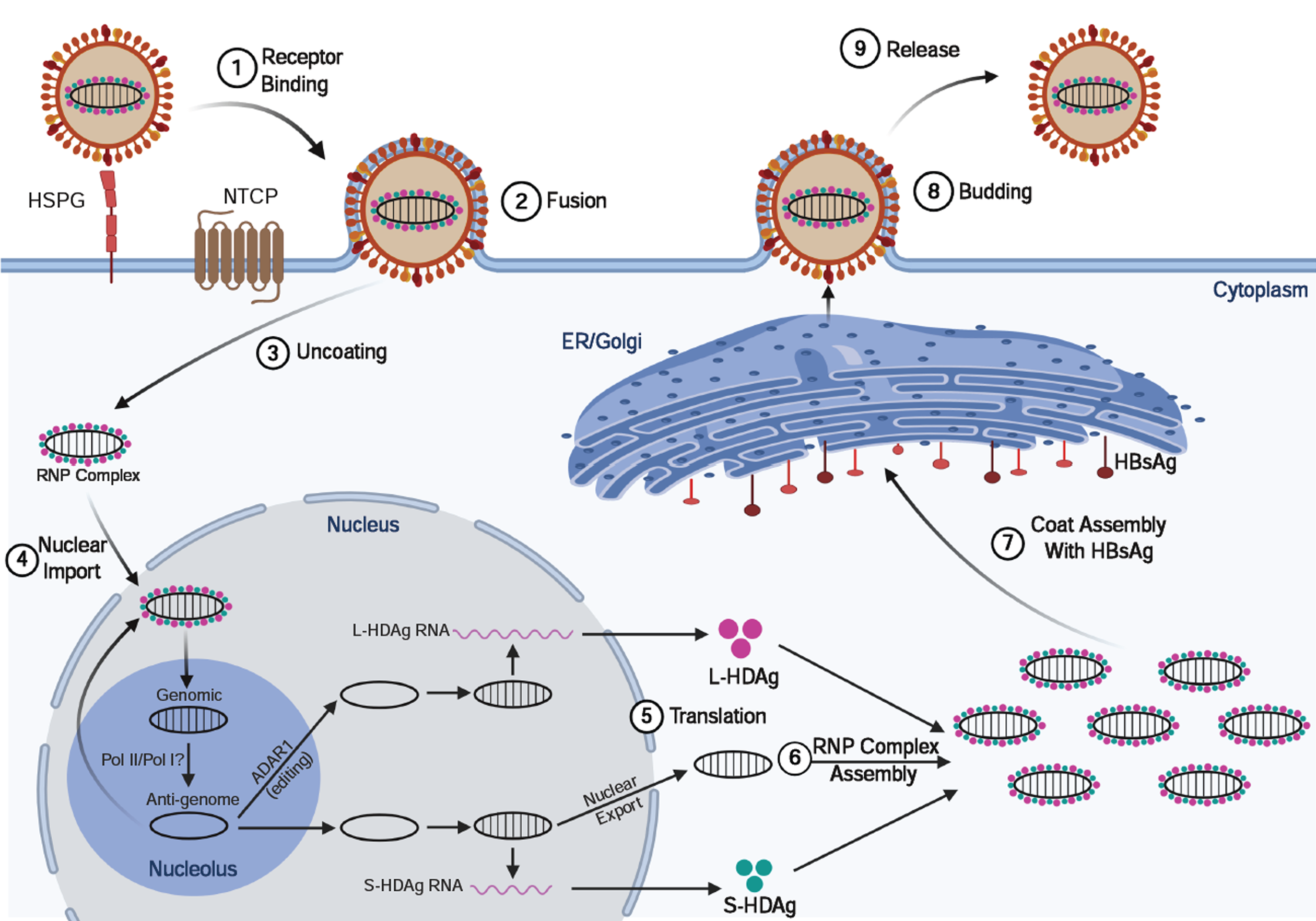
HDV is the smallest human infecting virus and the sole member of the Deltavirus genus[27]. HDV is characterized as a “satellite” or “defective” virus as it is dependent on HBV co-infection for viral assembly and persistence. HDV has an approximate 1.7 kb circular, single-stranded, negative-sense RNA genome that encodes for a single protein of two isoforms: The small and large delta antigens (S-HDAg and L-HDAg, respectively)[28]. Viral entry (Figure 2) occurs similarly to HBV due to HDV’s co-opted use of the envelope HBsAg protein[29]. Following viral entry, HDV uncoats in the cytoplasm and the ribonucleoprotein complex consisting of the HDV viral genome and HDAg complex is imported into the nucleus[30]. Rolling-circle replication occurs in the nucleolus using the host RNA polymerase II to produce antigenomic positive sense HDV RNA that serves as a template for genomic HDV RNA synthesis and protein production[31].
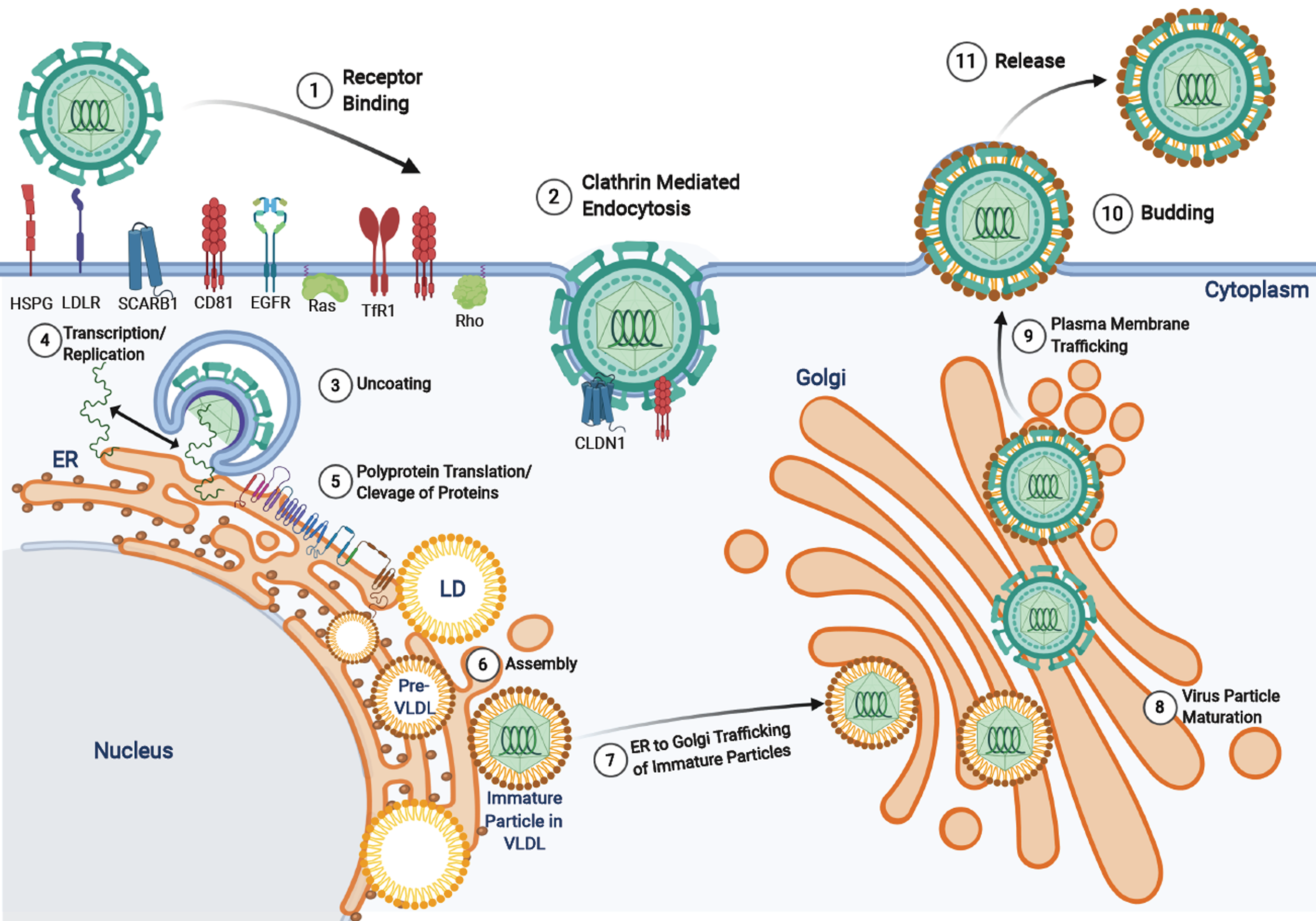
The HCV is part of the Flaviviridae family and the genus Hepacivirus. HCV is an enveloped, positive-sense, 9.6 kb single-stranded RNA virus with highly conserved 5’ and 3’ untranslated regions[34]. HCV primarily infects hepatocytes due to the expression of essential entry receptors and liver-specific cellular host factors (miRNA-122) required for viral replication[35]. However, extrahepatic manifestations have been observed in peripheral blood mononuclear cells, epithelial cells, kidneys and in the peripheral nervous system[36]. Through complex mechanisms, HCV particles interact with several receptors (see[37] for details) to induce conformational changes and proceeds to enter the cell (Figure 3) via clathrin-mediated endocytosis[37]. Endosomal acidification causes the fusion of the viral envelope to the endosome membrane, disassociation of the viral core, and release of the HCV RNA genome into the cytoplasm[37]. In the ER the viral RNA is replicated and translated from a single open reading frame using the 5’ untranslated regions internal ribosomal entry site. The translated product is an approximately 3000 amino acid polyprotein precursor that is cleaved by host and viral proteases to form ten proteins[38]. There are three structural proteins - core, E1 and E2 - and seven non-structural proteins p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B that have roles in polyprotein cleavage, viral replication, assembly, and release[39]. More recently, two isoforms of the core protein, known as the “mini-core” were discovered to be translated from an alternative open reading frame at amino acids 70 and 91 which preserve the c-terminal portion of the mature p21 core nucleocapsid but lack the N-terminus. The function of these mini-core proteins has yet to be elucidated, however, mutations in amino acid positions 70 and 91 are associated with increased risk of HCC, insulin resistance, and failure of interferon treatments[40,41]. Following viral replication and protein translation, the core protein assembles in the ER on a lipid droplet and recruits HCV viral RNA which is subsequently encapsidated[42]. The viral nucleocapsid is then processed through the ER-lumen and Golgi apparatus for maturation and the HCV particle associated with very low-density lipoproteins are released from the plasma membrane[38,43,44].
