Published online Oct 14, 2020. doi: 10.3748/wjg.v26.i38.5784
Peer-review started: May 27, 2020
First decision: July 29, 2020
Revised: August 12, 2020
Accepted: September 12, 2020
Article in press: September 12, 2020
Published online: October 14, 2020
Processing time: 139 Days and 15.1 Hours
The past decade has seen significant advances in endoscopic imaging and optical enhancements to aid early diagnosis. There is still a treatment gap due to the underdiagnosis of lesions of the oesophagus. Computer aided diagnosis may play an important role in the coming years in providing an adjunct to endoscopists in the early detection and diagnosis of early oesophageal cancers, therefore curative endoscopic therapy can be offered. Research in this area of artificial intelligence is expanding and the future looks promising. In this review article we will review current advances in artificial intelligence in the oesophagus and future directions for development.
Core Tip: Computer aided diagnosis of oesophageal pathology may potentially be an adjunct for the endoscopist which will improve the detection of early neoplasia in Barrett’s oesophagus and early squamous neoplasia such that curative endoscopic therapy can be offered. There are significant miss rates of oesophageal cancers despite advances in endoscopic imaging modalities and an artificial intelligence (AI) tool will off-set human factors associated with some miss rates. To fulfil the potential of this exciting area of AI certain criteria need to be met which we will expand upon. Once implemented this will have a significant impact on this field of endoscopy.
- Citation: Hussein M, González-Bueno Puyal J, Mountney P, Lovat LB, Haidry R. Role of artificial intelligence in the diagnosis of oesophageal neoplasia: 2020 an endoscopic odyssey. World J Gastroenterol 2020; 26(38): 5784-5796
- URL: https://www.wjgnet.com/1007-9327/full/v26/i38/5784.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i38.5784
The past decade has seen significant advances in endoscopic imaging and optical enhancements to aid early diagnosis. Oesophageal cancer (adenocarcinoma and squamous cell carcinoma) is associated with significant mortality[1]. As of 2018 oesophageal cancer was ranked seventh in the world in terms of cancer incidence and mortality, with 572000 new cases[2]. Oesophageal squamous cell carcinoma accounts for more than 90% of oesophageal cancers in china with an overall 5-year survival rate less than 20%[3].
Despite the technological advances there is still a treatment gap due to the underdiagnosis of lesions of the oesophagus[4]. A metanalysis of 24 studies showed that missed oesophageal cancers are found within a year of index endoscopy in a quarter of patients undergoing surveillance for Barrett’s oesophagus (BE)[5]. A large multicentre retrospective study of 123395 upper gastrointestinal (GI) endoscopies showed an overall missed oesophageal cancer rate of 6.4%. The interval between a negative endoscopy and the diagnosis was less than 2 years in most cases[6]. Multivariate analysis showed that one of the factors associated with the miss rate is a less experienced endoscopist.
Efforts are necessary to improve the detection of early neoplasia secondary to BE and early squamous cell neoplasia (ESCN) such that curative minimally invasive endoscopic therapy can be offered to patients. Computer aided diagnosis may play an important role in the coming years in providing an adjunct to endoscopists in the early detection and diagnosis of early oesophageal cancers.
In this review article we will review current advances in artificial intelligence in the oesophagus and future directions for development.
Machine learning is the use of mathematical models to capture structure in data[7]. The algorithms improve automatically through experience and do not need to be explicitly programmed[8]. The final trained models can be used to make prediction of oesophageal diagnosis. Machine learning is classified into supervised and unsupervised learning. During supervised learning, the model is trained with data containing pairs of inputs and outputs. It learns how to map the inputs and outputs and applies this to unseen data. In unsupervised learning the algorithm is given data inputs which are not directly linked to the outputs and therefore has to formulate its own structure and set of patterns from the inputs[9].
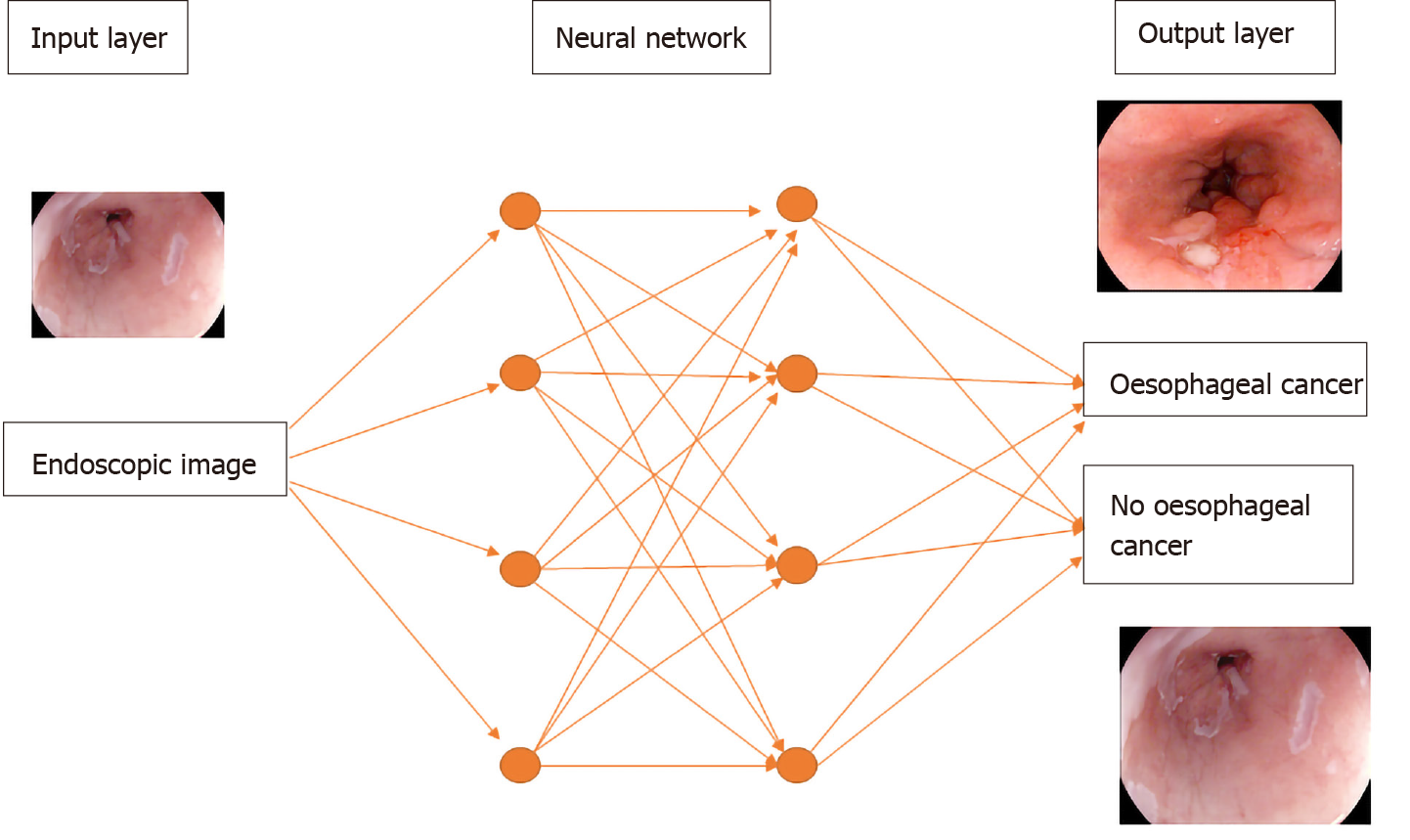
Deep learning is a subtype of machine learning in which the model, a neural network, is composed of several layers of neurons, similar to the human brain. This enables automatic learning of features, which is particularly useful in endoscopy where images and videos lack structure and are not easily processed into specific features[9]. A convolutional neural network (CNN) is a subtype of deep learning which can take an input endoscopic image and learn specific features (e.g., colour, size, pit pattern), process the complex information through many different layers and produce an output prediction (e.g., oesophageal dysplasia or no dysplasia) (Figure 1).
