Published online May 21, 2020. doi: 10.3748/wjg.v26.i19.2349
Peer-review started: November 18, 2019
First decision: January 19, 2020
Revised: February 20, 2010
Accepted: April 18, 2020
Article in press: April 18, 2020
Published online: May 21, 2020
Processing time: 174 Days and 21.7 Hours
Pancreatic cancer (PC) is one of the deadliest cancers worldwide. PC metastasis involves a complex set of events, including epithelial-mesenchymal transition (EMT), that increase tumor cell invasiveness. Recent evidence has shown that hypoxia is a major EMT regulator in pancreatic cancer cells and facilitates metastasis; however, the mechanisms remain elusive.
To investigate the role of miR-301a in hypoxia-induced EMT in PC cells.
Real-time PCR and Western blot analysis were used to detect the expression of miR-301a and EMT markers in PDAC cells cultured in hypoxic and normoxic conditions. Western blot analysis was used to detect the expression of EMT markers in PDAC cells with miR-301a overexpression. Wound healing assay and Transwell assay were used to detect the migration capabilities of PDAC cells with miR-301a overexpression and knockout. Luciferase assay was used to detect the miR-301a promoter and the 3’ untranslated region activity of TP63. Orthotopic PC mouse models were used to study the role of miR-301a in metastasis of PDAC cells in vivo. In situ hybridization assay was used to detect the expression of miR-301a in PDAC patient samples (adjacent paratumor and paired tumor tissues).
Hypoxic environment could directly promote the EMT of PC cells. The expression level of miR-301a was increased in a HIF2α dependent manner in hypoxia-cultured CFPAC-1 and BxPC-3 cells. Overexpression of miR-301a enhanced the hypoxia-induced EMT of PC cells, while knocking out miR-301a result in the suppression of hypoxia-induced EMT. TP63 was a direct target of miR-301a and involved in the metastatic process of PC cells. Furthermore, miR-301a upregulation facilitated PDAC distant metastasis and lymph node metastasis in vivo. Additionally, miR-301a overexpression was indicative of aggressive clinicopathological behaviors and poor prognosis.
The newly identified HIF-2α-miR301a-TP63 signaling pathway may play a crucial role in hypoxia-induced EMT in PDAC cells.
Core tip: In this study, we found that miR-301a expression was increased during the process of hypoxia-induced epithelial-mesenchymal transition (EMT) in pancreatic cancer (PC) cells. miR-301a overexpression facilitated hypoxia-induced EMT, while miR-301a knockout inhibited hypoxia-induced EMT in PC cells. The increased expression of miR-301a was transcriptionally regulated by HIF-2α. In addition, we identified a new target gene of miR-301a, namely, TP63, and confirmed that TP63 was involved in EMT and metastasis of PC cells. Collectively, our data suggest that the newly identified HIF-2α-miR301a-TP63 signaling pathway plays a crucial role in hypoxia-induced EMT in pancreatic ductal adenocarcinoma and that miR-301a may serve as a new prognostic biomarker and candidate miRNA for tumor diagnosis and treatment.
- Citation: Zhang KD, Hu B, Cen G, Yang YH, Chen WW, Guo ZY, Wang XF, Zhao Q, Qiu ZJ. MiR-301a transcriptionally activated by HIF-2α promotes hypoxia-induced epithelial-mesenchymal transition by targeting TP63 in pancreatic cancer. World J Gastroenterol 2020; 26(19): 2349-2373
- URL: https://www.wjgnet.com/1007-9327/full/v26/i19/2349.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i19.2349
The extremely malignant biological behavior of pancreatic ductal adenocarcinoma (PDAC) and the lack of effective treatment for it make this cancer the most lethal malignant tumor type[1]. Most patients are diagnosed with the cancer after it has metastasized to other organs[2]. The development of effective treatments for this disease is a complex and challenging problem due to our poor understanding of the molecular mechanisms underlying PDAC metastasis. Therefore, explorations of the molecular mechanism of tumor metastasis are urgently needed.