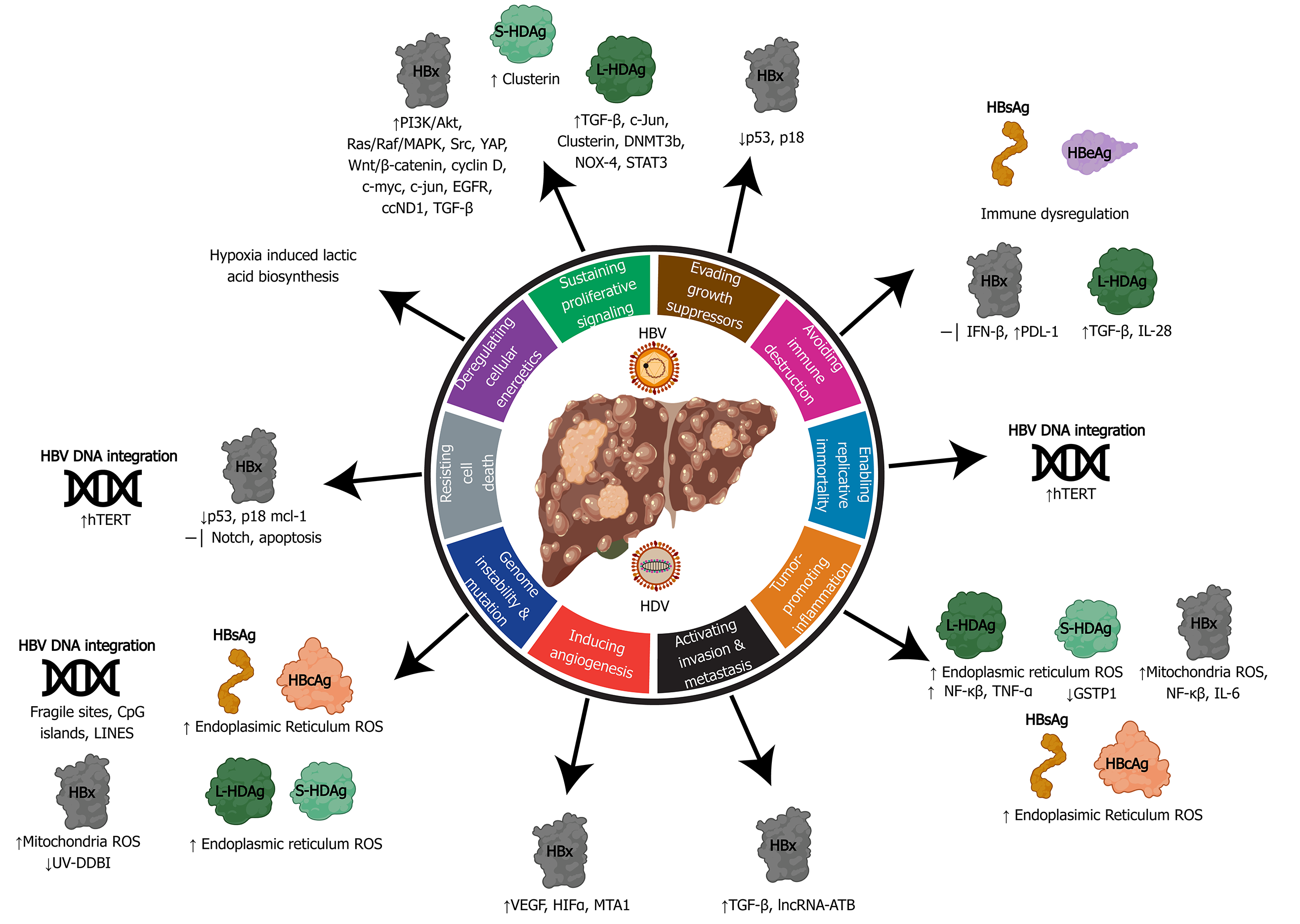
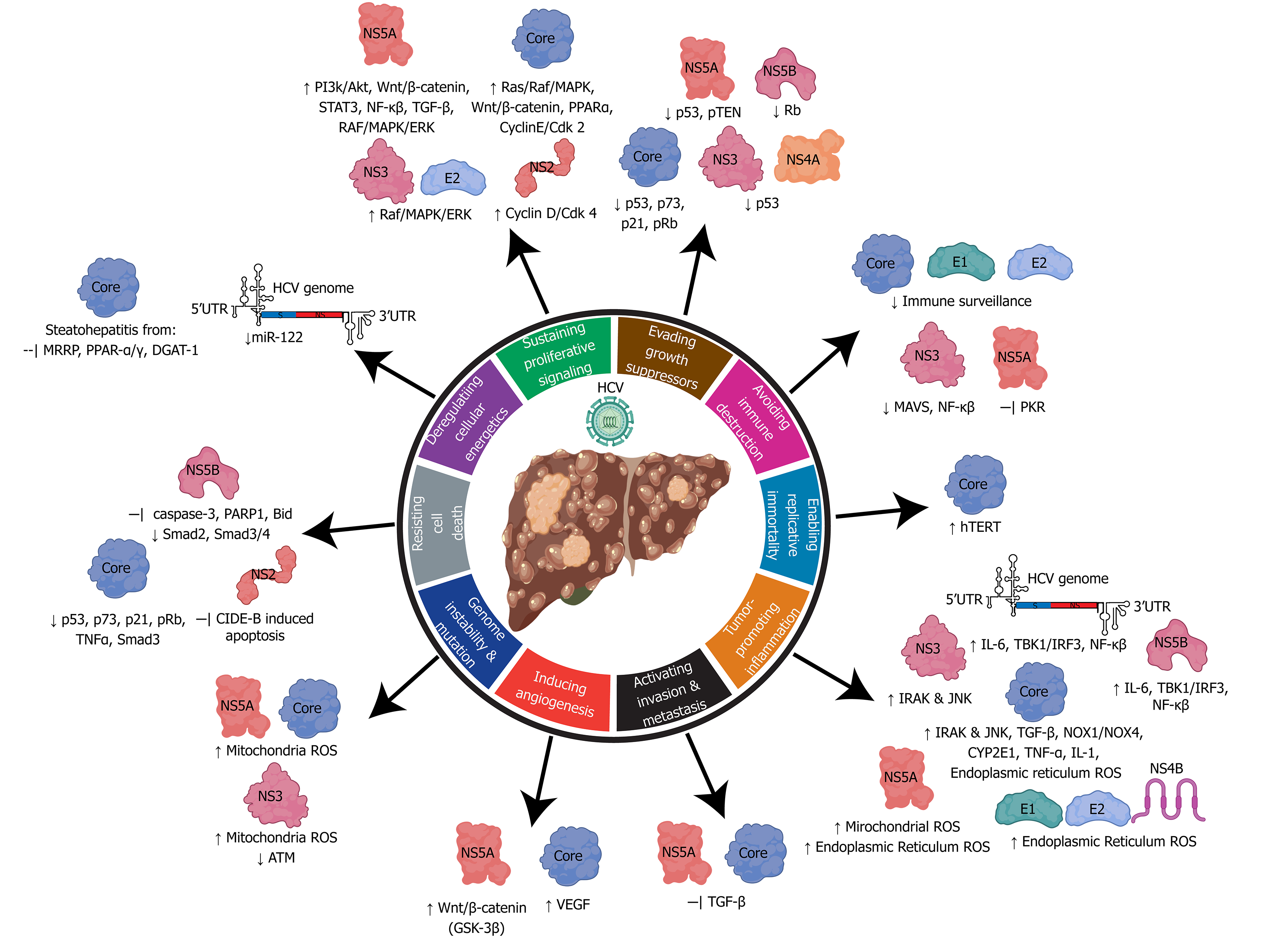
HBV, HCV, and HDV use several mechanisms to co-opt the infected cells which may unintentionally lead to HCC development. Commonly used mechanisms between all three viruses include: (1) Persistent liver inflammation and immune-mediated oxidative stress damage from a chronic viral infection; (2) Intracellular oxidative stress damage induced by viral proteins; and (3) Deregulation of cell signaling pathways by viral proteins (e.g. HBx, L-HDAg, S-HDAg, HCV core, NS3, and NS5A/B). HBV is the only hepatotropic DNA virus that also uses viral DNA integration to induce genome instability, which can lead to the creation of fusion gene products, and altered expression of oncogenes or tumor suppressors. In addition, HCV facilitates metabolic reprogramming leading to steatosis, which aids in the progression of fibrosis and HCC.
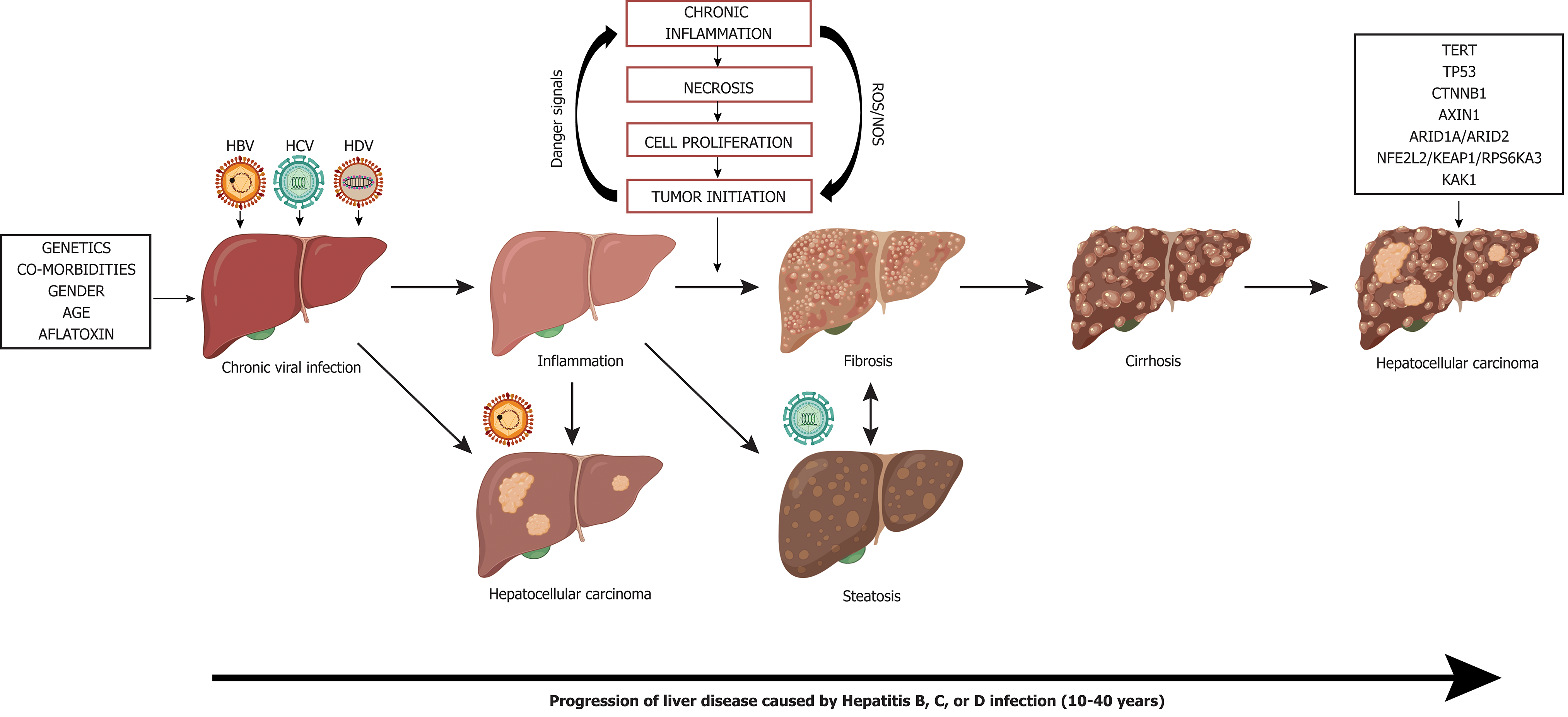
There are several viral traits that are common to human oncogenic viruses[45,46]: (1) Oncoviruses are ubiquitous in the environment and infection alone is not sufficient for cancer development. Although chronic infection with HBV/HCV/HDV results in higher rates of HCC development, not all persistently infected individuals develop liver cancer. Thus, this observation would suggest that the viruses by themselves are insufficient for cancer development. (2) Virally induced cancers are biological accidents as tumor formation is not an intentional outcome of viral infection. (3) Viral cancers appear in the context of persistent infections and occur many years to decades after initial exposure. Hepatic viruses have co-evolved with their hosts and hence, have evolved effective immune evasion strategies to establish long-term infection such as expression of viral proteins that interfere with innate interferon responses, inflammation, and adaptive immunity. (4) Most viral remnants within a tumor are non-infectious and tumors are non-permissive for viral replication. Active virion production is typically absent in transformed tumor cells. (5) All virally induced cancers have non-infectious co-factors that influence tumorigenesis. Host factors such as age, sex, genetics, environmental factors, and immunodeficiencies are associated with viral hepatitis-related HCC. (6) The immune system can play a deleterious or protective role, with some virus-associated cancers increasing with immu-nosuppression and others appearing during chronic inflammation. In the context of viral hepatitis induced HCC, the host antiviral immune response is unable to eliminate virally infected cells and instead causes immune-mediated damage. This phenomenon is evident in chronic infections where bouts of repeated hepatitis caused by the inflammation-necrosis-proliferation cycle leads to the production of reactive oxygen species (ROS) that promote genetic mutations, fibrosis, cirrhosis, and HCC.
The features of oncogenic viruses described above reflect the multifactored nature of virus-induced hepatocarcinogenesis. Human oncovirus infection alone is insufficient to directly drive cancer and viral infection provides only a portion of the oncogenic alterations. The combination of viral factors and other factors (i.e. host, environment, time) is generally required for the development of cancer[45]. The hallmarks of cancer outline developed by Hanahan and Weinberg[47] deconstruct the specifics of cellular deregulation into factors that contribute to cancer development[47]. This outline also explains the reliance on time progression to accumulate oncogenic mutations and the multistep nature of acquiring various hallmarks that eventually lead to cancer. By applying this system to viral-induced cancers, we can better understand how alterations in cellular processes induced by hepatitis viruses contribute to HCC development (depicted in Figure 4 and Figure 5).
Viral genotypes vary across the globe and play an important role in virus treatment response and assessing HCC risk. HBV has ten genotypes (A-J) which have a genetic divergence of > 8%. The HBV genotypes associated with the highest risk of HCC development are genotype C > B > F > D > A[7,48]. Some studies suggest that individuals infected with either HBV genotype B or C that have T1762/A1764 basal core promoter mutations have a higher risk of HCC development in younger individuals (< 50 years old) without cirrhosis[49]. In HBV genotype C infections, mutations/deletions in the preS region, enhancer II at position C1653T, and/or T173V in the basal core promoter can predict the development of HCC in 80% of cases[50]. Moreover, genotypes A and B have a better response to peg-IFN-α therapies, while there are no genotypic preferences for nucleos(t)ide analog treatments[51].
HCV has 6 major genotypes (1-6) that have a genetic divergence of 31%-35%. With HCV, studies linking genotype to the risk of developing HCC have inconsistent findings[7]. However more recently, a large cohort study of United States veteran concluded that HCV genotype 3 infections had an 80% higher risk of HCC development compared to genotype 1[52]. In a southeast Asian cohort, HCV genotype 6 is also associated with an increased is of HCC development[53]. With currently approved direct-acting antiviral treatments for all HCV genotypes, sustained virological response is observed in > 90% of treated individuals and reduces HCC risk in individuals without cirrhosis[54].
There are eight different HDV genotypes (1-8), which have a large genetic divergence ranging from 20%-40%[55]. There has not been a significant amount of research conducted to elucidate the effects of HDV genotypes on clinical outcomes. One study concluded that genotype 1 is associated with worse clinical disease including HCC than genotype 2[56]. Moreover, clinical outcomes of the disease are potentially regulated by both HBV and HDV genotypes. Due to the reliance of HDV on HBV co-infection, the only treatment option for HDV infection is peg-IFN-α, which is poorly tolerated and has < 30% response rate, highlighting the urgent need for improved therapies[57].
Non-resolving inflammation is a hallmark of cancer that significantly contributes to the development and progression of HCC[47]. Approximately 80% of HCC cases arise from hepatocyte injury and chronic inflammation resulting in cirrhosis[6,58]. HCC in chronic hepatitis B, C, or HBV/HDV co-infection patients occurs in the presence of cirrhosis[59,60]. In contrast, 10%-20% of HBV-related HCC can occur in the absence of cirrhosis and liver inflammation[61]. Under normal circumstances, the innate and adaptive immune responses are activated during an infection or tissue injury and immune cells are recruited to fight against the pathogen and induce wound healing. Following the elimination of the pathogen via cytolytic and non-cytolytic mechanisms, the damaged tissue is repaired through the wound-healing process[62]. However, the persistence of the inflammatory stimuli (e.g. chronic viral infection) or dysregulation of the immune regulatory mechanisms prevents complete wound-healing and causes non-resolving inflammation that may lead to liver complications resulting in autoimmunity, fibrosis, cirrhosis, metaplasia and/or tumor growth[62].