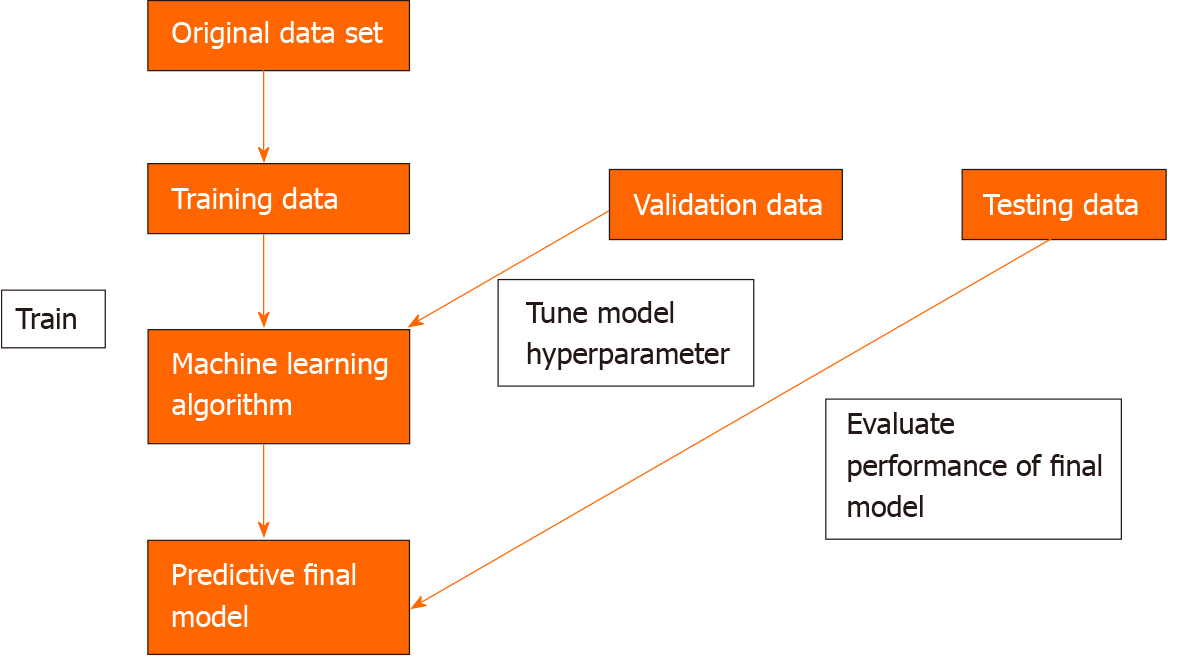
To develop a machine learning model, data needs to be split into 3 independent groups-training set, validation set and testing set. The training set is used to build a model using the oesophageal labels (e.g., dysplasia or no dysplasia). The validation set provides an unbiased evaluation of the model’s skill whilst tuning the hyper-parameters of the model, for example, the number of layers in the neural network. It is used to ensure that the model is not overfitting to the training data. Overfitting means that the model will perform well on the training data but not on the unseen testing data. The test set is used to evaluate the performance of the predictive final model[7] (Figure 2).
Endoscopic imaging has advanced into a new era with the development of high definition digital technology. A charge coupled device chip in standard white light endoscopy produces an image signal of 10000 to 400000 pixels displayed in a standard definition format. The chips in a high definition white light endoscope produce image signals of 850000 to 1.3 million pixels displayed in high definition[10]. This has improved our ability to pick up the most subtle oesophageal mucosal abnormalities by assessing mucosal pit patterns and vascularity to allow a timely diagnosis of dysplasia or early cancer.
There have been further advances in optical technology in the endoscope with chromoendoscopy such as narrow-band imaging (NBI), i-scan (Pentax, Hoya) and blue laser imaging (Fujinon), which have further improved early neoplasia detection and diagnosis in the oesophagus. Table 1 summarises some of the studies investigating the accuracy of these imaging modalities in detecting BE dysplasia by formulating classification systems based on mucosal pit pattern, colour and vascular architecture.
| I-scan optical enhancement | NBI | BLI | |
| Ref. | Everson et al[11] | Sharma et al[12] | Subramaniam et al[13] |
| Features assessed | Mucosal pit pattern, vessels | Mucosal pit pattern, vessels | Colour, mucosal pit patterns, vessels |
| Accuracy | Experts = 84%, non-experts = 76% | 85% | Experts = 95.2%, non-experts = 88.3% |
| Sensitivity | Experts = 77%, non-experts = 81% | 80% | Experts = 96%, non-experts = 95.7% |
| Specificity | Experts = 92%, non-experts = 70% | 88% | Experts = 94.4%, non-experts = 80.8% |
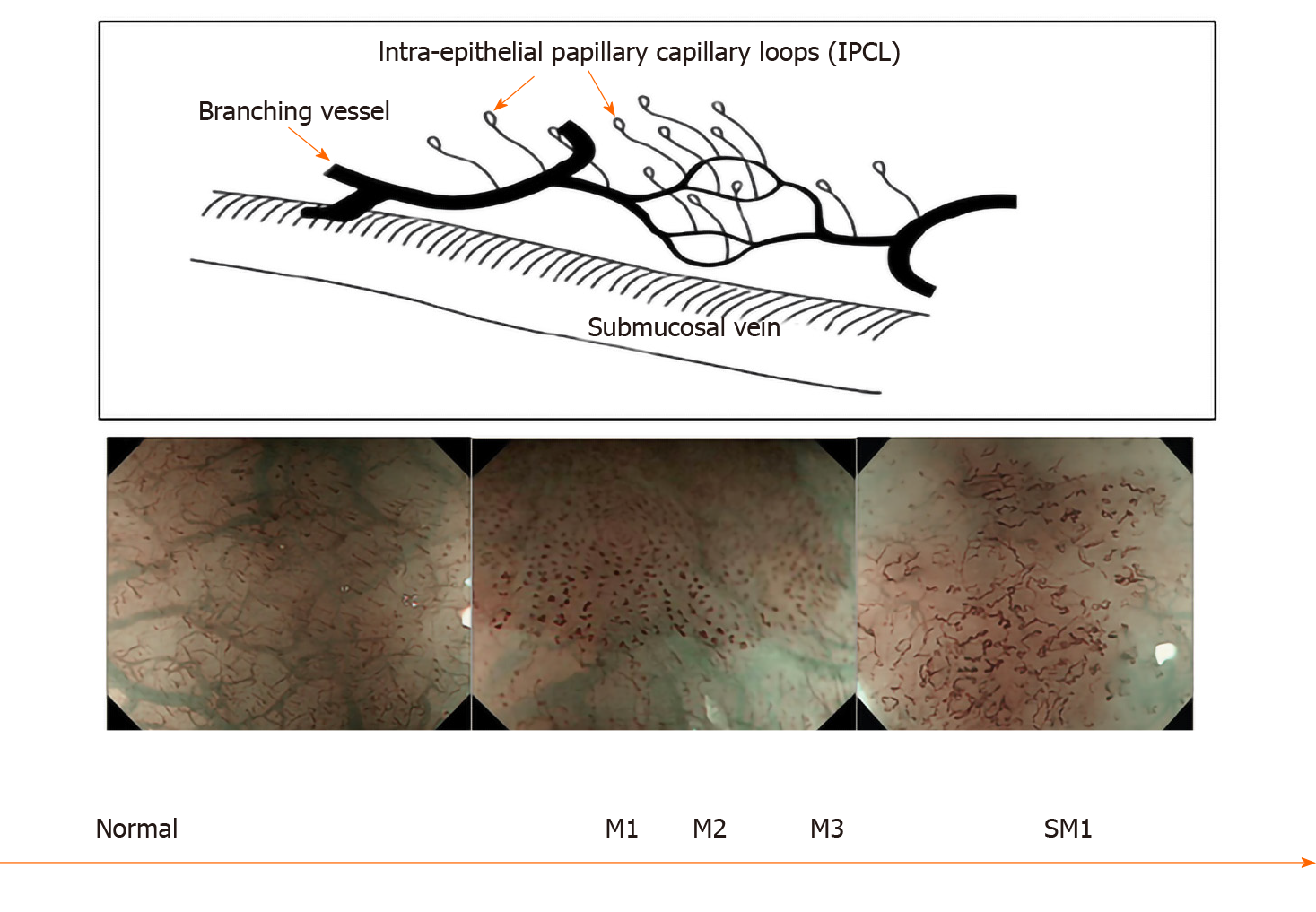
In squamous epithelium the microvascular vascular patterns of intrapapillary capillary loops (IPCL) is used to aid in the diagnosis of early squamous cell cancer (Figure 3)[14]. The classification systems that are currently used are based on magnification endoscopy assessment of IPCL patterns[15].