Epithelial-mesenchymal transition (EMT) is a process during which epithelial cells lose their cell polarity and gain migratory and invasive properties[3]. The tumor hypoxic microenvironment has previously been shown to induce EMT to promote the metastasis of many types of cancers, including pancreatic cancer (PC)[4], breast cancer[5], hepatocellular carcinoma (HCC)[6], and lung cancer[7]. Under hypoxic conditions, hypoxia-inducible factors (HIFs), namely, HIF-1α and HIF-2α, play critical roles in hypoxia-induced EMT by regulating EMT-related transcription factors (EMT-TFs), including Snail, Twist1, and ZEB1[8-10]. Although the roles of different EMT-TFs and the relevant signaling pathways in mediating EMT are well demonstrated, other regulators such as microRNAs (miRNAs) that are involved in the hypoxia-induced EMT process should be further investigated.
MiRNAs are small non-coding RNAs, containing approximately 22 nucleotides, that act as gene regulators via the cleavage and/or translational repression of target mRNAs[11,12]. MiRNAs are involved in many complex biological processes, such as drug resistance[13-15], tumor growth[16-18], invasion[19,20], and metastasis[21-24]. Several miRNAs, including miR-1236[25], miR-143-5p[26], and miRNA-34a[27], have been reported to participate in regulating hypoxia-induced EMT. In addition, miR-205 is remarkably induced by hypoxia in cervical and lung cancer cells[28]. Interestingly, miR-205 upregulation under hypoxia decreases epithelial marker E-cadherin, increases mesenchymal marker vimentin, and promotes a morphological transition from a typical cobblestone-like appearance to a mesenchymal-like structure[28]. Another study has revealed that the expression level of miR-187-3p in HCC significantly decreases under hypoxia and that miR-187-3p is involved in the promoting effects of hypoxia on the metastasis and EMT of HCC cells[29]. However, the miRNAs involved in hypoxia-induced EMT in PDAC cells have not been identified.
Several recent studies have shown that miR-301a functions as an oncogene in multiple human cancers, including HCC[30], PC[31], Ewing's sarcoma[32], gastric cancer[33], and malignant melanoma[34]. Our previous study has revealed that abnormally high expression levels of miR-301a are associated with lymph node metastasis, advanced pathological stage, and worse survival[35]. MiR-301a overexpression enhances the colony formation, invasion, and migration of PDAC cells in vitro as well as their tumorigenicity in vivo[35]. However, the underlying mechanisms of abnormal miR-301a expression and its role in EMT in PDAC cells remain unclear and require further exploration.
TP63, a member of the p53 family, not only induces transcription of canonical p53 targets but is also a master regulator of epithelial cells[36]. There are many isoforms of p63, including the major types TAp63 and ΔNp63[36]. These two isoforms exhibit unique biological functions by regulating their own distinct target genes. Data from studies of different cancer environments suggest that TAp63 has tumor suppressor type properties and that ∆Np63 has mainly oncogenic properties[37]. Expression of p63 in PC is not well recognized and its role in tumor progression needs further researching.
In this study, we showed that miR-301a is overexpressed under hypoxic conditions in PDAC cells and that miR-301a transcriptional induction is HIF2α dependent. The overexpression and knockout of miR-301a promote and inhibit hypoxia-induced EMT, respectively. Furthermore, we identified TP63 as a miR-301a target gene that is significantly downregulated under hypoxia. Functionally, we showed that ectopic miR-301a expression promotes metastasis in nude mice. Collectively, these data show that the newly identified HIF-2α-miR301a-TP63 signaling pathway that is induced by hypoxia plays crucial roles in hypoxia-induced EMT in PDAC.
The human PC cell lines Panc-1, BxPC-3, and CFPAC-1 were obtained from the American Type Culture Collection (ATCC, Manassas, VA, United States). PANC-1 cells were cultured in DMEM (HyClone, Logan, UT, United States) supplemented with 10% fetal bovine serum (FBS; GIBCO). BxPC-3 cells were cultured in RPMI 1640 (HyClone) supplemented with 10% FBS. CFPAC-1 cells were cultured in IMDM (GIBCO) supplemented with 10% FBS. All cell lines were cultured in an incubator at 37 °C in an atmosphere containing 5% CO2. For hypoxia treatment, cells were cultured in an incubator with 1% O2, 5% CO2, and 94% N2 at 37 °C (Thermo, Waltham, MA, United States) for the indicated time period. Before the experiments were conducted, the cell lines were tested and authenticated according to microscopic morphology, growth curve analysis, and mycoplasma detection per the ATCC cell line verification test recommendations.