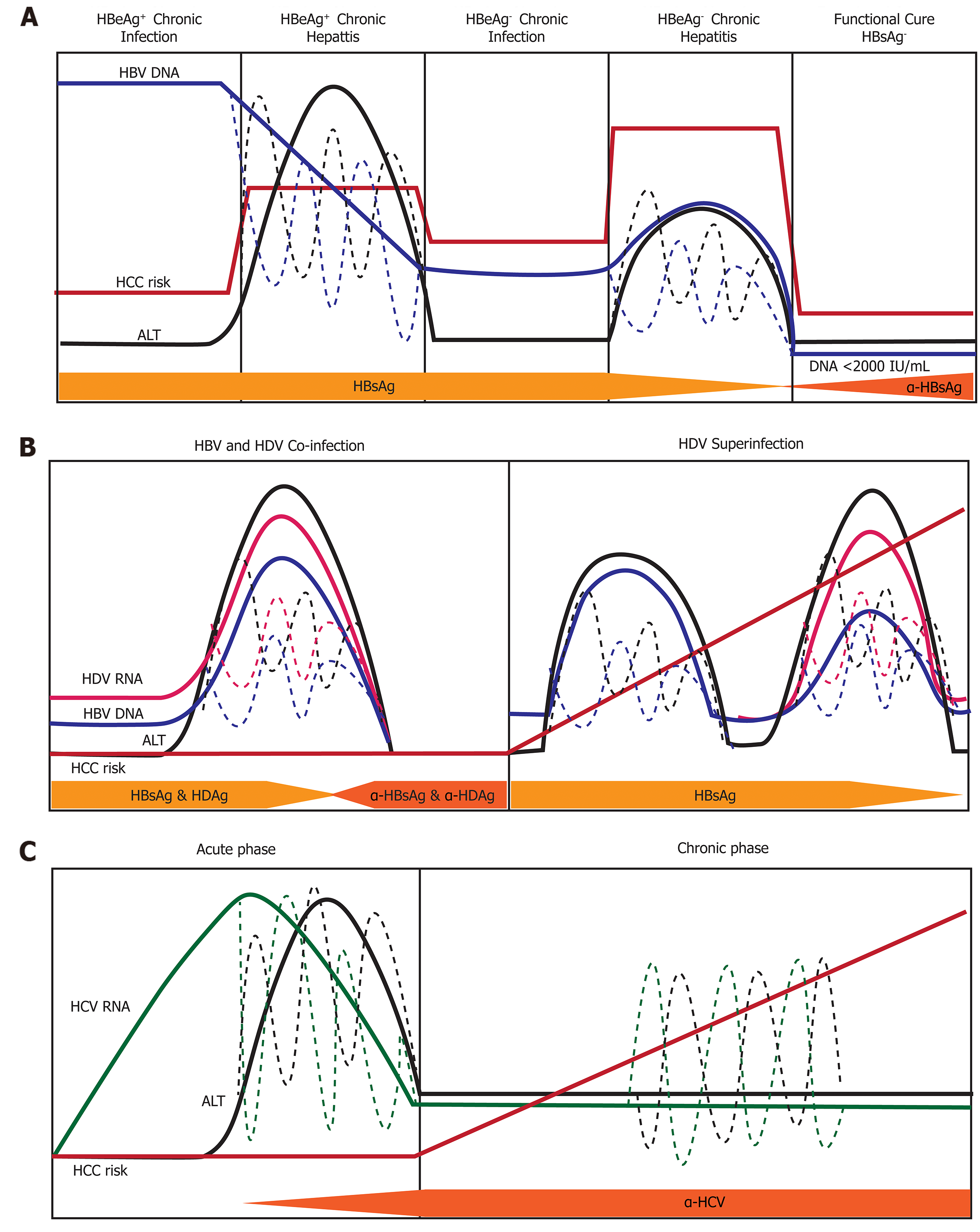
There are five clinical phases of chronic hepatitis B infection (Figure 6A) from the 2019 AASLD guidelines[63]: HBeAg+ chronic infection, HBeAg+ chronic hepatitis, HBeAg- chronic infection, HBeAg- chronic hepatitis, and a functional cure (HBsAg-). Each clinical phase is defined by a host immune response with respect to HBV viral activity. During the initial HBeAg+ chronic infection phase the host is immune response has a poorly activated HBV-specific CD8+ T-cell response[64]. Transition to the chronic hepatitis phase is characterized by increased activation of the adaptive immune response (e.g., HBV-specific CD8+ T-cells, pro-inflammatory cytokines) which causes decreased HBV DNA levels, liver inflammation, and variable/progressive liver fibrosis. Failure to completely clear HBV in the HBeAg+ chronic infection phase results in prolonged over-active immune cell-mediated damage that leads to rapid liver disease progression. Immune-mediated liver damage is facilitated by natural killer cells and T-cells through the release of ROS and proinflammatory cytokines which causes bouts of necroinflammation, hepatocyte regeneration/healing and remodeling of the liver microenvironment[65,66]. Constant necroinflammation and failed wound healing responses lead to prolonged oxidative stress exposure which can promote the rapid development of fibrosis, cirrhosis, and cell transformation (epigenetic alterations, oncogenic mutations, telomere shortening, and genomic instability)[67,68]. The 5-year cumulative HCC risk for CHB patients with cirrhosis ranges from 9.7%-15.5%[69]. However, 20% of HCC caused by HBV does not require liver cirrhosis, indicating there are other intrinsic viral associated factors that are responsible for transforming hepatocytes.
HDV infection occurs either in a co-infection model with HBV or as a superinfection from horizontal transmission in individuals with CHB (Figure 6B). The mechanisms used by HDV to modulate the immune system are different from that expressed by HBV and HCV due to the consistent presence of co-infection. The natural history of chronic HDV infection is also dynamic and can be characterized as[70]: (1) Suppressed HBV replication and active HDV replication with high ALT; (2) Slightly decreased HDV replication and HBV reactivation with moderate ALT; and (3) Late-stage disease where cirrhosis and HCC are caused by either HBV/HDV or remission resulting in a reduction of both HBV and HDV viral load. During initial infection, HDV evades IFN-α-mediated innate immune responses to promote cell survival and viral persistence[71]. Under normal cellular conditions, double-stranded RNA induces expression IFN-α which binds to the IFN receptor-associated JAK kinase tyrosine kinase-2(tyr-2). Dimerization of the tyr-2 receptor activates a JAK/STAT signaling cascade to produce innate antiviral proteins: myxovirus resistance A, 2’,5’-oligoadenylate synthase, and dsRNA-activated protein kinase[72]. HDV blocks phosphorylation of tyr-2 to prevent downstream signaling and impairs phosphorylation activity and nuclear accumulation of STAT1/STAT2[71].
The superinfection of HDV in patients with CHB has the most severe liver disease outcome, partially due to the pre-existing liver damage caused by HBV infection[73]. Moreover, superinfection with HDV leads to HBV viral load suppression through mechanisms that are not thoroughly understood[74]. Recognition of MHC-1 HDV antigens on infected cells by CD8+ T-cells mediates cellular killing. Released viral antigens are endocytosed by Kupffer cells (liver resident macrophages), B-lymphocytes, and dendritic cells and presented to CD4+ helper T-cells via MHC-II receptors. Clonal expansion of CD4+ T-cells releases IL-2, IL-10, and IFN-γ cytokines which stimulate immune-mediated killing of HDV infected cells, severe liver necrosis and progressive liver disease[75].
Infection from the HCV is usually acquired from horizontal transmission in adulthood, where 75%-80% of people develop a chronic infection from viral persistence[11]. Chronic hepatitis C (CHC) infected individuals have mild liver inflammation (Figure 6C), stable HCV RNA titers, and liver disease that progresses especially in the presence of other risk factors (age, male, obesity, diabetes alcohol, HIV or HBV co-infection)[76]. HCV infection activates intrinsic type I and III IFN responses which induces transcription of innate-antiviral IFN stimulated genes[77]. Adaptive viral-specific CD8+/CD4+ T-cells and natural killer cells facilitate the release of pro-inflammatory cytokines and growth factors while destroying of HCV infected hepatocytes by promoting the inflammation-necrosis-proliferation cycle[78]. Immune-mediated damage produces large amounts of ROS mediated DNA damage, lipid peroxidation, epigenetic modifications, mitochondrial alteration, senescence, and chromosomal translocation that lead to hepatocyte transformation[79]. Immune failure to remove all HCV infected cells causes the selection of viral escape mutants within a carrier population. These escape mutants prevent stimulation of CD4+/CD8+ T-cell responses, and aid in viral immune evasion, chronic infection, loss of immune regulation, and promotion of HCV-mediated HCC[80,81]. Moreover, persistent liver inflammation caused by immune cells over decades of infection can lead to the development of fibrosis, cirrhosis, and HCC. Approximately 10-20% of CHC patients develop cirrhosis in 20-30 years in the absence of treatment for hepatitis C, indicating the high risk for HCC development[82].
The liver is made up of approximately 80% parenchymal cells (i.e., hepatocytes) and 20% non-parenchymal cells (e.g., sinusoidal endothelial cells, hepatic stellate cells, and Kupffer cells)[83]. Infection with viral hepatitis primarily targets the large population of hepatocytes, leading to production of ROS. Release of ROS and pro-inflammatory cytokines by Kupffer cells/hepatocytes activate neighboring stellate cells and liver sinusoidal endothelial cells which are key players in the development of fibrosis[84,85] (Figure 7). Stellate cells and fibroblasts enhance collagen synthesis and alter the extracellular matrix which lead to remodeling of the liver microenvironment[86].
Progressive inflammation and fibrosis pave the way for disease progression to cirrhosis which is the largest risk factor for HCC development (Figure 7). Cirrhosis is irreversible and often individuals are asymptomatic, which makes diagnosis and treatment difficult[87]. Those who develop severe symptoms of cirrhosis, are likely to have advanced liver disease and HCC. This is especially problematic for the populations who are unaware of their infection status with HBV, HCV, and/or HDV because they are unable to seek treatment intervention to lower the risk of developing cirrhosis. During cirrhosis, altered blood flow can lead to a hypoxic environment for hepatocytes leading to altered molecular signaling and increased oxidative damage[88]. Cells within the context of cirrhosis have experienced a multitude of changes from inflammation mediated damage, repair, and regeneration. The hypoxic environment in the liver during cirrhosis can select for altered oncogenic cells and promote angiogenesis. For a comprehensive review of molecular mechanisms of host factors driving the progression of liver cirrhosis to HCC see (Fridland et al[88] and Kanda et
Although HBV uses reverse transcription for replication, unlike retroviruses, integration is not an essential step in the virus lifecycle and does not produce replication-competent virus[90]. During reverse transcription of the pgRNA, partially double-stranded rcDNA is formed 90% of the time[17]. The rcDNA is the genetic material that can be used to replenish the cccDNA pool and produce viable virions that can proceed to infect new hepatocytes. For 10% of cases, the reverse transcription process does not produce rcDNA and instead synthesizes double-stranded linear DNA (dslDNA)[17]. The HBV dslDNA can also be present in virions and repaired to produce cccDNA with a 16nt insertion that cannot produce pgRNA (unless it reverts to wild type cccDNA via homologous recombination)[91]. Integration of dslDNA is reported to occur in 1 of approximately 105-106 infected hepatocytes, and has been observed to occur early in infection (children as young as 5 mo old), and in patients who have acute HBV, CHB, and HCC[23,92]. The currently accepted mechanisms for HBV integration driving HCC include (reviewed by Tu et al[17]): (1) Chromosomal instability from HBV integrated DNA; (2) Insertional mutagenesis in proto-oncogenes and tumor suppressors; and (3) Expression of mutant HBV proteins from integration[17].
A key hallmark of cancer is genome instability. Hepatitis B virus can induce genome instability through viral integration into the host genome to cause cellular transformation (Figure 4). In non-HCC patients, HBV integration sites are randomly distributed through the genome and do not contain enriched sequence mutations. However, in CHB-HCC patients, HBV integration can be enriched in certain areas to cause chromosomal instability through integration near fragile sites: Intergenic regions, repetitive regions (e.g., LINEs), short interspaced nuclear elements, simple repeats, CpG islands, and telomeres[93]. Chromosomal rearrangements and gene copy number variations also contribute to chromosomal instability and are present in the majority of CHB associated HCC[94].
Next-generation sequencing studies that compare HBV integration sites between tumor and matched non-tumor tissues have found that HCC tumors generally have a greater number of integration events and increased integration frequency in coding or promoter regions. In non-tumorous HBV infected hepatocytes, recurrent integration in driver genes can promote hepatocyte clonal expansion[17]. In 10%-15% of HCC cases, recurrent integration of the enhancer II/core HBV promoter in/near telomerase reverse transcriptase (TERT) or myeloid/lymphoid or mixed-lineage leukemia 4 genes causes upregulation of these oncogenes[95,96]. The upregulation of these genes has been observed in early and late tumor development, which may indicate that integration in these genes may aid in cell transformation and HCC progression.
Integration of HBV dslDNA can lead to the persistent expression of mutant and truncated HBsAg, HBcAg and HBx proteins. High expression rates of these normal and mutated proteins are associated with ER and mitochondrial stress responses which can increase the risk of HCC[97]. These mutants have also been observed to stimulate hepatocyte expansion and may provide a proliferative advantage. In animal models, over-expression of mutant HBsAg and HBx show precancerous liver lesions and HCC[98]. Moreover, expression of C-terminal truncated HBx protein from integrated HBV induces stem-cell-like properties, cell transformation, tumor invasion, and inhibition of apoptosis[17,99].
The HBx protein (17 kDa) plays various roles in the HBV lifecycle and HCC development (Figure 4)[100]. HBx does not directly bind to DNA, instead, it interacts with other proteins to cause promiscuous transactivation of viral and cellular genes[101]. There are four main mechanisms used by HBx that contribute to HCC[102]: (1) Integration of HBx gene into the hepatocyte genome promoting genetic instability (Figure 4); (2) Interaction with the mitochondrial and other cellular proteins to induce oxidative stress; (3) Activation of cell survival signaling pathways and inactivation of tumor-suppressors; and (4) HBx induced epigenetic modifications such as DNA methylation, histone acetylation, and microRNA expression.
HBx modulates proto-oncogenic signaling pathways that are involved in inflammation and proliferation: Mitogen-activated protein kinase (MAPK)/ Ras/Raf/c-Jun, NF-κβ, JAK-STAT, protein kinase C, Src, survivin and PI3K cascades[101,103]. HBx has also been proposed to activate the Wnt/β-catenin pathway through the binding of Antigen presenting cells protein or inactivation of GSK-3β through Extracellular signal regulated kinase activation. These mechanisms result in the accumulation of β-catenin and increased transcription of pro-an-giogenic/metastatic factors[104,105]. HBx promotes genome instability through inhibition of UV-induced DNA damage repair pathways and S-phase progression by binding to UV-DDB1[106]. Hypoxic cirrhotic nodules expressing HBx promotes survival and growth through transcriptional activation/stabilization of HIF1α, which activates transcription of Ang-2 and vascular endothelial growth factor to promote angiogenesis and metastasis[107]. HBx also upregulates matrix metalloproteinases that digest fibrous capsules in tumors resulting in increased epithelial-mesenchymal transition (EMT) and metastasis[108]. HBx can also trans-activate cAMP-response element binding protein response element genes and Yes-associated protein, which are often over-expressed in HCC[109]. The tumor suppressor p53 can also be bound by HBx in the cytoplasm and prevent p53 nuclear localization. Binding of p53 by HBx causes deregulation of cell-cycle checkpoints, inhibition of p53 dependent apoptosis and DNA-repair pathways. Loss of p53 activity leads to genome instability and the deregulation of tumor suppressors[47].