The disordered and distorted mucosal and vascular patterns used to define the above classifications can be used to train the CNN to detect early cancer in the oesophagus.
BE is the only identifiable premalignant condition associated with invasive oesophageal adenocarcinoma. There is a linear progression from non-dysplastic BE, to low grade and high-grade dysplasia. Early neoplasia which is confined to the mucosa have significant eradication rates of > 80%[16].
The standard of care for endoscopic surveillance for patients with BE are random biopsies taken as part of the Seattle protocol where four-quadrant biopsies are taken every 2 cm of BE[17]. This method is not perfect and is associated with sampling error. The area of a 2 cm segment of BE is approximately 14 cm2, a single biopsy sample is approximately 0.125 cm2. Therefore, Seattle protocol biopsies will only cover 0.5 cm2 of the oesophageal mucosa which is 3.5% of the BE segment[18]. Dysplasia can often be focal and therefore easily missed. Studies have also shown that compliance with this protocol is poor and is worse on longer segments of BE[19].
The American Society for Gastrointestinal Endoscopy preservation and incorporation of valuable endoscopic innovations (PIVI) initiative was developed to direct endoscopic technology development. Any imaging technology with targeted biopsies in BE would need to achieve a threshold per patient sensitivity of at least 90% for the detection of high-grade dysplasia and intramucosal cancer. It would require a specificity of at least 80% in BE in order to eliminate the requirement for random mucosal biopsies during BE endoscopic surveillance. This would improve the cost and effectiveness of a surveillance programme. This is the minimum target an AI technology would need to meet in order to be able to be ready for prime time and a possible adjunct during a BE surveillance endoscopy[20].
An early study tested a computer algorithm developed based on 100 images from 44 patients with BE. It was trained using colour and texture filters. The algorithm diagnosed neoplastic lesions on a per image level with a sensitivity and specificity of 0.83. At the patient level a sensitivity and specificity of 0.86 and 0.87 was achieved respectively. This was the first study where a detection algorithm was developed for detecting BE lesions and compared with expert annotations[21].
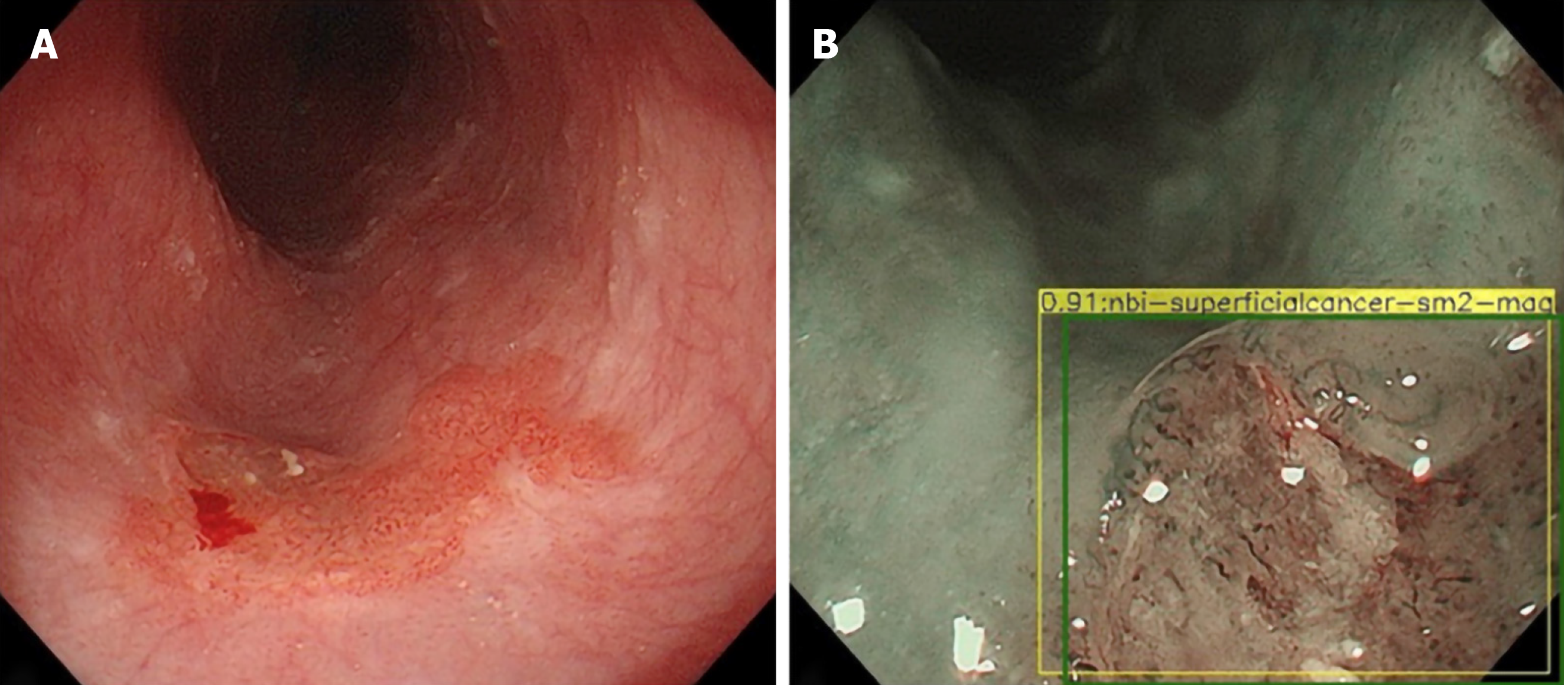
A recent study developed a hybrid ResNet-UNet model computer aided diagnosis system which classified images as containing neoplastic or non-dysplastic BE with a sensitivity and specificity of 90% and 88% respectively. It achieved higher accuracy than non-expert endoscopists[22].