Total RNA was extracted with the TRIzol Reagent (Invitrogen) according to the manufacturer’s instructions. The concentration and purity of the RNA were determined on a Nanodrop-2000 spectrophotometer (Thermo Scientific). Complementary DNA was generated with the ImProm-II Reverse Transcription System (Promega, United States). Real-time PCR was performed with SYBR Green PCR Mater Mixture Reagents (Applied Biosystems, United States) on the ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). The relative expression of miR-301a was normalized to the internal control U6, and the levels were calculated using the 2-ΔΔCt method.
Cells and tissues were lysed with 2 × SDS-lysis buffer. Equal amounts of protein were separated by SDS-PAGE and transferred onto nitrocellulose membranes, and the membranes were blocked and incubated overnight with monoclonal antibodies. An anti-β-actin antibody (CST) was used as the loading control. The antibodies against ZO-1, β-catenin, N-cadherin, Snail, and Slug were purchased from Cell Signaling Technologies, and the antibodies against E-cadherin, Fibronectin, Vimentin, and HIF-1α were purchased from BD Biosciences.
For the in vitro cell migration assays, PC cells were seeded into 6-well plates at 2 × 105 cells per well and incubated at 37 °C with 5% CO2 for 24 h to achieve full confluence before the wound was made. An approximately 0.4–0.5-mm line was scraped using the fine end of a sterile pipette tip. Then, the cells were washed gently with PBS and cultured for 24 h. Pictures of the scratches were taken with an inverted microscope and analyzed using ImageJ software. All experiments were repeated three times.
BxPC-3 and CFPAC-1 cells that were stably transfected with miR-301a or PANC-1 cells that were depleted of miR-301a (5 × 104 cells/well) were suspended in 200 μL of serum-free medium and added to the upper chambers of Transwell plates, and 700 μL of medium containing 10% FBS was added to the bottom chambers. After the plates were incubated at 37 °C for 48 h, the cells on the top sides of the Transwell membranes were wiped off carefully with cotton swabs, and the cells on the bottom sides of the Transwell membranes were fixed with 4% paraformaldehyde for 15 min and stained with 0.5% crystal violet for 3 h. The number of cells in three random fields on each membrane was counted.
Coverslip-grown cells were washed three times in prewarmed 1 × PBS and fixed in 4% paraformaldehyde solution for 10 min. Cells were blocked in 2% bovine serum albumin for 1 h and incubated with the appropriate diluted primary antibody overnight at 4 °C. Fluorescently labeled secondary antibodies were applied at 1:200 dilutions for 1 h at room temperature. The coverslips were washed with 1 × PBS before being mounted with Vectashield containing 4,6-diamidino-2-phenylindole (DAPI) onto slides. Images were captured with the Nikon Eclipse Ti (Nikon, Kanagawa, Japan).
For luciferase assay, 1 × 105 BxPC-3 cells or HEK293T cells per well were seeded into 24-well plates and incubated at 37 °C for 24 h. Then, the cells were co-transfected with 100 pmol/L negative control (NC) or miR-301a mimic and 100 ng of the wild-type or mutant TP63 3’ untranslated region (3’UTR) psiCHECK-2 plasmid. After the cells were cultured for 36 h, luciferase activity was measured with the Dual-Luciferase Reporter Assay System (Promega). Normalized data were calculated as the ratio of the Renilla/firefly luciferase activities.
To determine whether HIF-2Α could regulate miR-301a expression, we constructed the promoter of miR-301a using the pGL4.27 vector that carried the promoter luciferase assay.
To construct the miR-301a overexpression plasmid, the miR-301a expression cassette containing the miR-301a hairpin sequence and flanking regions was cloned from human genomic DNA isolated from CFPAC-1 cells and inserted into the lentivirus pLVX-IRES-Puro vector.
A partial wild-type sequence or the mutant sequence of the TP63 3’UTR was inserted between the XhoI and Not1 restriction sites into the 3’UTR of the hRluc gene in the psiCHECK2 vector.