HBx is an epigenetic regulator of DNA hyper or hypomethylation in proto-oncogenes and tumor suppressors, respectively[110]. Viral-induced upregulation of DNMTs causes aberrant hypermethylation of CpG islands in tumor suppressor, leading to gene silencing[111]. In one study, 82% of HCC tumors had at least one tumor suppressor gene inactivated by hypermethylation, indicating the important role of epigenetic modifications in cancer development[112]. HBx protein increases the transcription of methyl catalase DNMT1, which hypermethylates the tumor suppressor gene E-cadherin and INK4A[113]. Loss of INK4A leads to loss of cell-cycle regulation, and loss of E-cadherin promotes epithelial to mesenchymal transition which promotes invasion and metastasis. HBx has also shown to promote histone acetylation and deacetylation to alter the expression of cancer-related genes, microRNAs, and non-coding RNAs. Increased levels of miR-29a, miR143, miR-148a, and miR-602 by HBx promotes upregulation of genes involved in angiogenesis and metastasis[114]. There are several miRNAs that are downregulated by HBx, one of the most important being miR-122 a liver-specific miRNA that has an anti-tumorigenic role[115]. HBx also induces expression of long non-coding RNAs: LINE1 which upregulates Wnt/B-Catenin (promoting invasion and metastasis), HULC, UCA1 which inhibit tumor suppressors p18 and p27 (promoting G1/S cell cycle transition), and DBH-AS1 which activates extracellular signal-regulated kinase (ERK)/ p38/MAPK (anti-apoptosis)[114].
Individuals with CHB exhibit 1.5-4 times higher levels of oxidative stress (8-oxoguanine DNA products, lipid peroxidation, oxidation of proteins, decreased levels of anti-oxidant enzyme glutathione and higher oxidative forms) in the liver and plasma/sera compared to HBV negative individuals[116,117]. Extracellular oxidative stress can be immune mediated through the expression of pro-inflammatory cytokines or the release of ROS from cellular destruction. Intrinsic oxidative stress in the ER and mitochondria can be mediated by HBV associated proteins HBsAg, HBcAg, and HBx (Figure 4)[118]. These HBV associated proteins can be expressed from integrated HBV DNA or from the cccDNA minichromosome.
During the HBV lifecycle, secretory proteins such as the HBsAg and HBeAg are folded and assembled in the ER and transported through the Golgi[119]. High expression levels of secretory proteins or mutant HBV proteins that are misfolded can accumulate in the ER and cause activation of an unfolded protein response (UPR)[120]. The UPR induces inflammation, tissue damage from cell death, regeneration, and fibrosis (Figure 7). The wild-type and mutant LHBsAg and mutant HBcAg induce oxidative stress through protein accumulation in the ER membrane causing an UPR[121]. Excess LHBsAg leads to the blockage of HBsAg secretion, while mutant LHBsAg leads to ER stress, which may induce DNA damage and genomic instability[121-123]. A study of a Korean cohort with CHB genotype C suggested mutations in the HBcAg could upregulate ER stress resulting in ROS, increased pro-inflammatory cytokines, and increased intracellular Ca2+[124]. Activation of the UPR response by HBsAg and HBcAg causes release of hydrogen peroxide and calcium ions into the cytoplasm, enhancing ROS production[121]. HBV infection also reduces anti-oxidative stress response pathways (e.g. Nrf2/ARE, catalase, and HO-1)[125]. Moreover, HBsAg is able to promote cell transformation through immune dysregulation, upregulation of survival signaling pathways, activation of transcription factors (NF-kB, AP-1, STAT3), increased mutations through the generation of free-radicals, cell-cycle deregulation, release of pro-inflammatory cytokines, and activation of stellate cells[67,126,127].
The HBx protein can localize into several cellular compartments (mitochondria, cytoplasm, and nucleus) to aid in various roles including transcription, cell-cycle progression, protein-degradation, apoptosis, and genetic instability[100]. Localization of HBx on the outer mitochondrial membrane, causes reduced expression and activity of respiratory complex proteins I, II, IV and V in the electron transport. Reduced cellular respiration results in altered mitochondrial function, increased production of superoxide anions, and 8-oxoguanine DNA products[118,128,129].
HDV can indirectly mediate hepatocarcinogenesis through innate immune response modifications, induction of adaptive immune responses, epigenetic changes, lncRNA modifications and ROS production (Figure 4). The L-HDAg has an important role in facilitating many of these mechanisms through interaction with signaling pathways involved in pro-growth/survival, apoptosis, and wound healing[130,131]. Activation of the transforming growth factor β (TGF-β) and AP-1 pathways by L-HDAg binding of Smad3, STAT3, and c-jun promotes EMT, fibrosis, and cell-transformation[131,132].
HDV is also able to promote oxidative stress in the ER through L-HDAg’s interaction with NOX-4[133]. Activation of the NOX4 pathway causes the release of ROS which can activate STAT3 and NF-κβ signaling[133]. The L-HDAg can also promote pro-inflammatory NF-κβ activity through stimulation of TNF-α. The S-HDAg can directly bind to glutathione S-transferase P1 mRNA causing downregulation in expression, increased ROS, and apoptosis[134]. Moreover, epigenetic modifications such as histone H3 acetylation by small and large HDAg enhances clusterin gene expression[128]. Increased levels of clusterin and histone acetylation aid in HDV infected cell survival and are upregulated in cancerous cells[135,136].
Similar to HBV, individuals with chronic HCV infection experience significant decreases in antioxidant enzymes, and two to seven logs increase in liver and blood oxidative stress[137,138]. Prolonged oxidative stress results in increased levels of free oxygen radicals, DNA adduct formation (e.g. 8-oxoG), protein adducts, and lipid peroxidation[116,139,140].
In the ER, oxidative stress is mediated by HCV core, E1, E2, NS4B, and NS5A proteins (Figure 5). Viral glycoproteins E1/E2, and non-structural protein NS4B induce the unfolded protein response in the ER, which causes calcium release and production of hydrogen peroxide[141,142]. NS5A facilitates calcium uptake in the mitochondria and ER, causing release of hydrogen peroxide and organelle dysfunction[143]. HCV core-mediated binding of the mitochondria activates the mitochondrial calcium uniporter facilitating the uptake of ER released calcium ions[144]. Subsequently, an influx of calcium into the mitochondria directly effects the electron transport chain and leads to increased ROS production[144,145].
Enhanced expression of TGF-β1 from HCV core and NS5A upregulates the production of Nicotinamide adenine dinucleotide phosphate oxidases NOX1/NOX4 and cytochrome p450 2E1 oxidase (CYP2E1)[146,147]. CYP2E1 aids in metabolism of ethanol and drugs with the release of superoxide and hydrogen peroxide by-products[148]. Although CYP2E1 has an important metabolic role, high levels of expression induced by HCV core and NS5A lead to increased levels of ROS by-product accumulation[146,149]. In the context of HCV induced fibrosis, CYP2E1 expression levels are also increased which may imply that ROS have an important role in liver damage[150]. NOX1 expression and localization to the nuclear membrane is stimulated shortly after HCV infection and promotes release of superoxide ions into the cytoplasm[151]. NOX4 can be found on the nuclear or the ER membranes, which release hydrogen peroxide into the cytoplasm or nucleus promoting direct DNA damage[151].
Steatohepatitis is characterized by the presence of excess triglycerides in hepatocytes. HCV promotes steatohepatitis through enhancing lipogenesis, and impairing lipid degradation/export which may cause cellular lipotoxicity[152]. Approximately 40%-80% of CHC individuals have steatohepatitis due to viral pathogenesis, which is associated with the increased risk of HCC[153]. The HCV core protein is a key player in altering lipid metabolism through (Figure 5): (1) Decreasing lipid turnover of HCV core particles coated lipid droplets (LD); (2) Inhibition of LD mobility; (3) Inhibition of microsomal triglyceride transfer protein which prevents lipid export and degradation; and (4) Inhibition of peroxisome proliferator-activating receptor-α/γ; inhibition of diacylglycerol acetyltransferase 1[154,155]. Accumulation of free fatty acids causes severe mitochondrial and ER oxidative stress. The accumulation or ROS stimulates lipid peroxidation and activation of inflammatory signaling cascades such as TNF-α and IL-1 which can lead to the development of steatohepatitis and insulin resistance[156].
Activation of cell-survival and growth pathways are mediated by core, E2, NS2, NS3, NS4A NS5a, and NS5B proteins (Figure 5). To promote cell cycling and evasion of the G1/S checkpoint, NS5B binds to tumor suppressor Rb to facilitate proteasomal degradation and release of E2F to produce cell-cycle dependent genes[157]. NS2 activates the cyclin D/CDK 4 complex to induce the expression of cyclin E[158]. The core protein upregulates cyclin E/CDK 2 to promote cell cycle transition from G1 to S phase with checkpoint evasion, genome instability, and aberrant cell growth[159]. NS5A inactivates the tumor suppressor pTEN through binding, to cause proliferative growth and survival using the PI3K/Akt pathway[160]. HCV Core, E2, NS3, and NS5A interact with various proteins in the RAF/MAPK/ERK pathways to promote cellular proliferation[161-164]. The Wnt/β-catenin signaling pathway is activated by direct binding of β-catenin by NS5A or phospho-inactivation of GSK-3β by NS5A and core proteins[165-168]. Activation of Wnt target genes promotes proliferation, angiogenesis and EMT transfer. High quantities of β-catenin are associated with poor prognosis of HCC[169].
The inhibition of apoptosis contributes to the development of HCC through the growth of abnormal cells. The tumor suppressor protein, p53, is targeted by many HCV proteins to prevent apoptosis, DNA-repair, and senescence. NS5A directly binds to p53 causing inhibition, while NS2, NS3/4A interfere with the p53 pathway by inducing p53 delocalization from the nucleus to the cytoplasm or perinuclear regions[170-173]. There is some evidence that the HCV core protein is also able to bind to p53, however, this is debated, because high levels of core cause repression while low levels promote p53 activity[174]. To avoid cell death, HCV also has various protein mechanisms to inhibit TNF-α cytokine-mediated apoptosis: (1) The core protein activates FLICE, an inhibitor of TNF-α signaling[175]; (2) NS5A protein prevents TNF-α-mediated cell death by inhibiting activation of caspase-3 and PARP1 cleavage[176]; and (3) NS5A can also interact with intrinsic apoptosis regulator Bid to inhibit activation of apoptosis[177].
The expression of TGF- β signaling is antiproliferative and pro-apoptosis[178]. HCV NS5A binds directly to the TGF- β receptor 1 to block signaling, and as such prevents phosphorylation and nuclear localization of smad2 and the smad3/smad4 heterodimer[179]. Mutant core proteins derived from HCV tumors inhibit the TGF- β pathway through direct interaction with Smad3, which results in the inhibition of DNA-binding by the Smad3/4 heterodimer[180]. Inhibition of the TGF- β pathway promotes EMT which enhances fibrogenesis, tumor invasion, and metastasis[178].
As the progression of liver disease to HCC occurs, many common driver mutations that allow for selective growth advantage for tumor cells over normal cells can be identified (Figure 7). HCC tumors are highly heterogeneous within the same individual and these differences in tumor genetic profiles are amplified by single cell and next-generation sequencing. From sequence analysis several driver genes have been identified in the progression of HCC: TERT, tumor protein p53 (TP53), catenin beta 1 (CTNNB1), Wnt/β-catenin signaling protein AXIN1, chromatin remodeling genes ARID1A and ARID2, oxidative stress response genes NFE2L2 and KEAP1, RAS/MAPK signaling (RPS6KA3), and the JAK/STAT signaling cascade activator (KAK1). The most disrupted driver genes are described below, for a comprehensive overview refer to[181].
Regardless of geographic location, recurrent somatic mutations in the TERT promoter have been identified as the most common mutation in HCC (20.7%-59%)[181]. The TERT protein has an important role in maintaining telomere length by adding short-repeated TTAGGG nucleotides at the end of chromosomes[182]. Maintenance of the telomeres is important to avoid DNA damage, however normal adult cells do not express TERT and can only undergo 40-60 cycles of replication before senescence[183]. In HCC, activating mutations in the TERT promoter enable replicative immortality through the consistent addition of telomeric repeats which allow cells expressing TERT to replicate without entering senescence[182,183]. TERT mutations have been identified to occur early in malignant transformation and persist throughout tumor progression[184]
The tumor suppressor protein P53 is a critical protein commonly mutated in cancer, that is involved in cell-cycle arrest at the G1/S checkpoint and activation of apoptosis[185]. TP53 is most frequently mutated in its DNA binding domain, to prevent its role in activating TP53 responsive genes that aid in cell-cycle control[186]. Loss of p53 function aids cell transformation through constant cell cycling without DNA damage checkpoint regulation[5]. Mutational frequency of P53 in HCC is dependent on geographic locations, as mutational frequency requires aflatoxin exposure. High dietary aflatoxin exposure and endemic hepatitis B infection is associated with TP53 mutations located on R249S/V157F and poor prognosis[187,188].
High mutational frequency in the CTNNB1 gene which codes for the β-catenin transcription factor in the Wnt-pathway is associated with tumor progression and poor prognosis[5]. Activating mutations in CTNNB1 increase cytoplasmic accumulation of β-catenin without a Wnt signaling molecule[189]. Normally without a signaling molecule, β-catenin is expressed, bound by the destruction complex, phosphorylated by GSK3, and degraded by the proteasome[189]. However, in the presence of a Wnt ligand signal, β-catenin is not degraded and instead it is expressed to high levels and translocates into the nucleus and activates transcription factor TCF to transcribe Wnt target genes[190]. These target genes are involved in growth, proliferation, and EMT promoting metastasis[5,191]. In HCC, the tumor suppressor AXIN1 is the second most mutated gene (6.8%) in the Wnt-signaling pathway. AXIN1 is involved as a scaffold protein for the β-catenin destruction complex, and inactivating mutations prevent the complex from forming and destroying β-catenin, thus leading to increased β-catenin levels leading to tumor growth and proliferation[190,192]. The deregulation of Wnt/β-catenin signaling has been observed in 40%-70% of HCC patients. Moreover, β-catenin associated mutations occur in lower frequencies in HBV-related HCC, and are more common in HCV and alcohol-related HCC[181].