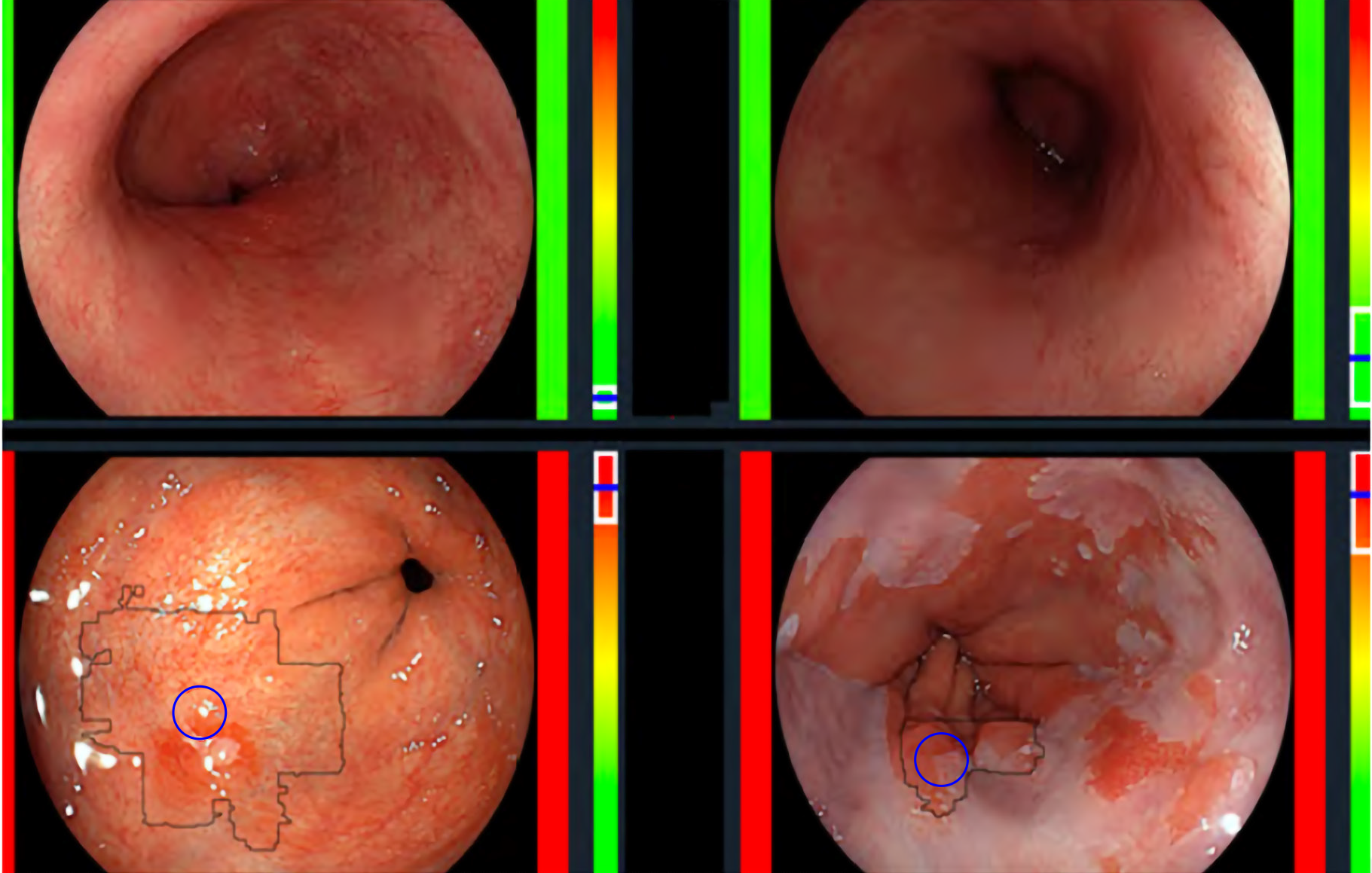
De Groof et al[23] performed one of the first studies to assess the accuracy of a computer-aided detection (CAD) system during live endoscopic procedures of 10 patients with BE Dysplasia and 10 patients without BE dysplasia. Three images were evaluated every 2 cm of BE by the CAD system. Sensitivity and specificity of the CAD system in per level analysis was 91% and 89% respectively (Figure 4).
Hussein et al[24] developed a CNN trained using a balanced data set of 73266 frames from BE videos of 39 patients. On an independent validation set of 189436 frames from 19 patients the CNN could detect dysplasia with a sensitivity of 88.3% and specificity of 80%. The annotations were created from and tested on frames from whole videos minimising selection bias.
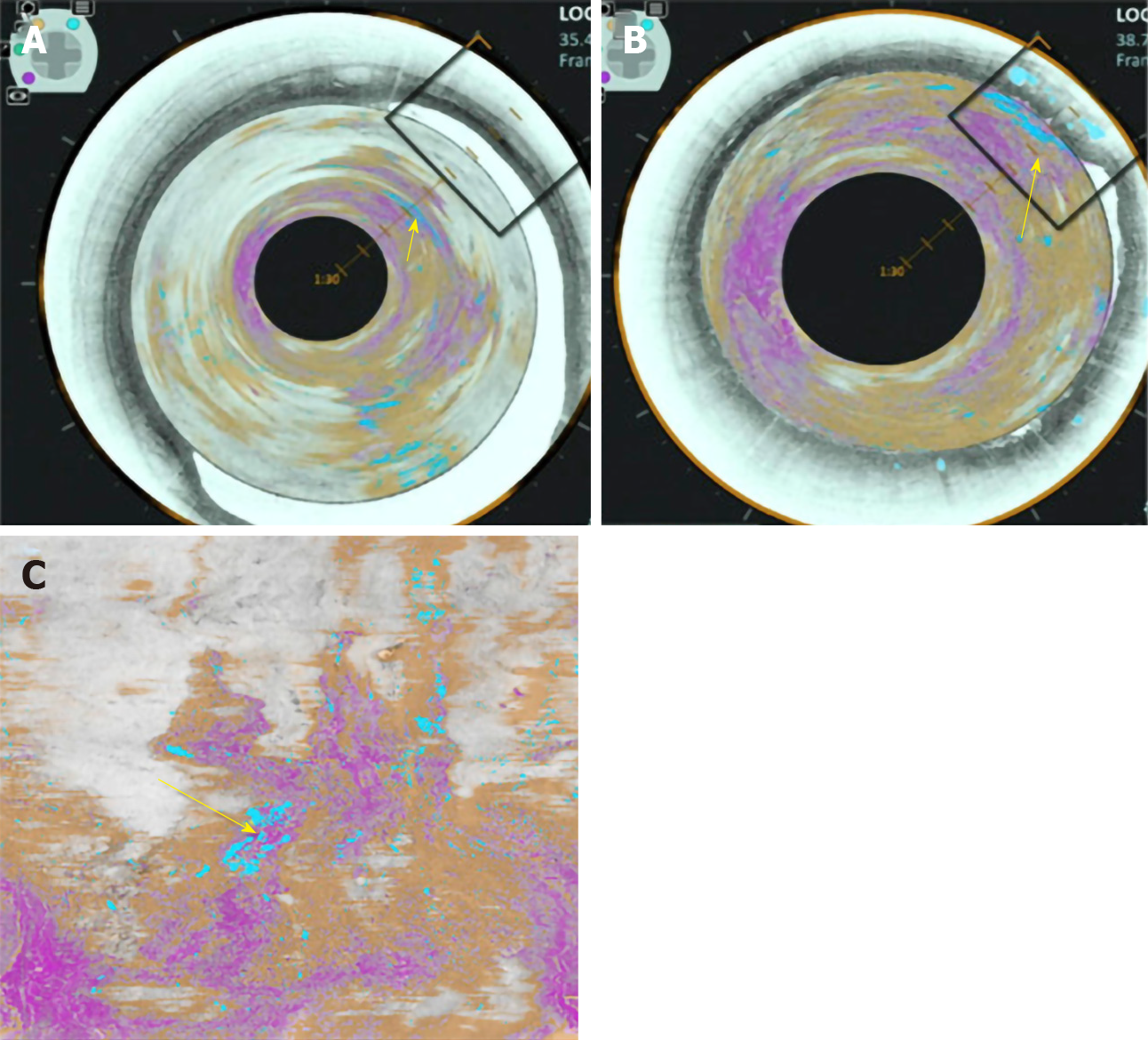
Volumetric laser endomicroscopy (VLE) is a wide field imaging technology used to aid endoscopists in the detection of dysplasia in BE. An infrared light produces a circumferential scan of 6cm segments of BE up to a depth of 3 mm allowing the oesophageal layer and submucosal layer with its associated vascular networks to be visualized[25]. The issue is there is large volumes of complex data which the endoscopist needs to interpret. An Artificial intelligence system called intelligent real-time image segmentation has been used to interpret the data produced from VLE. This software identifies 3 VLE features associated with histological evidence of dysplasia and displays the output with colour schemes. A hyper reflective surface (pink colour) suggests that there is increased surface maturation, cellular crowding and increased nuclear-to-cytoplasmic ratio. Hyporeflective structures (blue colour) suggests abnormal morphology of BE epithelial glands. A lack of layered architecture (orange colour) differentiates squamous epithelium from BE (Figure 5)[26]. A recent study analysed ex-vivo images from 29 BE patients with and without early cancer retrospectively. A CAD system which analysed multiple neighbouring VLE frames showed improved neoplasia detection in BE relative to single frame analysis with an AUC of 0.91[27].