The sgRNAs targeting miR-301a were designed with the online guide design tool (http://crispr.mit.edu). The top two highest scoring sgRNAs that targeted sequences close to the miR-301a stem-loop were selected for cloning into LentiCRISPRv2 vectors and were verified by DNA sequencing. Lentiviral particles were produced using LentiCRISPRv2 plasmids and the packaging plasmids pCMΔ8.9 and pMD2.G (3:2:1). After the PANC-1 cells were treated with lentiviral particles for 48 h, infected cells were selected with puromycin and then subcultured in 96-well plates using the limited dilution method. Monoclonal cells were selected, cultured, and expanded in vitro. PCR primers were designed according to the miR-301a gene sequence, and afterward, we collected genomeic DNA from the monoclonal cells for PCR rapid testing. Gene sequencing and comparative analysis were performed.
The oligonucleotide probe of miR-301a was purchased from Wuhan Boster Biological Technology. The sequence of miR-301a probe was 5'-TTTGACAATACTATTGCACT-3'. PDAC tissue slides were deparaffinized and digested with 20 µg/mL proteinase K in pre-warmed 50 mmol/L Tris for 20 min at 37 °C. The slides were rinsed five times in distilled water, immersed in ice-cold 20% (v/v) acetic acid for 20 s, dehydrated by washing for approximately 1 min per wash in 70% ethanol, 95% ethanol, and 100% ethanol, and air dried. Then, 100 µL of hybridization solution was added to each slide. After that, the slides were prehybridized in a hybridization solution at 57 °C for 2 h. Tissues were hybridized overnight in the presence of 10 ng 3’-5’ DIG-labeled miR-301a-3p LNA probes at 50 °C. Slides were washed twice stringently and transferred to a humidified chamber, followed by the addition of 200 µL blocking buffer to each section (MABT + 2% BSA) and blockage for 2 h at room temperature. The anti-label antibody at the required dilution in blocking buffer was added and incubated for 2 h at room temperature and an immunological reaction was carried out by using the rabbit antibody against digoxingenin and alkaline phosphatase, according to the manufacturer’s recommendation. Each side was assigned a score for intensity and staining positive pattern.
The mice used for PC orthotopic transplantation were 4-wk-old female BALB/c nude mice, which were purchased from the Shanghai Laboratory Animal Center (SLAC) and kept in the Department of Animal Science, Shanghai Jiaotong University School of Medicine. All animal experiments were approved by the Animal Ethics Committee of Shanghai Jiaotong University School of Medicine.
A total of 2 × 105 CFPAC-1- LUC cells that were stably transfected with pLVX-miR-301a or NC were subcutaneously injected into nude mice. Two weeks later, the tumor tissues were cut into pieces that were approximately 1 mm3 in size, and then, the pieces were inoculated into pancreases (orthotopic) of the nude mice.
Primary and metastatic neoplasms were monitored by measuring the firefly luciferase activity, and these measurements were performed by bioluminescence imaging on the sixth week. The mice were sacrificed and weighed on the 6th week. The primary tumors and abdominal metastases were weighed, and the ascites samples were collected. Metastases of the abdominal lymph nodes, liver, and lung tissues were observed and recorded. The tumor tissues were fixed with 4% paraformaldehyde for HE staining.
Three or more independent experiments were carried out for each assay in this study. All data are presented as the mean ± SD. Two-tailed Student’s t-test was used for statistical comparison in each group and to determine the correlation between miR-301a expression and the clinicopathological variables. The Kaplan–Meier method was used to evaluate the overall survival. A P value less than 0.05 was considered to be statistically significant (P < 0.05, P < 0.01, and P < 0.001).