The ARID1A and ARID2 genes encode for a key subunit component of the SWI/SNF chromatin remodeling complex[193]. The dysregulation of the SWI/SNF complex contributes to tumor heterogeneity, and drug treatment resistance[181]. ARID1A acts as a tumor suppressor gene that is expressed at high levels in normal livers to regulate DNA activity using nucleosomes to restrict cell proliferation[5]. Inactivating mutations in the ARID1A and ARID2 genes occur in around 10% of HCC cases and are associated with poor prognosis, increased cell proliferation, and migration/metastases of HCC cells[181,193,194].
The low 5-year survival rate for those diagnosed with HCC is largely due to the failure of early detection of small lesions and lack of medical therapy for advanced disease[195]. The best curative treatment option for HCC is liver transplantation, however, this is limited to those who are in the early stages of HCC and follow the Milan transplantation criteria or the Alberta HCC algorithm in Canada[196,197]. Other available treatment options during earlier stages of HCC include surgical resection (70% 5-year survival rate) and radiofrequency ablation therapy (40%-70% 5-year survival rate in tumors < 2 cm)[198,199]. During intermediate HCC disease stages, transarterial chemoembolization treatment can be offered (median survival rate 16-20 mo)[200]. In advanced disease, HCC is often quite unresponsive to most chemotherapeutics, and current chemotherapeutics (Sorafenib and Lenvatinib) that target overexpressed receptor-tyrosine-kinase pathways (e.g. vascular endothelial growth factor, MAPK, EGFR, RAS, IGF, PI3K/PTEN/Akt/mTOR, Wnt/beta-catenin) only increase median survival by three months[201]. In 2018, the monoclonal antibody nivolumab which targets the programed cell death 1 receptor on T-cells was approved as a second-line therapeutic for HCC. Nivolumab has a 20% response rate in initial phase II clinical trials and works by activating T-cells for the immune-mediated killing of tumors[196].
An important feat for the future of HCC treatment will be the development of effective immunotherapies. Nivolumab was the first monoclonal antibody approved for advanced-stage HCC treatment and there are many others that are currently in clinical trials. Since both chronic viral infection and cancer create an immunosuppressive environment to prevent cytotoxic killing, future immunotherapies could target regulatory (Treg) and resident memory T-cells (TRM) to reactivate the immune system against viral infection and HCC tumors[202]. Additionally, molecular pathways deregulated by HBV/HCV/HDV could be targeted by immunotherapies through inhibition of pathways involved in aberrant cell growth leading to HCC. Alternatively, tumors caused by viral etiologies can be targeted through training of the immune system to target viral particles and/or fusion proteins in HCC[203]. Tan et al[203], describes a CAR-T cell technology that can recognize HBV specific epitopes in HCC tumors. Since HBV-associated tumors do not contain actively replicating viruses and only express partially integrated/truncated proteins, T-cells can be designed to target these tumors associated antigens. One of the patients from this study treated with HBV specific CAR-T cells had a decreased tumor volume in 5 of 6 pulmonary metastases over the course of 1-year. Building on the study performed by Tan et al[203], another possibility to improve the persistence of CAR-T technology in viral related HCC could be through engineering a separate CAR-T receptor to recognize viral antigens to boost T-cell populations while targeting cancer-specific lesions. Although immunotherapeutics for solid tumors is in its infancy, this is likely the future for the development of better treatment options for HCC.
Chronic hepatitis B, C, and delta viral infections affect almost half a billion people worldwide. Decades-long persistent viral infection and immune-mediated damage cause significant changes in the liver microenvironment and are the strongest risk factors for the development of HCC. Current treatment options for HBV and HCV reduce HCC risk, but do not eliminate it. Moreover, the lack of an HBV virological cure and limited treatment options for HDV requires the exploration of more effective treatments.
HBV and HCV can manipulate pathways in ten hallmarks of cancer, which may explain how these viruses escalate risk of HCC development. HDV is not considered a “directly” oncogenic virus, due to HDV reliance on HBV to complete the viral life cycle, uses several mechanisms to aid the progression of liver disease and increase the risk of HCC[46]. Successful epidemiology studies, in-vitro cell culture studies, and animal studies have provided us with significant insight into the molecular mechanisms of interactions between host-viral interactions. Genomic analyses comparing HCC tumors to those of healthy tissue have provided us insight into driver mutations that aid in the progression of viral-mediated HCC and possible targets for future treatments. Although much progress has been made in the field, there is a lot that remains unknown due to the lack of cell-culture systems that can be used to study all viral genotypes, co-infections, and animal models that can be infected with these viruses to produce liver disease similar to humans. The development of stronger experimental models will provide us with further insight into the role these viruses play in promoting HCC development. Until we have better screening methods and more accessible and/or effective antiviral treatments, the rates of liver cancer will be steadily on the rise. Thus, we need to investigate commonly deregulated pathways in HCC to identify targets and develop more effective treatments to improve the survival rate for those diagnosed with this disease.
Viral life cycle figures were produced using BioRender Premium.
| 1. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56665] [Article Influence: 7083.1] [Reference Citation Analysis (135)] |
| 2. | Plummer M, de Martel C, Vignat J, Ferlay J, Bray F, Franceschi S. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob Health. 2016;4:e609-e616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1068] [Cited by in RCA: 1059] [Article Influence: 105.9] [Reference Citation Analysis (0)] |
| 3. | Ward EM, Sherman RL, Henley SJ, Jemal A, Siegel DA, Feuer EJ, Firth AU, Kohler BA, Scott S, Ma J, Anderson RN, Benard V, Cronin KA. Annual Report to the Nation on the Status of Cancer, Featuring Cancer in Men and Women Age 20-49 Years. J Natl Cancer Inst. 2019;111:1279-1297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 246] [Article Influence: 41.0] [Reference Citation Analysis (1)] |
| 4. | Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2184] [Cited by in RCA: 3116] [Article Influence: 445.1] [Reference Citation Analysis (17)] |
| 5. | Ding XX, Zhu QG, Zhang SM, Guan L, Li T, Zhang L, Wang SY, Ren WL, Chen XM, Zhao J, Lin S, Liu ZZ, Bai YX, He B, Zhang HQ. Precision medicine for hepatocellular carcinoma: driver mutations and targeted therapy. Oncotarget. 2017;8:55715-55730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (2)] |
| 6. | Lee DH, Lee JM. Primary malignant tumours in the non-cirrhotic liver. Eur J Radiol. 2017;95:349-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 53] [Article Influence: 5.9] [Reference Citation Analysis (1)] |
| 7. | El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264-1273.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2183] [Cited by in RCA: 2562] [Article Influence: 183.0] [Reference Citation Analysis (3)] |
| 8. | World Health Organization. Hepatitis D. 2019. Available from: https://www.who.int/news-room/fact-sheets/detail/hepatitis-d. |
| 9. | World Health Organization. HIV and hepatitis coinfections. 2018. Available from: https://www.who.int/hiv/topics/hepatitis. |
| 10. | Miao Z, Zhang S, Ou X, Li S, Ma Z, Wang W, Peppelenbosch MP, Liu J, Pan Q. Estimating the Global Prevalence, Disease Progression, and Clinical Outcome of Hepatitis Delta Virus Infection. J Infect Dis. 2020;221:1677-1687. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 259] [Article Influence: 43.2] [Reference Citation Analysis (0)] |
| 11. | World Health Organization. Global hepatitis report, 2017. 2019. Available from: https://www.who.int/hepatitis/publications/global-hepatitis-report2017. |
| 12. | Zampino R, Pisaturo MA, Cirillo G, Marrone A, Macera M, Rinaldi L, Stanzione M, Durante-Mangoni E, Gentile I, Sagnelli E, Signoriello G, Miraglia Del Giudice E, Adinolfi LE, Coppola N. Hepatocellular carcinoma in chronic HBV-HCV co-infection is correlated to fibrosis and disease duration. Ann Hepatol. 2015;14:75-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 13. | Grabowski J, Wedemeyer H. Hepatitis delta: immunopathogenesis and clinical challenges. Dig Dis. 2010;28:133-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (1)] |
| 14. | Joshi SS, Coffin CS. Hepatitis B and Pregnancy: Virologic and Immunologic Characteristics. Hepatol Commun. 2020;4:157-171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 15. | Papatheodoridis GV, Chan HL, Hansen BE, Janssen HL, Lampertico P. Risk of hepatocellular carcinoma in chronic hepatitis B: assessment and modification with current antiviral therapy. J Hepatol. 2015;62:956-967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 419] [Article Influence: 38.1] [Reference Citation Analysis (2)] |
| 16. | Kanwal F, Kramer J, Asch SM, Chayanupatkul M, Cao Y, El-Serag HB. Risk of Hepatocellular Cancer in HCV Patients Treated With Direct-Acting Antiviral Agents. Gastroenterology. 2017;153:996-1005.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 523] [Cited by in RCA: 718] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 17. | Tu T, Budzinska MA, Shackel NA, Urban S. HBV DNA Integration: Molecular Mechanisms and Clinical Implications. Viruses. 2017;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 200] [Cited by in RCA: 326] [Article Influence: 36.2] [Reference Citation Analysis (2)] |
| 18. | Coffin CS, Mulrooney-Cousins PM, van Marle G, Roberts JP, Michalak TI, Terrault NA. Hepatitis B virus quasispecies in hepatic and extrahepatic viral reservoirs in liver transplant recipients on prophylactic therapy. Liver Transpl. 2011;17:955-962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (1)] |
| 19. | Virlogeux V, Trépo C. Extrahepatic Manifestations of Chronic Hepatitis B Infection. Curr Hepatology Rep. 2018;17:156-165. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (2)] |
| 20. | Lau KC, Joshi SS, Gao S, Giles E, Swidinsky K, van Marle G, Bathe OF, Urbanski SJ, Terrault NA, Burak KW, Osiowy C, Coffin CS. Oncogenic HBV variants and integration are present in hepatic and lymphoid cells derived from chronic HBV patients. Cancer Lett. 2020;480:39-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 21. | Seeger C, Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000;64:51-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1094] [Cited by in RCA: 1122] [Article Influence: 43.2] [Reference Citation Analysis (2)] |
| 22. | Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. Elife. 2012;1:e00049. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1280] [Cited by in RCA: 1648] [Article Influence: 117.7] [Reference Citation Analysis (1)] |
| 23. | Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology. 2015;479-480:672-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 569] [Cited by in RCA: 655] [Article Influence: 59.5] [Reference Citation Analysis (1)] |
| 24. | Nassal M. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut. 2015;64:1972-1984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 727] [Article Influence: 66.1] [Reference Citation Analysis (0)] |
| 25. | Summers J, Mason WS. Replication of the genome of a hepatitis B--like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1108] [Cited by in RCA: 1118] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 26. | Turton KL, Meier-Stephenson V, Badmalia MD, Coffin CS, Patel TR. Host Transcription Factors in Hepatitis B Virus RNA Synthesis. Viruses. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 66] [Article Influence: 11.0] [Reference Citation Analysis (2)] |
| 27. | Wang KS, Choo QL, Weiner AJ, Ou JH, Najarian RC, Thayer RM, Mullenbach GT, Denniston KJ, Gerin JL, Houghton M. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature. 1986;323:508-514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 537] [Cited by in RCA: 541] [Article Influence: 13.5] [Reference Citation Analysis (1)] |
| 28. | Weiner AJ, Choo QL, Wang KS, Govindarajan S, Redeker AG, Gerin JL, Houghton M. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 delta and p27 delta. J Virol. 1988;62:594-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 181] [Article Influence: 4.8] [Reference Citation Analysis (1)] |
| 29. | Sureau C, Guerra B, Lanford RE. Role of the large hepatitis B virus envelope protein in infectivity of the hepatitis delta virion. J Virol. 1993;67:366-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 116] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 30. | Cunha C, Monjardino J, Cheng D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus RNA in the nucleus of human cells. RNA. 1998;4:680-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 1.0] [Reference Citation Analysis (4)] |
| 31. | Lai MM. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 213] [Article Influence: 6.9] [Reference Citation Analysis (1)] |
| 32. | Wong SK, Lazinski DW. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc Natl Acad Sci. 2002;99:15118-15123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 156] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 33. | Sureau C, Negro F. The hepatitis delta virus: Replication and pathogenesis. J Hepatol. 2016;64:S102-S116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 205] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 34. | Lindenbach BD, Rice CM. Unravelling hepatitis C virus replication from genome to function. Nature. 2005;436:933-938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 602] [Cited by in RCA: 606] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 35. | Ploss A, Evans MJ. Hepatitis C virus host cell entry. Curr Opin Virol. 2012;2:14-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (1)] |
| 36. | Cacoub P, Comarmond C, Domont F, Savey L, Desbois AC, Saadoun D. Extrahepatic manifestations of chronic hepatitis C virus infection. Ther Adv Infect Dis. 2016;3:3-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 126] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 37. | Ding Q, von Schaewen M, Ploss A. The impact of hepatitis C virus entry on viral tropism. Cell Host Microbe. 2014;16:562-568. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (1)] |
| 38. | Morozov VA, Lagaye S. Hepatitis C virus: Morphogenesis, infection and therapy. World J Hepatol. 2018;10:186-212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (14)] |
| 39. | Kim CW, Chang KM. Hepatitis C virus: virology and life cycle. Clin Mol Hepatol. 2013;19:17-25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 106] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 40. | Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Arase Y, Ikeda K, Kumada H. Amino acid substitutions in the hepatitis C virus core region are the important predictor of hepatocarcinogenesis. Hepatology. 2007;46:1357-1364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 96] [Article Influence: 5.1] [Reference Citation Analysis (1)] |
| 41. | Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Saitoh S, Arase Y, Ikeda K, Kumada H. Amino acid substitutions in the hepatitis C virus core region of genotype 1b are the important predictor of severe insulin resistance in patients without cirrhosis and diabetes mellitus. J Med Virol. 2009;81:1032-1039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 2.4] [Reference Citation Analysis (1)] |