Table 2 provides a summary of all the studies investigating the development of deep learning algorithms for the diagnosis of early neoplasia in BE.
| Ref. | Year | Endoscopic processor | Study design | Study aim | Algorithm used | No. of patients | No. of BE images | Sensitivity | Specificity |
| Van der Sommen et al[21] | 2016 | WLE Fujinon | Retrospective | Assess feasibility of computer system to detect early neoplasia in BE | Machine learning, specific textures and colour filters | 44 | 100 (60 dysplasia, 40 NDBE) | 83% (per image), 86% (per patient) | 83% (per image), 87% (per patient) |
| Sweger et al[28] | 2017 | VLE | Retrospective | Assess feasibility of computer algorithm to identify BE dysplasia on ex vivo VLE images | Several machine learning methods; discriminant analysis, support vector machine, AdaBoost, random forest, K-nearest neighbors | 19 | 60 (30 dysplasia, 30 NDBE) | 90% | 93% |
| Ebigbo et al[29] | 2018 | WLE, NBI, Olympus | Retrospective | Detection of early oesophageal cancer | Deep CNN with a residual net architecture | 50 with early neoplasia | 248 | 97% (WLE), 94% (NBI) | 88% (WLE), 80% (NBI) |
| de Groof et al[30] | 2019 | WLE, Fujinon | Prospective | Develop CAD to detect early neoplasia in BE | Supervised Machine learning. Trained on colour and texture features | 60 | 60 (40 dysplasia, 20 NDBE) | 95% | 85% |
| de Groof et al[22] | 2020 | WLE Fujinon, WLE Olympus | Retrospective, Prospective | Develop and validate deep learning CAD to improve detection of early neoplasia in BE | CNN pretrained on GastroNet. Hybrid ResNet/U-Net model | 669 | 1704 (899 dysplasia, 805 NDBE) | 90% | 88% |
| Hashimoto et al[31] | 2020 | WLE, Olympus | Retrospective | Assess if CNN can aid in detecting early neoplasia in BE | CNN pretrained on image net and based on Xception architecture and YOLO v2 | 100 | 1832 (916 dysplasia, 916 NDBE) | 96.4% | 94.2% |
| de Groof et al[23] | 2020 | WLE, Fujinon | Prospective | Evaluate CAD assessment of early neoplasia during live endoscopy | CNN pretrained on GastroNet; hybrid ResNet/U-Net Model | 20 | - | 91% | 89% |
| Struyvenberg MR et al[27] | 2020 | VLE | Prospective | Evaluate feasibility of automatic data extraction followed by CAD using mutiframe approach to detect to dysplasia in BE | CAD multiframe analysis with principal component analysis | 29 | - | - | - |
With advances in endoscopic therapy in recent years ESCN confined to the mucosal layer can be curatively resected endoscopically with a < 2% incidence of local lymph node metastasis. IPCL are the microvascular features which can be endoscopically used to help classify and identify ESCN and if there is a degree of invasion in the muscularis mucosa and submucosal tissue[16].
Lugols chromoendoscopy is a screening method for identifying ESCN during an upper GI endoscopy. However, despite a sensitivity of > 90%, it is associated with a low specificity of approximately 70%[32]. There is also a risk of allergic reaction with iodine staining. Advanced endoscopic imaging with NBI has a high accuracy for detecting ESCN however a randomised control trial showed its specificity was approximately 50%[33]. Computer assisted detection systems have been developed to try and overcome many of these issues which aid endoscopists in detecting early ESCN lesions.
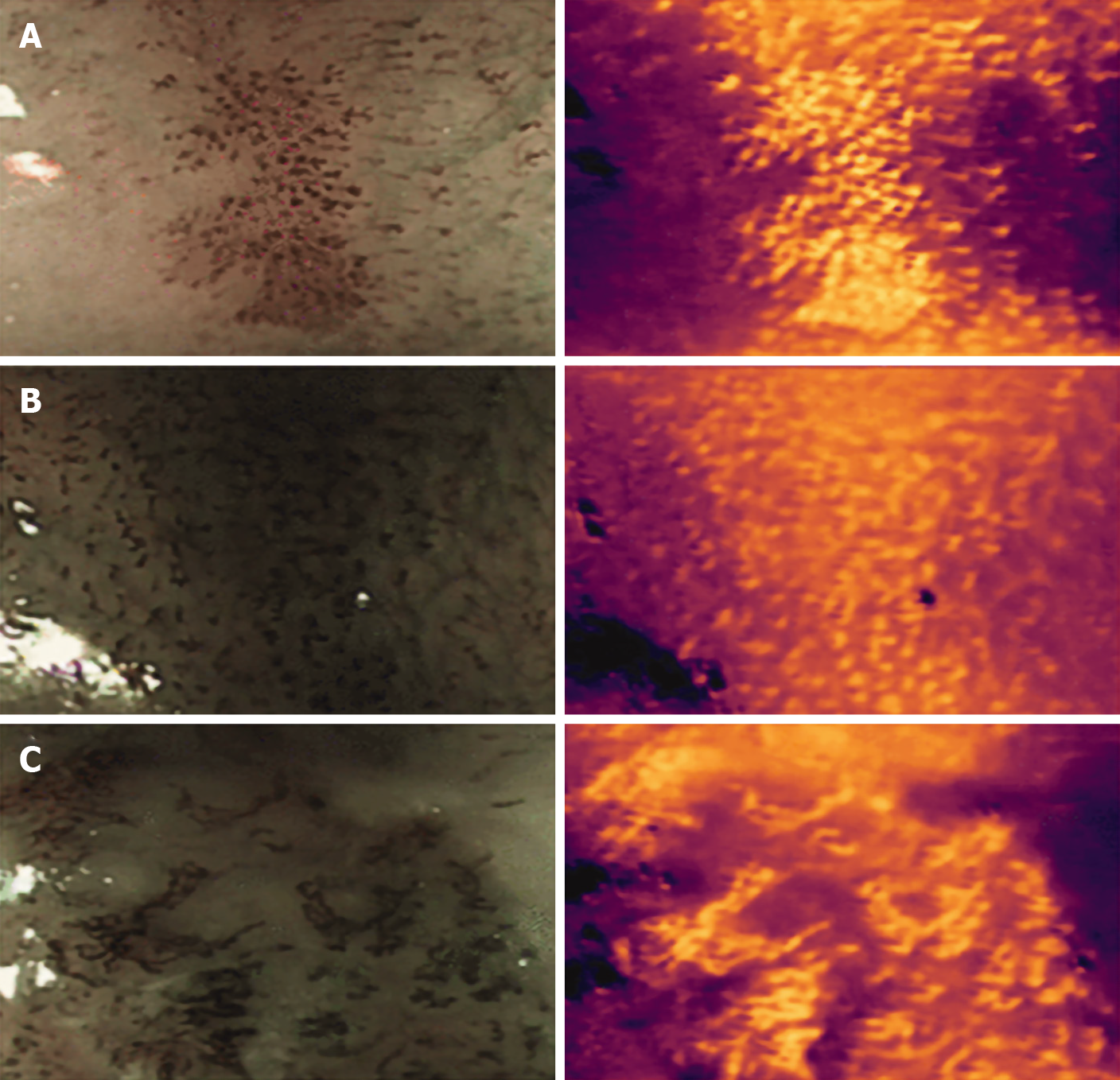
Everson et al[16] developed a CNN trained with 7046 sequential high definition magnification endoscopy with NBI. These were classified by experts using the IPCL patterns and based on the Japanese Endoscopic Society classification. The CNN was able to accurately classify abnormal IPCL patterns with a sensitivity and specificity of 89% and 98% respectively. The diagnostic prediction times were between 26 and 37 ms (Figure 6).
Nakagawa et al[34] developed a deep learning-based AI algorithm using over 14000 magnified and non-magnified endoscopic images from 804 patients. This was able to predict the depth of invasion of ESCN with a sensitivity of 90.1% and specificity of 95.8% (Figure 7).