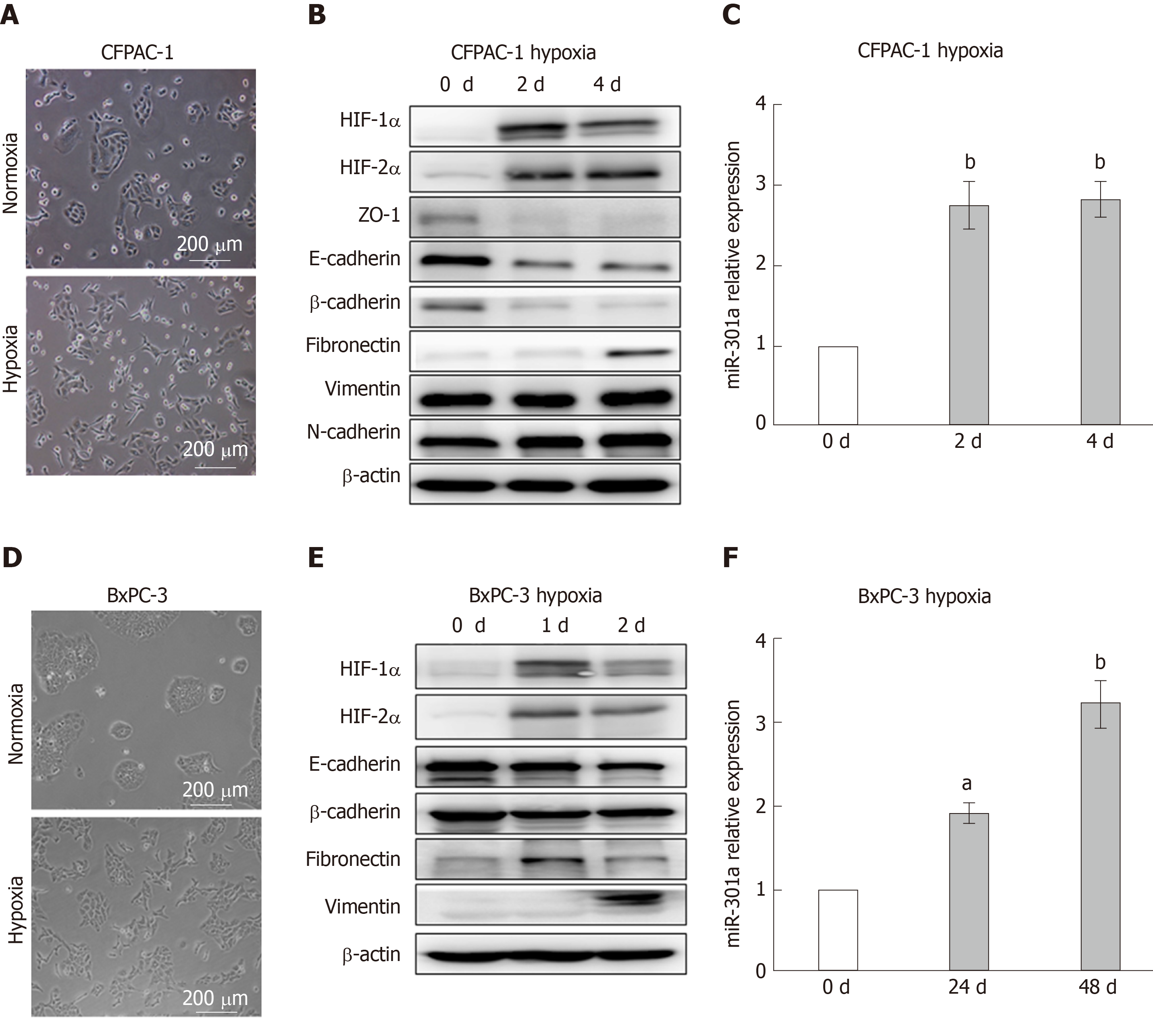
To investigate the effect of hypoxia on in EMT in PC cells, we used two PC cell lines with obvious epithelial phenotypes, namely, CFPAC-1 and BxPC-3. These two cell lines have epithelial morphology and polarization with apical microvilli, tight junctions, and gap junctions. After the CFPAC-1 and BxPC-3 cells were cultured under normoxia or hypoxia (oxygen concentration 1%) for 48 h (Figure 1C and F), the morphological changes were evaluated under an inverted phase-contrast microscope. We observed that CFPAC-1 and BxPC-3 cells cultured in normoxia exhibited a typical epithelial morphology, whereas these cells exhibited a typical mesenchymal morphology after culturing in hypoxia for 48 h (Figure 1A and D). We further determined the expression levels of HIF-1α, HIF-2α, and EMT markers in the CFPAC-1 and BxPC-3 cells cultured in normoxia or hypoxia. The results showed that the expression levels of HIF-1α, HIF-2α, and the mesenchymal markers (Fibronectin and Vimentin) in CFPAC-1 and BxPC-3 cells cultured under hypoxia were significantly higher than those in cells cultured under normoxia. In contrast, the expression levels of the epithelial markers (E-cadherin and β-catenin) were markedly decreased in the cells cultured under hypoxia (Figure 1B and E). These results strongly indicated that the hypoxic environment could directly promote EMT in PC cells. We further examined the expression of miR-301a in hypoxia- and normoxia-cultured PC cells by qRT-PCR. We found that the expression level of miR-301a was significantly higher in hypoxia-cultured CFPAC-1 and BxPC-3 cells than in normoxia-cultured cells.