| 42. | Moriishi K, Matsuura Y. Exploitation of lipid components by viral and host proteins for hepatitis C virus infection. Front Microbiol. 2012;3:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (2)] |
| 43. | Grassi G, Di Caprio G, Fimia GM, Ippolito G, Tripodi M, Alonzi T. Hepatitis C virus relies on lipoproteins for its life cycle. World J Gastroenterol. 2016;22:1953-1965. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 40] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (2)] |
| 44. | Scheel TK, Rice CM. Understanding the hepatitis C virus life cycle paves the way for highly effective therapies. Nat Med. 2013;19:837-849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 431] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 45. | Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology. 2009;392:1-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 276] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 46. | Chang Y, Moore PS, Weiss RA. Human oncogenic viruses: nature and discovery. Philos Trans R Soc Lond B Biol Sci. 2017;372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 47. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51728] [Cited by in RCA: 48738] [Article Influence: 3249.2] [Reference Citation Analysis (12)] |
| 48. | Livingston SE, Simonetti JP, McMahon BJ, Bulkow LR, Hurlburt KJ, Homan CE, Snowball MM, Cagle HH, Williams JL, Chulanov VP. Hepatitis B virus genotypes in Alaska Native people with hepatocellular carcinoma: preponderance of genotype F. J Infect Dis. 2007;195:5-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 193] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 49. | Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003;124:327-334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 414] [Cited by in RCA: 428] [Article Influence: 18.6] [Reference Citation Analysis (2)] |
| 50. | Liu S, Zhang H, Gu C, Yin J, He Y, Xie J, Cao G. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101:1066-1082. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 279] [Cited by in RCA: 332] [Article Influence: 19.5] [Reference Citation Analysis (1)] |
| 51. | Lin CL, Kao JH. Hepatitis B virus genotypes and variants. Cold Spring Harb Perspect Med. 2015;5:a021436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 193] [Article Influence: 17.5] [Reference Citation Analysis (1)] |
| 52. | Kanwal F, Kramer JR, Ilyas J, Duan Z, El-Serag HB. HCV genotype 3 is associated with an increased risk of cirrhosis and hepatocellular cancer in a national sample of U.S. Veterans with HCV. Hepatology. 2014;60:98-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 248] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 53. | Lee MH, Hsiao TI, Subramaniam SR, Le AK, Vu VD, Trinh HN, Zhang J, Jin M, Wong VW, Wong GL, Nguyen MH. HCV Genotype 6 Increased the Risk for Hepatocellular Carcinoma Among Asian Patients With Liver Cirrhosis. Am J Gastroenterol. 2017;112:1111-1119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 54. | Zoratti MJ, Siddiqua A, Morassut RE, Zeraatkar D, Chou R, van Holten J, Xie F, Druyts E. Pangenotypic direct acting antivirals for the treatment of chronic hepatitis C virus infection: A systematic literature review and meta-analysis. EClinicalMedicine. 2020;18:100237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 71] [Article Influence: 11.8] [Reference Citation Analysis (2)] |
| 55. | Botelho-Souza LF, Souza Vieira D, de Oliveira Dos Santos A, Cunha Pereira AV, Villalobos-Salcedo JM. Characterization of the Genotypic Profile of Hepatitis Delta Virus: Isolation of HDV Genotype-1 in the Western Amazon Region of Brazil. Intervirology. 2015;58:166-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (3)] |
| 56. | Su CW, Huang YH, Huo TI, Shih HH, Sheen IJ, Chen SW, Lee PC, Lee SD, Wu JC. Genotypes and viremia of hepatitis B and D viruses are associated with outcomes of chronic hepatitis D patients. Gastroenterology. 2006;130:1625-1635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 163] [Article Influence: 8.2] [Reference Citation Analysis (1)] |
| 57. | Hughes SA, Wedemeyer H, Harrison PM. Hepatitis delta virus. Lancet. 2011;378:73-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 371] [Cited by in RCA: 406] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 58. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3846] [Cited by in RCA: 4301] [Article Influence: 226.4] [Reference Citation Analysis (2)] |
| 59. | Puigvehí M, Moctezuma-Velázquez C, Villanueva A, Llovet JM. The oncogenic role of hepatitis delta virus in hepatocellular carcinoma. JHEP Rep. 2019;1:120-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 62] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 60. | Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, Pracht M, Yokosuka O, Rosmorduc O, Breder V, Gerolami R, Masi G, Ross PJ, Song T, Bronowicki JP, Ollivier-Hourmand I, Kudo M, Cheng AL, Llovet JM, Finn RS, LeBerre MA, Baumhauer A, Meinhardt G, Han G; RESORCE Investigators. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2160] [Cited by in RCA: 2839] [Article Influence: 315.4] [Reference Citation Analysis (1)] |
| 61. | Chayanupatkul M, Omino R, Mittal S, Kramer JR, Richardson P, Thrift AP, El-Serag HB, Kanwal F. Hepatocellular carcinoma in the absence of cirrhosis in patients with chronic hepatitis B virus infection. J Hepatol. 2017;66:355-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 107] [Article Influence: 11.9] [Reference Citation Analysis (1)] |
| 62. | Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3548] [Cited by in RCA: 4584] [Article Influence: 254.7] [Reference Citation Analysis (0)] |
| 63. | Terrault NA, Lok ASF, McMahon BJ, Chang KM, Hwang JP, Jonas MM, Brown RS Jr, Bzowej NH, Wong JB. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67:1560-1599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2290] [Cited by in RCA: 3090] [Article Influence: 386.3] [Reference Citation Analysis (1)] |
| 64. | Kennedy PTF, Sandalova E, Jo J, Gill U, Ushiro-Lumb I, Tan AT, Naik S, Foster GR, Bertoletti A. Preserved T-cell function in children and young adults with immune-tolerant chronic hepatitis B. Gastroenterology. 2012;143:637-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 252] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 65. | Yang P, Markowitz GJ, Wang XF. The hepatitis B virus-associated tumor microenvironment in hepatocellular carcinoma. Natl Sci Rev. 2014;1:396-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 66. | Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1202] [Cited by in RCA: 1225] [Article Influence: 58.3] [Reference Citation Analysis (0)] |
| 67. | Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093-5107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 374] [Cited by in RCA: 375] [Article Influence: 16.3] [Reference Citation Analysis (1)] |
| 68. | Mani SKK, Andrisani O. Hepatitis B Virus-Associated Hepatocellular Carcinoma and Hepatic Cancer Stem Cells. Genes (Basel). 2018;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 69. | Varbobitis I, Papatheodoridis GV. The assessment of hepatocellular carcinoma risk in patients with chronic hepatitis B under antiviral therapy. Clin Mol Hepatol. 2016;22:319-326. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 78] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 70. | Wu JC, Chen TZ, Huang YS, Yen FS, Ting LT, Sheng WY, Tsay SH, Lee SD. Natural history of hepatitis D viral superinfection: significance of viremia detected by polymerase chain reaction. Gastroenterology. 1995;108:796-802. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 108] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 71. | Pugnale P, Pazienza V, Guilloux K, Negro F. Hepatitis delta virus inhibits alpha interferon signaling. Hepatology. 2009;49:398-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 78] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 72. | Larner A, Reich NC. Interferon signal transduction. Biotherapy. 1996;8:175-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.0] [Reference Citation Analysis (1)] |
| 73. | Smedile A, Farci P, Verme G, Caredda F, Cargnel A, Caporaso N, Dentico P, Trepo C, Opolon P, Gimson A, Vergani D, Williams R, Rizzetto M. Influence of delta infection on severity of hepatitis B. Lancet. 1982;2:945-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 283] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 74. | Wu JC, Chen PJ, Kuo MY, Lee SD, Chen DS, Ting LP. Production of hepatitis delta virus and suppression of helper hepatitis B virus in a human hepatoma cell line. J Virol. 1991;65:1099-1104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 119] [Article Influence: 3.4] [Reference Citation Analysis (4)] |
| 75. | Nisini R, Paroli M, Accapezzato D, Bonino F, Rosina F, Santantonio T, Sallusto F, Amoroso A, Houghton M, Barnaba V. Human CD4+ T-cell response to hepatitis delta virus: identification of multiple epitopes and characterization of T-helper cytokine profiles. J Virol. 1997;71:2241-2251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 83] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 76. | Thein HH, Yi Q, Dore GJ, Krahn MD. Estimation of stage-specific fibrosis progression rates in chronic hepatitis C virus infection: a meta-analysis and meta-regression. Hepatology. 2008;48:418-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 598] [Cited by in RCA: 629] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 77. | Mihm S. Activation of Type I and Type III Interferons in Chronic Hepatitis C. J Innate Immun. 2015;7:251-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 1.4] [Reference Citation Analysis (2)] |
| 78. | Lechner F, Wong DK, Dunbar PR, Chapman R, Chung RT, Dohrenwend P, Robbins G, Phillips R, Klenerman P, Walker BD. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191:1499-1512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1003] [Cited by in RCA: 1004] [Article Influence: 38.6] [Reference Citation Analysis (1)] |
| 79. | Bartsch H, Nair J. Chronic inflammation and oxidative stress in the genesis and perpetuation of cancer: role of lipid peroxidation, DNA damage, and repair. Langenbecks Arch Surg. 2006;391:499-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 355] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 80. | Chang KM, Rehermann B, McHutchison JG, Pasquinelli C, Southwood S, Sette A, Chisari FV. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J Clin Invest. 1997;100:2376-2385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 241] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 81. | Wölfl M, Rutebemberwa A, Mosbruger T, Mao Q, Li HM, Netski D, Ray SC, Pardoll D, Sidney J, Sette A, Allen T, Kuntzen T, Kavanagh DG, Kuball J, Greenberg PD, Cox AL. Hepatitis C virus immune escape via exploitation of a hole in the T cell repertoire. J Immunol. 2008;181:6435-6446. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 82. | Westbrook RH, Dusheiko G. Natural history of hepatitis C. J Hepatol. 2014;61:S58-S68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 552] [Cited by in RCA: 630] [Article Influence: 52.5] [Reference Citation Analysis (1)] |
| 83. | Werner M, Driftmann S, Kleinehr K, Kaiser GM, Mathé Z, Treckmann JW, Paul A, Skibbe K, Timm J, Canbay A, Gerken G, Schlaak JF, Broering R. All-In-One: Advanced preparation of Human Parenchymal and Non-Parenchymal Liver Cells. PLoS One. 2015;10:e0138655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (1)] |
| 84. | Zhou WC, Zhang QB, Qiao L. Pathogenesis of liver cirrhosis. World J Gastroenterol. 2014;20:7312-7324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 335] [Cited by in RCA: 436] [Article Influence: 36.3] [Reference Citation Analysis (10)] |
| 85. | Li JT, Liao ZX, Ping J, Xu D, Wang H. Molecular mechanism of hepatic stellate cell activation and antifibrotic therapeutic strategies. J Gastroenterol. 2008;43:419-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 122] [Article Influence: 6.8] [Reference Citation Analysis (1)] |
| 86. | Xu L, Hui AY, Albanis E, Arthur MJ, O'Byrne SM, Blaner WS, Mukherjee P, Friedman SL, Eng FJ. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54:142-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 686] [Cited by in RCA: 878] [Article Influence: 41.8] [Reference Citation Analysis (1)] |
| 87. | Bruix J, Reig M, Sherman M. Evidence-Based Diagnosis, Staging, and Treatment of Patients With Hepatocellular Carcinoma. Gastroenterology. 2016;150:835-853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 1315] [Article Influence: 131.5] [Reference Citation Analysis (3)] |
| 88. | Fridland A, Verhoef V. Mechanism for ara-CTP catabolism in human leukemic cells and effect of deaminase inhibitors on this process. Semin Oncol. 1987;14:262-268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 89] [Cited by in RCA: 120] [Article Influence: 15.0] [Reference Citation Analysis (2)] |
| 89. | Kanda T, Goto T, Hirotsu Y, Moriyama M, Omata M. Molecular Mechanisms Driving Progression of Liver Cirrhosis towards Hepatocellular Carcinoma in Chronic Hepatitis B and C Infections: A Review. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 206] [Article Influence: 29.4] [Reference Citation Analysis (1)] |
| 90. | Ringelhan M, McKeating JA, Protzer U. Viral hepatitis and liver cancer. Philos Trans R Soc Lond B Biol Sci. 2017;372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 161] [Cited by in RCA: 249] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 91. | Yang W, Summers J. Infection of ducklings with virus particles containing linear double-stranded duck hepatitis B virus DNA: illegitimate replication and reversion. J Virol. 1998;72:8710-8717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 51] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 92. | Yaginuma K, Kobayashi H, Kobayashi M, Morishima T, Matsuyama K, Koike K. Multiple integration site of hepatitis B virus DNA in hepatocellular carcinoma and chronic active hepatitis tissues from children. J Virol. 1987;61:1808-1813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 97] [Article Influence: 2.5] [Reference Citation Analysis (1)] |
| 93. | Yan H, Yang Y, Zhang L, Tang G, Wang Y, Xue G, Zhou W, Sun S. Characterization of the genotype and integration patterns of hepatitis B virus in early- and late-onset hepatocellular carcinoma. Hepatology. 2015;61:1821-1831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 75] [Article Influence: 6.8] [Reference Citation Analysis (1)] |
| 94. | Jiang Z, Jhunjhunwala S, Liu J, Haverty PM, Kennemer MI, Guan Y, Lee W, Carnevali P, Stinson J, Johnson S, Diao J, Yeung S, Jubb A, Ye W, Wu TD, Kapadia SB, de Sauvage FJ, Gentleman RC, Stern HM, Seshagiri S, Pant KP, Modrusan Z, Ballinger DG, Zhang Z. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res. 2012;22:593-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 239] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 95. | Dong H, Zhang L, Qian Z, Zhu X, Zhu G, Chen Y, Xie X, Ye Q, Zang J, Ren Z, Ji Q. Identification of HBV-MLL4 Integration and Its Molecular Basis in Chinese Hepatocellular Carcinoma. PLoS One. 2015;10:e0123175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 96. | Sung WK, Zheng H, Li S, Chen R, Liu X, Li Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, Mulawadi FH, Wong KF, Liu AM, Poon RT, Fan ST, Chan KL, Gong Z, Hu Y, Lin Z, Wang G, Zhang Q, Barber TD, Chou WC, Aggarwal A, Hao K, Zhou W, Zhang C, Hardwick J, Buser C, Xu J, Kan Z, Dai H, Mao M, Reinhard C, Wang J, Luk JM. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765-769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 745] [Article Influence: 53.2] [Reference Citation Analysis (0)] |