Guo et al[3] trained a CAD system using 6473 NBI images for real time automated diagnosis of ESCN. The deep learning model was able to detect early ESCN on still NBI images with a sensitivity of 98% and specificity of 95%. On analysis of videos the per frame sensitivity was 60.8% on non-magnified images and 96.1% on magnified images. The per lesion sensitivity was 100%. This model had high sensitivity and specificity in both still images and real time video setting the scene for the development of better models for real time detection of early ESCN.
Endocytoscopy uses a high-power fixed-focus objective lens attached to the endoscope to give ultra-high magnification images. The area of interest is stained to allow identification of cellular structures like in standard histopathology techniques. This allows the endoscopist to characterise ESCN and make a real time histological diagnosis[35].
Kumagai et al[36] developed a CNN trained using more than 4000 endocytoscopic images of the oesophagus (malignant and non-malignant). The AI was able to diagnose esophageal squamous cell carcinoma with a sensitivity of 92.6%. This provides a potential AI tool which can aid the endoscopist by making an in vivo histological diagnosis. This would allow endoscopists to make a clinical decision in the same endoscopic session regarding resection of the early oesophageal cancer which would potentially save on costs by replacing the need for protocol biopsies
Table 3 provides a summary of all the studies investigating the development of deep learning algorithms for the diagnosis of ESCN.
| Ref. | Year | Endoscopic processor | Study design | Study aim | Algorithm used | No. of patients | No. of images | Sensitivity | Specificity |
| Shin et al[37] | 2015 | High resolution micro-endoscopy | Retrospective | Differentiate neoplastic and non-neoplastic squamous oesophageal mucosa | Quantitative image analysis. Two-class linear discriminant analysis to develop classifier | 177 | 375 | 87% | 97% |
| Quang et al[38] | 2016 | High resolution micro-endoscopy | Retrospective | Differentiate neoplastic and non-neoplastic squamous oesophageal mucosa | Two-class linear discriminant analysis to develop classifier | 3 | - | 95% | 91% |
| Horie et al[39] | 2018 | WLE, NBI, Olympus | Retrospective | Ability of AI to detect oesophageal cancer | Deep CNN (Multibox detector architecture) | 481 | - | 97% | - |
| Everson et al[16] | 2019 | Magnified NBI, Olympus | Retrospective | Develop AI system to classify IPCL patterns as normal/abnormal in endoscopically resectable lesions real time | CNN, explicit class activation maps generated to depict area of interest for CNN | 17 | 7046 | 89% | 98% |
| Nakagawa et al[34] | 2019 | Magnified and non-magnified, NBI, BLI, Olympus, Fujifilm | Retrospective | Predict depth of invasion of ESCN | Deep CNN (multibox detector architecture) | 959 | 15,252 | 90.1% | 95.8% |
| Kumagai et al[36] | 2019 | ECS | Retrospective | Deep learning AI to analyse ECS images as possible replacement of biopsy-based histology | CNN constructed based on GoogLeNet | - | 6235 | 92.6% | 89.3% |
| Zhao et al[40] | 2019 | ME NBI, Olympus | Retrospective | Classification of IPCLs to improve ESCN detection | A double-labeling fully convolutional network | 219 | - | 87% | 84.1% |
| Guo et al[3] | 2020 | ME and non-ME NBI, olympus | Retrospective | Develop a CAD for real-time diagnosis of ESCN | Model based on SegNet architecture | 2672 | 13144 images (4250 malignant, 8894 non-cancerous), 168865 video frames | Images = 98.04%, non-magnified video = 60.8%, magnified video = 96.1% | Images = 95.03%, non-magnified /magnified video = 99.9% |
| Tokai et al[41] | 2020 | WLE, NBI, Olympus | Retrospective | Ability of AI to measure squamous cell cancer depth | Deep CNN | - | 2044 | 84.1% | 73.3% |
| Ohmori et al[42] | 2020 | Magnified and non-magnified, WLE, NBI, BLI, Olympus, Fujifilm | Retrospective | Detect Oesophageal squamous cell cancer | CNN | - | 11806 non- magnified images, 11483 magnified images | Non-ME WLE = 90%, non-ME NBI/BLI = 100%, ME = 98% | Non-ME WLE = 76%, non-ME NBI/BLI = 63%, ME = 56% |
In digital pathology tissue slides are scanned as high-resolution images as each slide contains a large volume of cells. The cellular structure needs be visible to the histopathologist in order to identify areas of abnormality[43]. Histopathological analysis often requires a lot of time, high costs and often manual annotation of areas of interest by the histopathologists. There is also a possible miss rate of areas of early oesophageal dysplasia as the area can be focal. There is also suboptimal interobserver agreement among expert GI histopathologists in certain histological diagnosis such as low-grade dysplasia in BE[44].
A novel AI system to detect and delineate areas of early oesophageal cancer on histology slides could be a key adjunct to histopathologists and help improve detection and delineation of early oesophageal cancer.
Tomita et al[43] developed a convolutional attention-based mechanism to classify microscopic images into normal oesophageal tissue, BE with no dysplasia, BE with dysplasia and oesophageal adenocarcinoma using 123 histological images. Classification accuracy of the model was 0.85 in the BE-no dysplasia group, 0.89 in the BE with dysplasia group, and 0.88 in the oesophageal adenocarcinoma group.
The inspection time of the oesophagus and clear mucosal views have an impact on the quality of an oesophagoscopy and the yield of early oesophageal neoplasia detection. Assessment should take place with the oesophagus partially insufflated between peristaltic waves. An overly insufflated oesophagus can flatten a lesion which can in turn be missed[45]. The British Society of Gastroenterology consensus guidelines on the Quality of upper GI endoscopy recommends adequate mucosal visualisation achieved by a combination of aspiration, adequate air insufflation and use of mucosal cleansing techniques. They recommend that the quality of mucosal visualisation and the inspection time during a Barrett’s surveillance endoscopy should be reported[46].
Chen et al[47] investigated their AI system, ENDOANGEL, which provides prompting of blind spots during upper GI endoscopy, informs the endoscopist of the inspection time and gives a grading score of the percentage of the mucosa that is visualised.
Computer aided diagnosis of oesophageal pathology may potentially be a key adjunct for the endoscopist which will improve the detection of early neoplasia in BE and ESCN such that curative endoscopic therapy can be offered. There are significant miss rates of oesophageal cancers despite advances in endoscopic imaging modalities and an AI tool will off-set the human factors associated with some of these miss rates.
At the same time its key that AI systems avoid ‘overfitting’ where it performs well on training data but underperforms when exposed to new data. It needs to be able to detect early oesophageal cancer in low- and high-quality frames during real time endoscopy. This requires high volumes of both low- and high-quality training data tested on low- and high-quality testing data to reflect the real world setting during an endoscopy.
Further research is required on the use of AI in quality control in the oesophagus in order to allow endoscopists to meet the quality indicators necessary during a surveillance endoscopy as set out in many of the international guidelines. This will ensure a minimum standard of endoscopy is met.