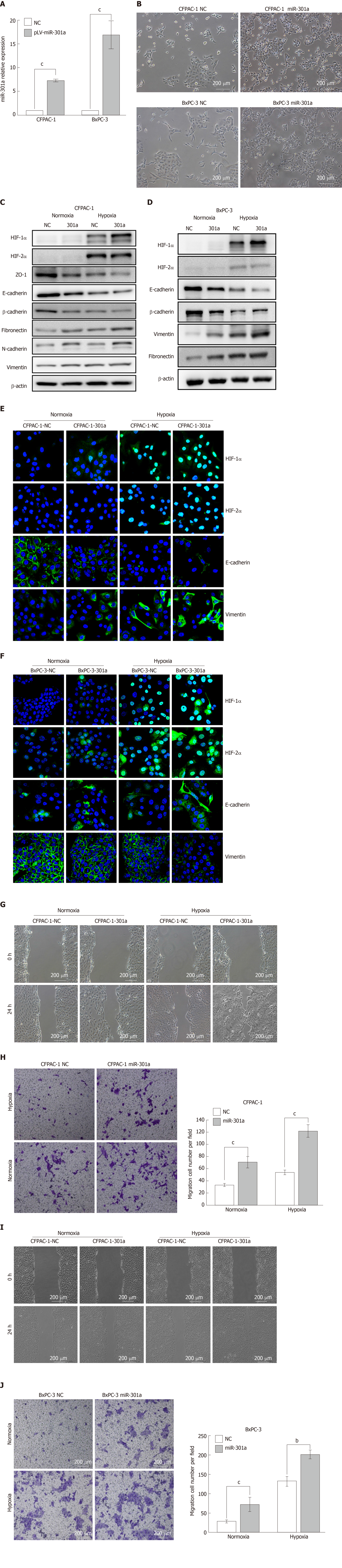
To investigate the role of miR-301a in hypoxia-induced EMT, we constructed PC cell lines that stably expressed miR-301a (Figure 2A) and evaluated the cell morphology under an inverted phase-contrast microscope. We found that miR-301a overexpression strongly promoted the transition of the pancreatic cells from an epithelial morphology to a mesenchymal morphology (Figure 2B). Next, we performed cell immunofluorescence and Western blot analyses to detect the protein expression of HIF-1α, HIF-2α, and the EMT markers. The results showed that the upregulation of miR-301a in CFPAC-1 and BxPC-3 cells increased the expression of the mesenchymal markers (Fibronectin and Vimentin) but decreased the expression of the epithelial markers (E-cadherin and β-catenin) under both normoxia and hypoxia (Figure 2C-F). Notably, HIF-1α protein expression was significantly higher in the miR-301a-overexpressing cells than in the control cells under hypoxia (Figure 2C and D). In addition, the immunofluorescence assays showed that HIF-1α protein accumulation in the nuclei of the tumor cells was more obvious in the miR-301a-overexpressing group than in the control group (Figure 2E and F). Because cells undergoing EMT generally exhibit elevated cell motility, we further examined the effects of elevated miR-301a expression on PC cell migration with migration assays and wound healing assays. As expected, miR-301a upregulation strongly promoted the migration of CFPAC-1 and BxPC-3 cells under both normoxia and hypoxia (Figure 2G-J). These results strongly indicated that miR-301a expression directly correlated with the EMT phenotype and metastatic potential of PC cells.