| 97. | Wang HC, Wu HC, Chen CF, Fausto N, Lei HY, Su IJ. Different types of ground glass hepatocytes in chronic hepatitis B virus infection contain specific pre-S mutants that may induce endoplasmic reticulum stress. Am J Pathol. 2003;163:2441-2449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 197] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 98. | Chisari FV, Filippi P, Buras J, McLachlan A, Popper H, Pinkert CA, Palmiter RD, Brinster RL. Structural and pathological effects of synthesis of hepatitis B virus large envelope polypeptide in transgenic mice. Proc Natl Acad Sci USA. 1987;84:6909-6913. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 262] [Cited by in RCA: 273] [Article Influence: 7.0] [Reference Citation Analysis (3)] |
| 99. | Ng KY, Chai S, Tong M, Guan XY, Lin CH, Ching YP, Xie D, Cheng AS, Ma S. C-terminal truncated hepatitis B virus X protein promotes hepatocellular carcinogenesis through induction of cancer and stem cell-like properties. Oncotarget. 2016;7:24005-24017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (1)] |
| 100. | Ma J, Sun T, Park S, Shen G, Liu J. The role of hepatitis B virus X protein is related to its differential intracellular localization. Acta Biochim Biophys Sin (Shanghai). 2011;43:583-588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (1)] |
| 101. | Murakami S. Hepatitis B virus X protein: a multifunctional viral regulator. J Gastroenterol. 2001;36:651-660. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 261] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 102. | Shlomai A, de Jong YP, Rice CM. Virus associated malignancies: the role of viral hepatitis in hepatocellular carcinoma. Semin Cancer Biol. 2014;26:78-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 141] [Article Influence: 11.8] [Reference Citation Analysis (1)] |
| 103. | Feitelson MA, Duan LX. Hepatitis B virus X antigen in the pathogenesis of chronic infections and the development of hepatocellular carcinoma. Am J Pathol. 1997;150:1141-1157. [PubMed] |
| 104. | Cha MY, Kim CM, Park YM, Ryu WS. Hepatitis B virus X protein is essential for the activation of Wnt/beta-catenin signaling in hepatoma cells. Hepatology. 2004;39:1683-1693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 192] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 105. | Hsieh A, Kim HS, Lim SO, Yu DY, Jung G. Hepatitis B viral X protein interacts with tumor suppressor adenomatous polyposis coli to activate Wnt/β-catenin signaling. Cancer Lett. 2011;300:162-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 111] [Article Influence: 7.4] [Reference Citation Analysis (1)] |
| 106. | Martin-Lluesma S, Schaeffer C, Robert EI, van Breugel PC, Leupin O, Hantz O, Strubin M. Hepatitis B virus X protein affects S phase progression leading to chromosome segregation defects by binding to damaged DNA binding protein 1. Hepatology. 2008;48:1467-1476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 88] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 107. | Yoo YG, Oh SH, Park ES, Cho H, Lee N, Park H, Kim DK, Yu DY, Seong JK, Lee MO. Hepatitis B virus X protein enhances transcriptional activity of hypoxia-inducible factor-1alpha through activation of mitogen-activated protein kinase pathway. J Biol Chem. 2003;278:39076-39084. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 151] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 108. | Martin-Vilchez S, Lara-Pezzi E, Trapero-Marugán M, Moreno-Otero R, Sanz-Cameno P. The molecular and pathophysiological implications of hepatitis B X antigen in chronic hepatitis B virus infection. Rev Med Virol. 2011;21:315-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 109. | Zhang T, Zhang J, You X, Liu Q, Du Y, Gao Y, Shan C, Kong G, Wang Y, Yang X, Ye L, Zhang X. Hepatitis B virus X protein modulates oncogene Yes-associated protein by CREB to promote growth of hepatoma cells. Hepatology. 2012;56:2051-2059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 110. | Tian Y, Yang W, Song J, Wu Y, Ni B. Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol Cell Biol. 2013;33:2810-2816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 117] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 111. | Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042-2054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2489] [Cited by in RCA: 2459] [Article Influence: 106.9] [Reference Citation Analysis (0)] |
| 112. | Yang B, Guo M, Herman JG, Clark DP. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol. 2003;163:1101-1107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 288] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 113. | Jung SY, Kim YJ. C-terminal region of HBx is crucial for mitochondrial DNA damage. Cancer Lett. 2013;331:76-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 54] [Article Influence: 3.9] [Reference Citation Analysis (1)] |
| 114. | Zhang B, Han S, Feng B, Chu X, Chen L, Wang R. Hepatitis B virus X protein-mediated non-coding RNA aberrations in the development of human hepatocellular carcinoma. Exp Mol Med. 2017;49:e293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 46] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 115. | Liang HW, Wang N, Wang Y, Wang F, Fu Z, Yan X, Zhu H, Diao W, Ding Y, Chen X, Zhang CY, Zen K. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J Hepatol. 2016;64:278-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 103] [Article Influence: 10.3] [Reference Citation Analysis (1)] |
| 116. | Kitada T, Seki S, Iwai S, Yamada T, Sakaguchi H, Wakasa K. In situ detection of oxidative DNA damage, 8-hydroxydeoxyguanosine, in chronic human liver disease. J Hepatol. 2001;35:613-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 89] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 117. | Swietek K, Juszczyk J. Reduced glutathione concentration in erythrocytes of patients with acute and chronic viral hepatitis. J Viral Hepat. 1997;4:139-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 64] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 118. | Ivanov AV, Valuev-Elliston VT, Tyurina DA, Ivanova ON, Kochetkov SN, Bartosch B, Isaguliants MG. Oxidative stress, a trigger of hepatitis C and B virus-induced liver carcinogenesis. Oncotarget. 2017;8:3895-3932. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 136] [Cited by in RCA: 132] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 119. | Tong S, Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol. 2016;64:S4-S16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 242] [Cited by in RCA: 328] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 120. | Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3842] [Cited by in RCA: 4722] [Article Influence: 337.3] [Reference Citation Analysis (1)] |
| 121. | Hsieh YH, Su IJ, Wang HC, Chang WW, Lei HY, Lai MD, Chang WT, Huang W. Pre-S mutant surface antigens in chronic hepatitis B virus infection induce oxidative stress and DNA damage. Carcinogenesis. 2004;25:2023-2032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 258] [Article Influence: 11.7] [Reference Citation Analysis (1)] |
| 122. | Xu Z, Jensen G, Yen TS. Activation of hepatitis B virus S promoter by the viral large surface protein via induction of stress in the endoplasmic reticulum. J Virol. 1997;71:7387-7392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 122] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 123. | Wang HC, Chang WT, Chang WW, Wu HC, Huang W, Lei HY, Lai MD, Fausto N, Su IJ. Hepatitis B virus pre-S2 mutant upregulates cyclin A expression and induces nodular proliferation of hepatocytes. Hepatology. 2005;41:761-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 107] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 124. | Lee H, Kim H, Lee SA, Won YS, Kim HI, Inn KS, Kim BJ. Upregulation of endoplasmic reticulum stress and reactive oxygen species by naturally occurring mutations in hepatitis B virus core antigen. J Gen Virol. 2015;96:1850-1854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 125. | Peiffer KH, Akhras S, Himmelsbach K, Hassemer M, Finkernagel M, Carra G, Nuebling M, Chudy M, Niekamp H, Glebe D, Sarrazin C, Zeuzem S, Hildt E. Intracellular accumulation of subviral HBsAg particles and diminished Nrf2 activation in HBV genotype G expressing cells lead to an increased ROI level. J Hepatol. 2015;62:791-798. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (1)] |
| 126. | Schlüter V, Rabe C, Meyer M, Koshy R, Caselmann WH. Intracellular accumulation of middle hepatitis B surface protein activates gene transcription. Dig Dis. 2001;19:352-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.3] [Reference Citation Analysis (1)] |
| 127. | Kojima T. Immune electron microscopic study of hepatitis B virus associated antigens in hepatocytes. Gastroenterol Jpn. 1982;17:559-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 128. | Lee YI, Hwang JM, Im JH, Lee YI, Kim NS, Kim DG, Yu DY, Moon HB, Park SK. Human hepatitis B virus-X protein alters mitochondrial function and physiology in human liver cells. J Biol Chem. 2004;279:15460-15471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 150] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 129. | McClain SL, Clippinger AJ, Lizzano R, Bouchard MJ. Hepatitis B virus replication is associated with an HBx-dependent mitochondrion-regulated increase in cytosolic calcium levels. J Virol. 2007;81:12061-12065. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 75] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 130. | Majumdar A, Curley SA, Wu X, Brown P, Hwang JP, Shetty K, Yao ZX, He AR, Li S, Katz L, Farci P, Mishra L. Hepatic stem cells and transforming growth factor β in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2012;9:530-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 125] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 131. | Choi SH, Jeong SH, Hwang SB. Large hepatitis delta antigen modulates transforming growth factor-beta signaling cascades: implication of hepatitis delta virus-induced liver fibrosis. Gastroenterology. 2007;132:343-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 59] [Article Influence: 3.1] [Reference Citation Analysis (1)] |
| 132. | Shih HH, Sheen IJ, Su CW, Peng WL, Lin LH, Wu JC. Hepatitis D virus isolates with low replication and epithelial-mesenchymal transition-inducing activity are associated with disease remission. J Virol. 2012;86:9044-9054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 133. | Williams V, Brichler S, Khan E, Chami M, Dény P, Kremsdorf D, Gordien E. Large hepatitis delta antigen activates STAT-3 and NF-κB via oxidative stress. J Viral Hepat. 2012;19:744-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 134. | Chen M, Du D, Zheng W, Liao M, Zhang L, Liang G, Gong M. Small hepatitis delta antigen selectively binds to target mRNA in hepatic cells: a potential mechanism by which hepatitis D virus downregulates glutathione S-transferase P1 and induces liver injury and hepatocarcinogenesis. Biochem Cell Biol. 2019;97:130-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (1)] |
| 135. | Liao FT, Lee YJ, Ko JL, Tsai CC, Tseng CJ, Sheu GT. Hepatitis delta virus epigenetically enhances clusterin expression via histone acetylation in human hepatocellular carcinoma cells. J Gen Virol. 2009;90:1124-1134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 2.3] [Reference Citation Analysis (1)] |
| 136. | Balantinou E, Trougakos IP, Chondrogianni N, Margaritis LH, Gonos ES. Transcriptional and posttranslational regulation of clusterin by the two main cellular proteolytic pathways. Free Radic Biol Med. 2009;46:1267-1274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 137. | Valgimigli M, Valgimigli L, Trerè D, Gaiani S, Pedulli GF, Gramantieri L, Bolondi L. Oxidative stress EPR measurement in human liver by radical-probe technique. Free Radic Res. 2002;36:939-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 79] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 138. | Barbaro G, Di Lorenzo G, Ribersani M, Soldini M, Giancaspro G, Bellomo G, Belloni G, Grisorio B, Barbarini G. Serum ferritin and hepatic glutathione concentrations in chronic hepatitis C patients related to the hepatitis C virus genotype. J Hepatol. 1999;30:774-782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 78] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 139. | Venturini D, Simão AN, Barbosa DS, Lavado EL, Narciso VE, Dichi I, Dichi JB. Increased oxidative stress, decreased total antioxidant capacity, and iron overload in untreated patients with chronic hepatitis C. Dig Dis Sci. 2010;55:1120-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (2)] |
| 140. | Ikegami T, Honda A, Miyazaki T, Kohjima M, Nakamuta M, Matsuzaki Y. Increased serum oxysterol concentrations in patients with chronic hepatitis C virus infection. Biochem Biophys Res Commun. 2014;446:736-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (1)] |
| 141. | Chan SW, Egan PA. Hepatitis C virus envelope proteins regulate CHOP via induction of the unfolded protein response. FASEB J. 2005;19:1510-1512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 123] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 142. | Zheng Y, Gao B, Ye L, Kong L, Jing W, Yang X, Wu Z, Ye L. Hepatitis C virus non-structural protein NS4B can modulate an unfolded protein response. J Microbiol. 2005;43:529-536. [PubMed] |
| 143. | Dionisio N, Garcia-Mediavilla MV, Sanchez-Campos S, Majano PL, Benedicto I, Rosado JA, Salido GM, Gonzalez-Gallego J. Hepatitis C virus NS5A and core proteins induce oxidative stress-mediated calcium signalling alterations in hepatocytes. J Hepatol. 2009;50:872-882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 93] [Article Influence: 5.5] [Reference Citation Analysis (1)] |
| 144. | Li Y, Boehning DF, Qian T, Popov VL, Weinman SA. Hepatitis C virus core protein increases mitochondrial ROS production by stimulation of Ca2+ uniporter activity. FASEB J. 2007;21:2474-2485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 133] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 145. | Korenaga M, Wang T, Li Y, Showalter LA, Chan T, Sun J, Weinman SA. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J Biol Chem. 2005;280:37481-37488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 323] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 146. | Smirnova OA, Ivanova ON, Bartosch B, Valuev-Elliston VT, Mukhtarov F, Kochetkov SN, Ivanov AV. Hepatitis C Virus NS5A Protein Triggers Oxidative Stress by Inducing NADPH Oxidases 1 and 4 and Cytochrome P450 2E1. Oxid Med Cell Longev. 2016;2016:8341937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 45] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 147. | Boudreau HE, Emerson SU, Korzeniowska A, Jendrysik MA, Leto TL. Hepatitis C virus (HCV) proteins induce NADPH oxidase 4 expression in a transforming growth factor beta-dependent manner: a new contributor to HCV-induced oxidative stress. J Virol. 2009;83:12934-12946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 112] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 148. | Lieber CS. Cytochrome P-4502E1: its physiological and pathological role. Physiol Rev. 1997;77:517-544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 649] [Cited by in RCA: 619] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 149. | Ivanov AV, Smirnova OA, Petrushanko IY, Ivanova ON, Karpenko IL, Alekseeva E, Sominskaya I, Makarov AA, Bartosch B, Kochetkov SN, Isaguliants MG. HCV core protein uses multiple mechanisms to induce oxidative stress in human hepatoma Huh7 cells. Viruses. 2015;7:2745-2770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (1)] |
| 150. | Nakai K, Tanaka H, Hanada K, Ogata H, Suzuki F, Kumada H, Miyajima A, Ishida S, Sunouchi M, Habano W, Kamikawa Y, Kubota K, Kita J, Ozawa S, Ohno Y. Decreased expression of cytochromes P450 1A2, 2E1, and 3A4 and drug transporters Na+-taurocholate-cotransporting polypeptide, organic cation transporter 1, and organic anion-transporting peptide-C correlates with the progression of liver fibrosis in chronic hepatitis C patients. Drug Metab Dispos. 2008;36:1786-1793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 93] [Article Influence: 5.2] [Reference Citation Analysis (2)] |