Research in this area of AI is expanding and the future looks promising. To fulfil this potential the following is required: (1) Further development is needed to improve the performance of AI technology in the oesophagus to detect early cancer/dysplasia in BE or ESCN during real time endoscopy; (2) High quality clinical evidence from randomised control trials; and (3) Guidelines from clinical bodies or national institutes. Once implemented this will have a significant impact on this field of endoscopy.
| 1. | Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359-E386. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20108] [Cited by in RCA: 20718] [Article Influence: 1883.5] [Reference Citation Analysis (23)] |
| 2. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53206] [Cited by in RCA: 56652] [Article Influence: 7081.5] [Reference Citation Analysis (134)] |
| 3. | Guo L, Xiao X, Wu C, Zeng X, Zhang Y, Du J, Bai S, Xie J, Zhang Z, Li Y, Wang X, Cheung O, Sharma M, Liu J, Hu B. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc. 2020;91:41-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 161] [Article Influence: 26.8] [Reference Citation Analysis (1)] |
| 4. | Menon S, Trudgill N. How commonly is upper gastrointestinal cancer missed at endoscopy? A meta-analysis. Endosc Int Open. 2014;2:E46-E50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 243] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 5. | Visrodia K, Singh S, Krishnamoorthi R, Ahlquist DA, Wang KK, Iyer PG, Katzka DA. Magnitude of Missed Esophageal Adenocarcinoma After Barrett's Esophagus Diagnosis: A Systematic Review and Meta-analysis. Gastroenterology. 2016;150:599-607.e7; quiz e14-5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 148] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 6. | Rodríguez de Santiago E, Hernanz N, Marcos-Prieto HM, De-Jorge-Turrión MÁ, Barreiro-Alonso E, Rodríguez-Escaja C, Jiménez-Jurado A, Sierra-Morales M, Pérez-Valle I, Machado-Volpato N, García-Prada M, Núñez-Gómez L, Castaño-García A, García García de Paredes A, Peñas B, Vázquez-Sequeiros E, Albillos A. Rate of missed oesophageal cancer at routine endoscopy and survival outcomes: A multicentric cohort study. United European Gastroenterol J. 2019;7:189-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 7. | van der Sommen F, de Groof J, Struyvenberg M, van der Putten J, Boers T, Fockens K, Schoon EJ, Curvers W, de With P, Mori Y, Byrne M, Bergman JJGHM. Machine learning in GI endoscopy: practical guidance in how to interpret a novel field. Gut. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 8. | Sharma P, Pante A, Gross S. Artificial intelligence in endoscopy. Gastrointest Endosc. 2020;91:925-931. [RCA] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Alagappan M, Brown JRG, Mori Y, Berzin TM. Artificial intelligence in gastrointestinal endoscopy: The future is almost here. World J Gastrointest Endosc. 2018;10:239-249. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 106] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (4)] |
| 10. | Hussein M, Lovat L, Haidry R. Advances in diagnostic and therapeutic endoscopy. Medicine. 2019;47:440-447. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 11. | Everson MA, Lovat LB, Graham DG, Bassett P, Magee C, Alzoubaidi D, Fernández-Sordo JO, Sweis R, Banks MR, Wani S, Esteban JM, Ragunath K, Bisschops R, Haidry RJ. Virtual chromoendoscopy by using optical enhancement improves the detection of Barrett's esophagus-associated neoplasia. Gastrointest Endosc. 2019;89:247-256.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 12. | Sharma P, Bergman JJ, Goda K, Kato M, Messmann H, Alsop BR, Gupta N, Vennalaganti P, Hall M, Konda V, Koons A, Penner O, Goldblum JR, Waxman I. Development and Validation of a Classification System to Identify High-Grade Dysplasia and Esophageal Adenocarcinoma in Barrett's Esophagus Using Narrow-Band Imaging. Gastroenterology. 2016;150:591-598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 169] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 13. | Subramaniam S, Kandiah K, Schoon E, Aepli P, Hayee B, Pischel A, Stefanovic M, Alkandari A, Coron E, Omae M, Baldaque-Silva F, Maselli R, Bisschops R, Sharma P, Repici A, Bhandari P. Development and validation of the international Blue Light Imaging for Barrett's Neoplasia Classification. Gastrointest Endosc. 2020;91:310-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 14. | Inoue H, Kaga M, Ikeda H, Sato C, Sato H, Minami H, Santi EG, Hayee B, Eleftheriadis N. Magnification endoscopy in esophageal squamous cell carcinoma: a review of the intrapapillary capillary loop classification. Ann Gastroenterol. 2015;28:41-48. [PubMed] |
| 15. | Arima M, Tada M, Arima H. Evaluation of microvascular patterns of superficial esophageal cancers by magnifying endoscopy. Esophagus. 2005;2:191-197. [DOI] [Full Text] |
| 16. | Everson M, Herrera L, Li W, Luengo IM, Ahmad O, Banks M, Magee C, Alzoubaidi D, Hsu HM, Graham D, Vercauteren T, Lovat L, Ourselin S, Kashin S, Wang HP, Wang WL, Haidry RJ. Artificial intelligence for the real-time classification of intrapapillary capillary loop patterns in the endoscopic diagnosis of early oesophageal squamous cell carcinoma: A proof-of-concept study. United European Gastroenterol J. 2019;7:297-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 70] [Article Influence: 10.0] [Reference Citation Analysis (1)] |
| 17. | Sehgal V, Rosenfeld A, Graham DG, Lipman G, Bisschops R, Ragunath K, Rodriguez-Justo M, Novelli M, Banks MR, Haidry RJ, Lovat LB. Machine Learning Creates a Simple Endoscopic Classification System that Improves Dysplasia Detection in Barrett's Oesophagus amongst Non-expert Endoscopists. Gastroenterol Res Pract. 2018;2018:1872437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 18. | Tschanz ER. Do 40% of patients resected for barrett esophagus with high-grade dysplasia have unsuspected adenocarcinoma? Arch Pathol Lab Med. 2005;129:177-180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 19. | Abrams JA, Kapel RC, Lindberg GM, Saboorian MH, Genta RM, Neugut AI, Lightdale CJ. Adherence to biopsy guidelines for Barrett's esophagus surveillance in the community setting in the United States. Clin Gastroenterol Hepatol. 2009;7:736-742; quiz 710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 283] [Cited by in RCA: 267] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 20. | Sharma P, Savides TJ, Canto MI, Corley DA, Falk GW, Goldblum JR, Wang KK, Wallace MB, Wolfsen HC; ASGE Technology and Standards of Practice Committee. The American Society for Gastrointestinal Endoscopy PIVI (Preservation and Incorporation of Valuable Endoscopic Innovations) on imaging in Barrett's Esophagus. Gastrointest Endosc. 2012;76:252-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 21. | van der Sommen F, Zinger S, Curvers WL, Bisschops R, Pech O, Weusten BL, Bergman JJ, de With PH, Schoon EJ. Computer-aided detection of early neoplastic lesions in Barrett's esophagus. Endoscopy. 2016;48:617-624. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 129] [Article Influence: 12.9] [Reference Citation Analysis (3)] |
| 22. | de Groof AJ, Struyvenberg MR, van der Putten J, van der Sommen F, Fockens KN, Curvers WL, Zinger S, Pouw RE, Coron E, Baldaque-Silva F, Pech O, Weusten B, Meining A, Neuhaus H, Bisschops R, Dent J, Schoon EJ, de With PH, Bergman JJ. Deep-Learning System Detects Neoplasia in Patients With Barrett's Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology. 2020;158:915-929.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 239] [Article Influence: 39.8] [Reference Citation Analysis (1)] |
| 23. | de Groof AJ, Struyvenberg MR, Fockens KN, van der Putten J, van der Sommen F, Boers TG, Zinger S, Bisschops R, de With PH, Pouw RE, Curvers WL, Schoon EJ, Bergman JJGHM. Deep learning algorithm detection of Barrett's neoplasia with high accuracy during live endoscopic procedures: a pilot study (with video). Gastrointest Endosc. 2020;91:1242-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 97] [Article Influence: 16.2] [Reference Citation Analysis (1)] |
| 24. | Hussein M, Juana Gonzales-Bueno P, Brandao P, Toth D, Seghal V, Everson MA, Lipman G, Ahmad OF, Kader R, Esteban JM, Bisschops R, Banks M, Mountney P, Stoyanov D, Lovat L, Haidry R. Deep Neural Network for the detection of early neoplasia in Barrett’s oesophagus. Gastrointest Endosc. 2020;91:AB250 (Abstract). [DOI] [Full Text] |