To examine the effects of the loss of endogenous miR-301a on PC cell biology, CRISPR/Cas9 technology was used to remove the nucleotide fragments encompassing miR-301a from the PC cell line PANC-1. Agarose gel electrophoresis of DNA was performed to visualize the amplification products that included the miR-301a flanking regions after total genomic DNA was isolated from 11 independent deletion clones and 1 non-targeted (NT) clone. We observed that the amplification products of the NO.2 clone were noticeably shorter than the others (Figure 3A). Sanger sequencing demonstrated that an approximately 169-bp fragment encompassing miR-301a from the NO.2 clone was removed (Figure 3B). In addition, qRT-PCR demonstrated a significant downregulation of miR-301a in the edited cells under both normoxia and hypoxia (Figure 3C). Next, we investigated the effects of miR-301a knockout on EMT and migration of PANC-1 cells in vitro. We observed that the PANC-1 cells showed obvious mesenchymal morphology changes in the hypoxic environment. After miR-301a was knocked down, the mesenchymal morphological changes of the PC cells observed under hypoxia were blocked (Figure 3E). Meanwhile, we detected the expression of EMT-related markers and the HIF-1α and HIF-2α proteins in the miR-301a-deleted cells by Western blot. The results showed that the HIF-1α and HIF-2α protein levels were significantly increased in the PANC-1 cells cultured under hypoxia compared with the cells cultured under normoxia (Figure 3D). Notably, the HIF-1α protein levels were significantly decreased after the miR-301a deletion compared with the control group under hypoxia. In addition, the knockout of miR-301a greatly promoted the expression of the epithelial markers (E-cadherin and ZO-1) in PANC-1 cells and inhibited the expression of the mesenchymal markers (Fibronectin and Vimentin) (Figure 3D). As expected, in the wound healing and Transwell migration assays, the miR-301a knockout significantly reduced PANC-1 cell migration under both normoxia and hypoxia (Figure 3F and G). These findings are consistent with the results of the miR-301a overexpression experiments. Together with the data from miR-301a overexpression experiments, these results indicate that miR-301a is a key regulator of EMT in PC cells.
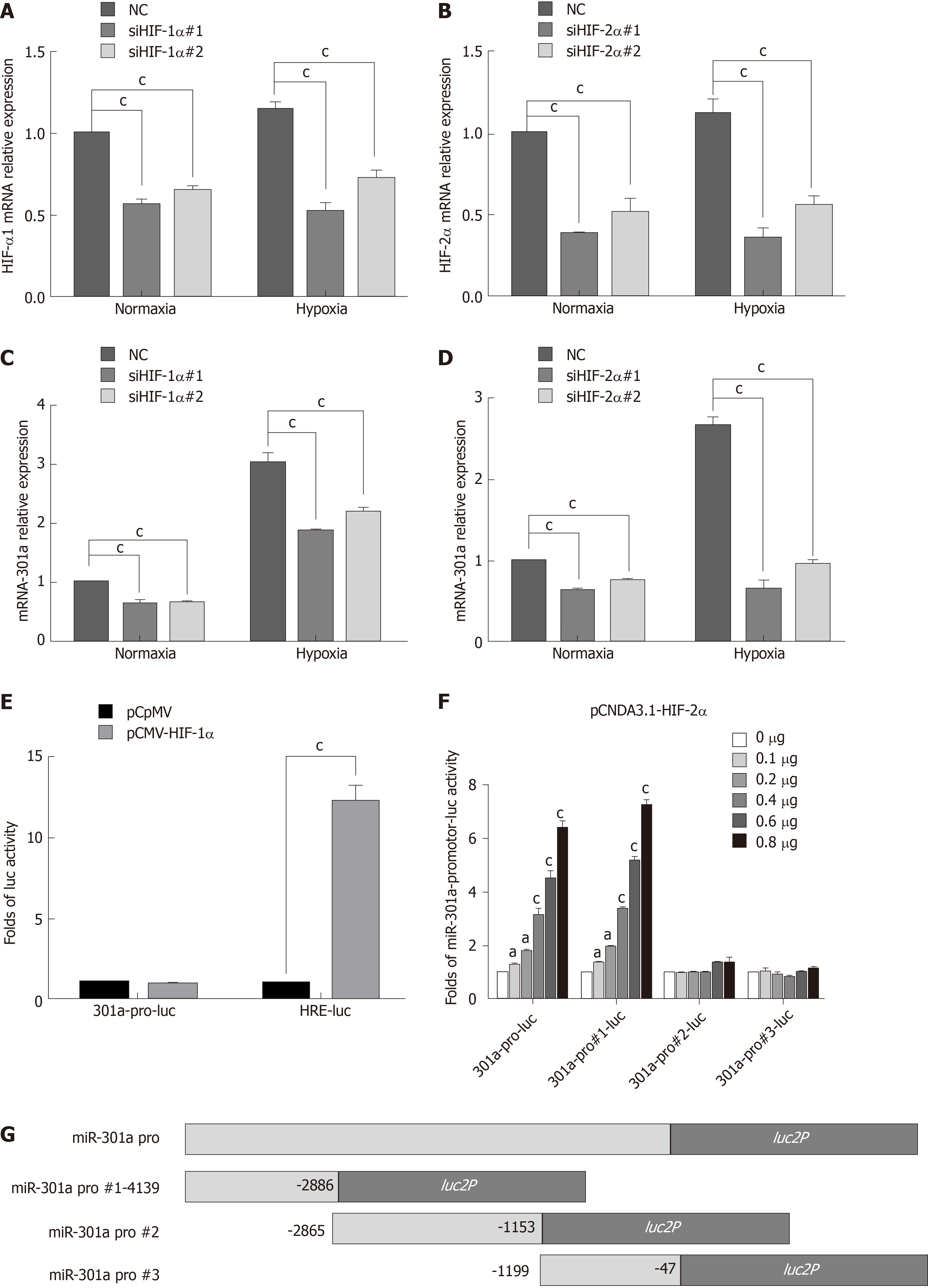
The molecular mechanism of miR-301a overexpression under hypoxia remains unclear. Because most of the genes altered during hypoxia were regulated by HIF-1α and HIF-2α, we knocked down HIF-1α or HIF-2α by siRNA interference and detected miR-301a expression by qRT-PCR. The results showed that miR-301a expression was significantly decreased after the knockdown of HIF-1α or HIF-2α under normoxia or hypoxia, and that miR-301a downregulation was especially notable after HIF-2α knockdown (Figure 4A-D). To determine whether miR-301a was transcriptionally activated by HIF-1α or HIF-2α, we cloned a 4139-bp DNA fragment that was upstream of miR-301a