| 151. | de Mochel NS, Seronello S, Wang SH, Ito C, Zheng JX, Liang TJ, Lambeth JD, Choi J. Hepatocyte NAD(P)H oxidases as an endogenous source of reactive oxygen species during hepatitis C virus infection. Hepatology. 2010;52:47-59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 150] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 152. | Syed GH, Amako Y, Siddiqui A. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol Metab. 2010;21:33-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 258] [Cited by in RCA: 292] [Article Influence: 18.3] [Reference Citation Analysis (4)] |
| 153. | Pekow JR, Bhan AK, Zheng H, Chung RT. Hepatic steatosis is associated with increased frequency of hepatocellular carcinoma in patients with hepatitis C-related cirrhosis. Cancer. 2007;109:2490-2496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 143] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 154. | Tu T, Bühler S, Bartenschlager R. Chronic viral hepatitis and its association with liver cancer. Biol Chem. 2017;398:817-837. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 88] [Article Influence: 9.8] [Reference Citation Analysis (1)] |
| 155. | Chang ML. Metabolic alterations and hepatitis C: From bench to bedside. World J Gastroenterol. 2016;22:1461-1476. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 90] [Cited by in RCA: 101] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 156. | Patel A, Harrison SA. Hepatitis C virus infection and nonalcoholic steatohepatitis. Gastroenterol Hepatol (N Y). 2012;8:305-312. [PubMed] |
| 157. | Munakata T, Nakamura M, Liang Y, Li K, Lemon SM. Down-regulation of the retinoblastoma tumor suppressor by the hepatitis C virus NS5B RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 2005;102:18159-18164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 110] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 158. | Bittar C, Shrivastava S, Bhanja Chowdhury J, Rahal P, Ray RB. Hepatitis C virus NS2 protein inhibits DNA damage pathway by sequestering p53 to the cytoplasm. PLoS One. 2013;8:e62581. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 159. | Cho JW, Baek WK, Suh SI, Yang SH, Chang J, Sung YC, Suh MH. Hepatitis C virus core protein promotes cell proliferation through the upregulation of cyclin E expression levels. Liver. 2001;21:137-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 160. | Cheng D, Zhang L, Yang G, Zhao L, Peng F, Tian Y, Xiao X, Chung RT, Gong G. Hepatitis C virus NS5A drives a PTEN-PI3K/Akt feedback loop to support cell survival. Liver Int. 2015;35:1682-1691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 161. | Hayashi J, Aoki H, Kajino K, Moriyama M, Arakawa Y, Hino O. Hepatitis C virus core protein activates the MAPK/ERK cascade synergistically with tumor promoter TPA, but not with epidermal growth factor or transforming growth factor alpha. Hepatology. 2000;32:958-961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 107] [Article Influence: 4.1] [Reference Citation Analysis (1)] |
| 162. | Zhao LJ, Wang L, Ren H, Cao J, Li L, Ke JS, Qi ZT. Hepatitis C virus E2 protein promotes human hepatoma cell proliferation through the MAPK/ERK signaling pathway via cellular receptors. Exp Cell Res. 2005;305:23-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 89] [Article Influence: 4.2] [Reference Citation Analysis (1)] |
| 163. | Bürckstümmer T, Kriegs M, Lupberger J, Pauli EK, Schmittel S, Hildt E. Raf-1 kinase associates with Hepatitis C virus NS5A and regulates viral replication. FEBS Lett. 2006;580:575-580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 2.9] [Reference Citation Analysis (1)] |
| 164. | Feng DY, Sun Y, Cheng RX, Ouyang XM, Zheng H. Effect of hepatitis C virus nonstructural protein NS3 on proliferation and MAPK phosphorylation of normal hepatocyte line. World J Gastroenterol. 2005;11:2157-2161. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 21] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (1)] |
| 165. | Liu J, Ding X, Tang J, Cao Y, Hu P, Zhou F, Shan X, Cai X, Chen Q, Ling N, Zhang B, Bi Y, Chen K, Ren H, Huang A, He TC, Tang N. Enhancement of canonical Wnt/β-catenin signaling activity by HCV core protein promotes cell growth of hepatocellular carcinoma cells. PLoS One. 2011;6:e27496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 102] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 166. | Street A, Macdonald A, McCormick C, Harris M. Hepatitis C virus NS5A-mediated activation of phosphoinositide 3-kinase results in stabilization of cellular beta-catenin and stimulation of beta-catenin-responsive transcription. J Virol. 2005;79:5006-5016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 115] [Article Influence: 5.5] [Reference Citation Analysis (2)] |
| 167. | Park CY, Choi SH, Kang SM, Kang JI, Ahn BY, Kim H, Jung G, Choi KY, Hwang SB. Nonstructural 5A protein activates beta-catenin signaling cascades: implication of hepatitis C virus-induced liver pathogenesis. J Hepatol. 2009;51:853-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 168. | Milward A, Mankouri J, Harris M. Hepatitis C virus NS5A protein interacts with beta-catenin and stimulates its transcriptional activity in a phosphoinositide-3 kinase-dependent fashion. J Gen Virol. 2010;91:373-381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 40] [Article Influence: 2.4] [Reference Citation Analysis (1)] |
| 169. | Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989-5005. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 608] [Cited by in RCA: 695] [Article Influence: 43.4] [Reference Citation Analysis (1)] |
| 170. | Deng L, Nagano-Fujii M, Tanaka M, Nomura-Takigawa Y, Ikeda M, Kato N, Sada K, Hotta H. NS3 protein of Hepatitis C virus associates with the tumour suppressor p53 and inhibits its function in an NS3 sequence-dependent manner. J Gen Virol. 2006;87:1703-1713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 75] [Article Influence: 3.8] [Reference Citation Analysis (1)] |
| 171. | Lai CK, Jeng KS, Machida K, Cheng YS, Lai MM. Hepatitis C virus NS3/4A protein interacts with ATM, impairs DNA repair and enhances sensitivity to ionizing radiation. Virology. 2008;370:295-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 73] [Article Influence: 3.8] [Reference Citation Analysis (2)] |
| 172. | Majumder M, Ghosh AK, Steele R, Ray R, Ray RB. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J Virol. 2001;75:1401-1407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 209] [Article Influence: 8.4] [Reference Citation Analysis (1)] |
| 173. | Lan KH, Sheu ML, Hwang SJ, Yen SH, Chen SY, Wu JC, Wang YJ, Kato N, Omata M, Chang FY, Lee SD. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene. 2002;21:4801-4811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 177] [Article Influence: 7.4] [Reference Citation Analysis (1)] |
| 174. | Kao CF, Chen SY, Chen JY, Wu Lee YH. Modulation of p53 transcription regulatory activity and post-translational modification by hepatitis C virus core protein. Oncogene. 2004;23:2472-2483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 99] [Article Influence: 4.5] [Reference Citation Analysis (1)] |
| 175. | Saito K, Meyer K, Warner R, Basu A, Ray RB, Ray R. Hepatitis C virus core protein inhibits tumor necrosis factor alpha-mediated apoptosis by a protective effect involving cellular FLICE inhibitory protein. J Virol. 2006;80:4372-4379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 78] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 176. | Ghosh AK, Majumder M, Steele R, Meyer K, Ray R, Ray RB. Hepatitis C virus NS5A protein protects against TNF-alpha mediated apoptotic cell death. Virus Res. 2000;67:173-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 2.6] [Reference Citation Analysis (2)] |
| 177. | Simonin Y, Disson O, Lerat H, Antoine E, Binamé F, Rosenberg AR, Desagher S, Lassus P, Bioulac-Sage P, Hibner U. Calpain activation by hepatitis C virus proteins inhibits the extrinsic apoptotic signaling pathway. Hepatology. 2009;50:1370-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 178. | de Caestecker MP, Piek E, Roberts AB. Role of transforming growth factor-beta signaling in cancer. J Natl Cancer Inst. 2000;92:1388-1402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 406] [Article Influence: 15.6] [Reference Citation Analysis (1)] |
| 179. | Choi SH, Hwang SB. Modulation of the transforming growth factor-beta signal transduction pathway by hepatitis C virus nonstructural 5A protein. J Biol Chem. 2006;281:7468-7478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 71] [Article Influence: 3.6] [Reference Citation Analysis (3)] |
| 180. | Pavio N, Battaglia S, Boucreux D, Arnulf B, Sobesky R, Hermine O, Brechot C. Hepatitis C virus core variants isolated from liver tumor but not from adjacent non-tumor tissue interact with Smad3 and inhibit the TGF-beta pathway. Oncogene. 2005;24:6119-6132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 67] [Article Influence: 3.2] [Reference Citation Analysis (1)] |
| 181. | Niu ZS, Niu XJ, Wang WH. Genetic alterations in hepatocellular carcinoma: An update. World J Gastroenterol. 2016;22:9069-9095. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 123] [Cited by in RCA: 135] [Article Influence: 13.5] [Reference Citation Analysis (3)] |
| 182. | Nault JC, Zucman-Rossi J. TERT promoter mutations in primary liver tumors. Clin Res Hepatol Gastroenterol. 2016;40:9-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 183. | Low KC, Tergaonkar V. Telomerase: central regulator of all of the hallmarks of cancer. Trends Biochem Sci. 2013;38:426-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 198] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 184. | Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, Calatayud AL, Pinyol R, Pelletier L, Balabaud C, Laurent A, Blanc JF, Mazzaferro V, Calvo F, Villanueva A, Nault JC, Bioulac-Sage P, Stratton MR, Llovet JM, Zucman-Rossi J. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505-511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1006] [Cited by in RCA: 1409] [Article Influence: 128.1] [Reference Citation Analysis (0)] |
| 185. | Pollutri D, Gramantieri L, Bolondi L, Fornari F. TP53/MicroRNA Interplay in Hepatocellular Carcinoma. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 186. | Pavletich NP, Chambers KA, Pabo CO. The DNA-binding domain of p53 contains the four conserved regions and the major mutation hot spots. Genes Dev. 1993;7:2556-2564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 387] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 187. | Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1212] [Cited by in RCA: 1251] [Article Influence: 65.8] [Reference Citation Analysis (1)] |
| 188. | Gouas D, Shi H, Hainaut P. The aflatoxin-induced TP53 mutation at codon 249 (R249S): biomarker of exposure, early detection and target for therapy. Cancer Lett. 2009;286:29-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 99] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 189. | Song X, Wang S, Li L. New insights into the regulation of Axin function in canonical Wnt signaling pathway. Protein Cell. 2014;5:186-193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 190. | Lee HC, Kim M, Wands JR. Wnt/Frizzled signaling in hepatocellular carcinoma. Front Biosci. 2006;11:1901-1915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 122] [Article Influence: 6.1] [Reference Citation Analysis (1)] |
| 191. | Voronkov A, Krauss S. Wnt/beta-catenin signaling and small molecule inhibitors. Curr Pharm Des. 2013;19:634-664. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 176] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 192. | Taniguchi K, Roberts LR, Aderca IN, Dong X, Qian C, Murphy LM, Nagorney DM, Burgart LJ, Roche PC, Smith DI, Ross JA, Liu W. Mutational spectrum of beta-catenin, AXIN1, and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene. 2002;21:4863-4871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 358] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 193. | He F, Li J, Xu J, Zhang S, Xu Y, Zhao W, Yin Z, Wang X. Decreased expression of ARID1A associates with poor prognosis and promotes metastases of hepatocellular carcinoma. J Exp Clin Cancer Res. 2015;34:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 82] [Article Influence: 7.5] [Reference Citation Analysis (1)] |
| 194. | Zhao J, Chen J, Lin H, Jin R, Liu J, Liu X, Meng N, Cai X. The Clinicopathologic Significance of BAF250a (ARID1A) Expression in Hepatocellular Carcinoma. Pathol Oncol Res. 2016;22:453-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (1)] |
| 195. | Jelic S, Sotiropoulos GC; ESMO Guidelines Working Group. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21 Suppl 5:v59-v64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 168] [Article Influence: 10.5] [Reference Citation Analysis (1)] |
| 196. | Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, Roberts LR, Heimbach JK. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68:723-750. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2121] [Cited by in RCA: 3435] [Article Influence: 429.4] [Reference Citation Analysis (3)] |
| 197. | Burak KW, Kneteman NM. An evidence-based multidisciplinary approach to the management of hepatocellular carcinoma (HCC): the Alberta HCC algorithm. Can J Gastroenterol. 2010;24:643-650. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 198. | Forner A, Llovet JM, Bruix J. Chemoembolization for intermediate HCC: is there proof of survival benefit? J Hepatol. 2012;56:984-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 93] [Article Influence: 6.6] [Reference Citation Analysis (2)] |
| 199. | Yao FY, Kerlan RK Jr, Hirose R, Davern TJ 3rd, Bass NM, Feng S, Peters M, Terrault N, Freise CE, Ascher NL, Roberts JP. Excellent outcome following down-staging of hepatocellular carcinoma prior to liver transplantation: an intention-to-treat analysis. Hepatology. 2008;48:819-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 385] [Cited by in RCA: 411] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 200. | Golfieri R, Cappelli A, Cucchetti A, Piscaglia F, Carpenzano M, Peri E, Ravaioli M, D'Errico-Grigioni A, Pinna AD, Bolondi L. Efficacy of selective transarterial chemoembolization in inducing tumor necrosis in small (<5 cm) hepatocellular carcinomas. Hepatology. 2011;53:1580-1589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 221] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 201. | Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S, Kim JS, Luo R, Feng J, Ye S, Yang TS, Xu J, Sun Y, Liang H, Liu J, Wang J, Tak WY, Pan H, Burock K, Zou J, Voliotis D, Guan Z. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3854] [Cited by in RCA: 4739] [Article Influence: 263.3] [Reference Citation Analysis (0)] |
| 202. | Lim CJ, Lee YH, Pan L, Lai L, Chua C, Wasser M, Lim TKH, Yeong J, Toh HC, Lee SY, Chan CY, Goh BK, Chung A, Heikenwälder M, Ng IO, Chow P, Albani S, Chew V. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus-related hepatocellular carcinoma. Gut. 2019;68:916-927. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 263] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 203. | Tan AT, Yang N, Lee Krishnamoorthy T, Oei V, Chua A, Zhao X, Tan HS, Chia A, Le Bert N, Low D, Tan HK, Kumar R, Irani FG, Ho ZZ, Zhang Q, Guccione E, Wai LE, Koh S, Hwang W, Chow WC, Bertoletti A. Use of Expression Profiles of HBV-DNA Integrated Into Genomes of Hepatocellular Carcinoma Cells to Select T Cells for Immunotherapy. Gastroenterology. 2019;156:1862-1876.e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 114] [Article Influence: 16.3] [Reference Citation Analysis (2)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/Licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: Canada
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kanda T S-Editor: Zhang L L-Editor: A P-Editor: Zhang YL