| 25. | Houston T, Sharma P. Volumetric laser endomicroscopy in Barrett's esophagus: ready for primetime. Transl Gastroenterol Hepatol. 2020;5:27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 26. | Trindade AJ, McKinley MJ, Fan C, Leggett CL, Kahn A, Pleskow DK. Endoscopic Surveillance of Barrett's Esophagus Using Volumetric Laser Endomicroscopy With Artificial Intelligence Image Enhancement. Gastroenterology. 2019;157:303-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 27. | Struyvenberg MR, van der Sommen F, Swager AF, de Groof AJ, Rikos A, Schoon EJ, Bergman JJ, de With PHN, Curvers WL. Improved Barrett's neoplasia detection using computer-assisted multiframe analysis of volumetric laser endomicroscopy. Dis Esophagus. 2020;33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (1)] |
| 28. | Swager AF, van der Sommen F, Klomp SR, Zinger S, Meijer SL, Schoon EJ, Bergman JJGHM, de With PH, Curvers WL. Computer-aided detection of early Barrett's neoplasia using volumetric laser endomicroscopy. Gastrointest Endosc. 2017;86:839-846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 105] [Article Influence: 11.7] [Reference Citation Analysis (1)] |
| 29. | Ebigbo A, Mendel R, Probst A, Manzeneder J, Souza LA, Papa JP, Palm C, Messmann H. Computer-aided diagnosis using deep learning in the evaluation of early oesophageal adenocarcinoma. Gut. 2019;68:1143-1145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 106] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 30. | de Groof J, van der Sommen F, van der Putten J, Struyvenberg MR, Zinger S, Curvers WL, Pech O, Meining A, Neuhaus H, Bisschops R, Schoon EJ, de With PH, Bergman JJ. The Argos project: The development of a computer-aided detection system to improve detection of Barrett's neoplasia on white light endoscopy. United European Gastroenterol J. 2019;7:538-547. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 89] [Article Influence: 12.7] [Reference Citation Analysis (1)] |
| 31. | Hashimoto R, Requa J, Dao T, Ninh A, Tran E, Mai D, Lugo M, El-Hage Chehade N, Chang KJ, Karnes WE, Samarasena JB. Artificial intelligence using convolutional neural networks for real-time detection of early esophageal neoplasia in Barrett's esophagus (with video). Gastrointest Endosc. 2020;91:1264-1271.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 164] [Article Influence: 27.3] [Reference Citation Analysis (1)] |
| 32. | Mori Y, Kudo SE, Mohmed HEN, Misawa M, Ogata N, Itoh H, Oda M, Mori K. Artificial intelligence and upper gastrointestinal endoscopy: Current status and future perspective. Dig Endosc. 2019;31:378-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 93] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 33. | Muto M, Minashi K, Yano T, Saito Y, Oda I, Nonaka S, Omori T, Sugiura H, Goda K, Kaise M, Inoue H, Ishikawa H, Ochiai A, Shimoda T, Watanabe H, Tajiri H, Saito D. Early detection of superficial squamous cell carcinoma in the head and neck region and esophagus by narrow band imaging: a multicenter randomized controlled trial. J Clin Oncol. 2010;28:1566-1572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 427] [Cited by in RCA: 546] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 34. | Nakagawa K, Ishihara R, Aoyama K, Ohmori M, Nakahira H, Matsuura N, Shichijo S, Nishida T, Yamada T, Yamaguchi S, Ogiyama H, Egawa S, Kishida O, Tada T. Classification for invasion depth of esophageal squamous cell carcinoma using a deep neural network compared with experienced endoscopists. Gastrointest Endosc. 2019;90:407-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 113] [Article Influence: 16.1] [Reference Citation Analysis (1)] |
| 35. | Singh R, Sathananthan D, Tam W, Ruszkiewicz. Endocytoscopy for diagnosis of gastrointestinal neoplasia: The Experts approach. Video Journal and Encyclopedia of GI Endoscopy. 2013;1:18-19. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 36. | Kumagai Y, Takubo K, Kawada K, Aoyama K, Endo Y, Ozawa T, Hirasawa T, Yoshio T, Ishihara S, Fujishiro M, Tamaru JI, Mochiki E, Ishida H, Tada T. Diagnosis using deep-learning artificial intelligence based on the endocytoscopic observation of the esophagus. Esophagus. 2019;16:180-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (1)] |
| 37. | Shin D, Protano MA, Polydorides AD, Dawsey SM, Pierce MC, Kim MK, Schwarz RA, Quang T, Parikh N, Bhutani MS, Zhang F, Wang G, Xue L, Wang X, Xu H, Anandasabapathy S, Richards-Kortum RR. Quantitative analysis of high-resolution microendoscopic images for diagnosis of esophageal squamous cell carcinoma. Clin Gastroenterol Hepatol. 2015;13:272-279.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 67] [Article Influence: 6.1] [Reference Citation Analysis (2)] |
| 38. | Quang T, Schwarz RA, Dawsey SM, Tan MC, Patel K, Yu X, Wang G, Zhang F, Xu H, Anandasabapathy S, Richards-Kortum R. A tablet-interfaced high-resolution microendoscope with automated image interpretation for real-time evaluation of esophageal squamous cell neoplasia. Gastrointest Endosc. 2016;84:834-841. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 62] [Article Influence: 6.2] [Reference Citation Analysis (1)] |
| 39. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 285] [Article Influence: 40.7] [Reference Citation Analysis (4)] |
| 40. | Zhao YY, Xue DX, Wang YL, Zhang R, Sun B, Cai YP, Feng H, Cai Y, Xu JM. Computer-assisted diagnosis of early esophageal squamous cell carcinoma using narrow-band imaging magnifying endoscopy. Endoscopy. 2019;51:333-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 87] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 41. | Tokai Y, Yoshio T, Aoyama K, Horie Y, Yoshimizu S, Horiuchi Y, Ishiyama A, Tsuchida T, Hirasawa T, Sakakibara Y, Yamada T, Yamaguchi S, Fujisaki J, Tada T. Application of artificial intelligence using convolutional neural networks in determining the invasion depth of esophageal squamous cell carcinoma. Esophagus. 2020;17:250-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 87] [Article Influence: 14.5] [Reference Citation Analysis (2)] |
| 42. | Ohmori M, Ishihara R, Aoyama K, Nakagawa K, Iwagami H, Matsuura N, Shichijo S, Yamamoto K, Nagaike K, Nakahara M, Inoue T, Aoi K, Okada H, Tada T. Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest Endosc. 2020;91:301-309.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 105] [Article Influence: 17.5] [Reference Citation Analysis (1)] |
| 43. | Tomita N, Abdollahi B, Wei J, Ren B, Suriawinata A, Hassanpour S. Attention-Based Deep Neural Networks for Detection of Cancerous and Precancerous Esophagus Tissue on Histopathological Slides. JAMA Netw Open. 2019;2:e1914645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 109] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 44. | Vennalaganti P, Kanakadandi V, Goldblum JR, Mathur SC, Patil DT, Offerhaus GJ, Meijer SL, Vieth M, Odze RD, Shreyas S, Parasa S, Gupta N, Repici A, Bansal A, Mohammad T, Sharma P. Discordance Among Pathologists in the United States and Europe in Diagnosis of Low-Grade Dysplasia for Patients With Barrett's Esophagus. Gastroenterology. 2017;152:564-570.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 107] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 45. | Everson MA, Ragunath K, Bhandari P, Lovat L, Haidry R. How to Perform a High-Quality Examination in Patients With Barrett's Esophagus. Gastroenterology. 2018;154:1222-1226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 46. | Beg S, Ragunath K, Wyman A, Banks M, Trudgill N, Pritchard DM, Riley S, Anderson J, Griffiths H, Bhandari P, Kaye P, Veitch A. Quality standards in upper gastrointestinal endoscopy: a position statement of the British Society of Gastroenterology (BSG) and Association of Upper Gastrointestinal Surgeons of Great Britain and Ireland (AUGIS). Gut. 2017;66:1886-1899. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 243] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 47. | Chen D, Wu L, Li Y, Zhang J, Liu J, Huang L, Jiang X, Huang X, Mu G, Hu S, Hu X, Gong D, He X, Yu H. Comparing blind spots of unsedated ultrafine, sedated, and unsedated conventional gastroscopy with and without artificial intelligence: a prospective, single-blind, 3-parallel-group, randomized, single-center trial. Gastrointest Endosc. 2020;91:332-339.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United Kingdom
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hori K S-Editor: Gao CC L-Editor: A P-Editor: Ma YJ