and placed this fragment upstream of Luc. The dual luciferase reporter gene assays showed that HIF-1α significantly increased the luciferase activity of the HRE-Luc group (positive control) but had no effect on the miR-301a promotor group (Figure 4E). Meanwhile, transfections with different concentrations of the HIF-2α plasmids significantly increased the activity of the miR-301a promoter in a dose-dependent manner. We further constructed three truncations of the miR-301a promotor (Figure 4G). As a result, the 4139-2866-bp DNA fragment upstream of miR-301a was able to drive the expression of Luc, indicating that the 1.2-kb DNA fragment was functional as a promoter (Figure 4F).
MiRNAs exert their functions by regulating the expression of their target genes. To search for putative targets of miR-301a, we used bioinformatics prediction software, namely, TargetScan (http://targetscan.org) and miRBase (http://www.mirbase.org), and identified a common set of four candidate genes whose 3’UTRs contained at least one putative miR-301a binding sequence and whose functions were associated with EMT and metastasis. P63 is a nuclear transcription factor of the p53 family and regulates crucial events in the formation of epithelial structures. Unlike p53, the p63 gene is rarely mutated in human cancer but is often deregulated. However, the mechanism of p63 downregulation remains unclear, and the role of p63 in PC requires further study. Our study found that p63 was dramatically downregulated under hypoxia. In addition, p63 was predicted to be a candidate target gene of miR-301a. Therefore, we speculated that hypoxia-induced EMT was regulated by p63 downregulation. To validate TP63 as a direct target of miR-301a and to determine the exact binding sites, we cloned the wild-type or mutant 3’UTR of TP63 into the downstream region of the Renilla luciferase gene in the psiCHECK vector, with a firefly luciferase coding gene as the internal control. MiR-301a, but not miR-NC, significantly suppressed the luciferase activities of the reporter genes containing the 3’UTR of TP63. In addition, neither miR-301a nor miR-NC had effects on the luciferase activity of the Luc-TP63-WT plasmid. Consistently, the ectopic expression of miR-301a markedly reduced TP63 expression at the protein level in CFPAC-1 and BxPC-3 cells (Figure 5A-D). In addition, miR-301a knockout evidently promoted TP63 expression in PANC-1 cells (Figure 5E). To examine the role of TP63 in miR-301a-regulated PC cell migration, we upregulated and downregulated TP63 in miR-301a stable expression cell lines and in miR-301a deletion cell lines, respectively. Our results showed that TP63 upregulation greatly blocked the miR-301a-induced migration enhancement and that TP63 downregulation greatly increased the migration of the miR-301a knockout cells. These findings show that TP63 can partially reverse miR-301a-induced migration, suggesting that TP63 is a functional mediator of miR-301a in PC cells.
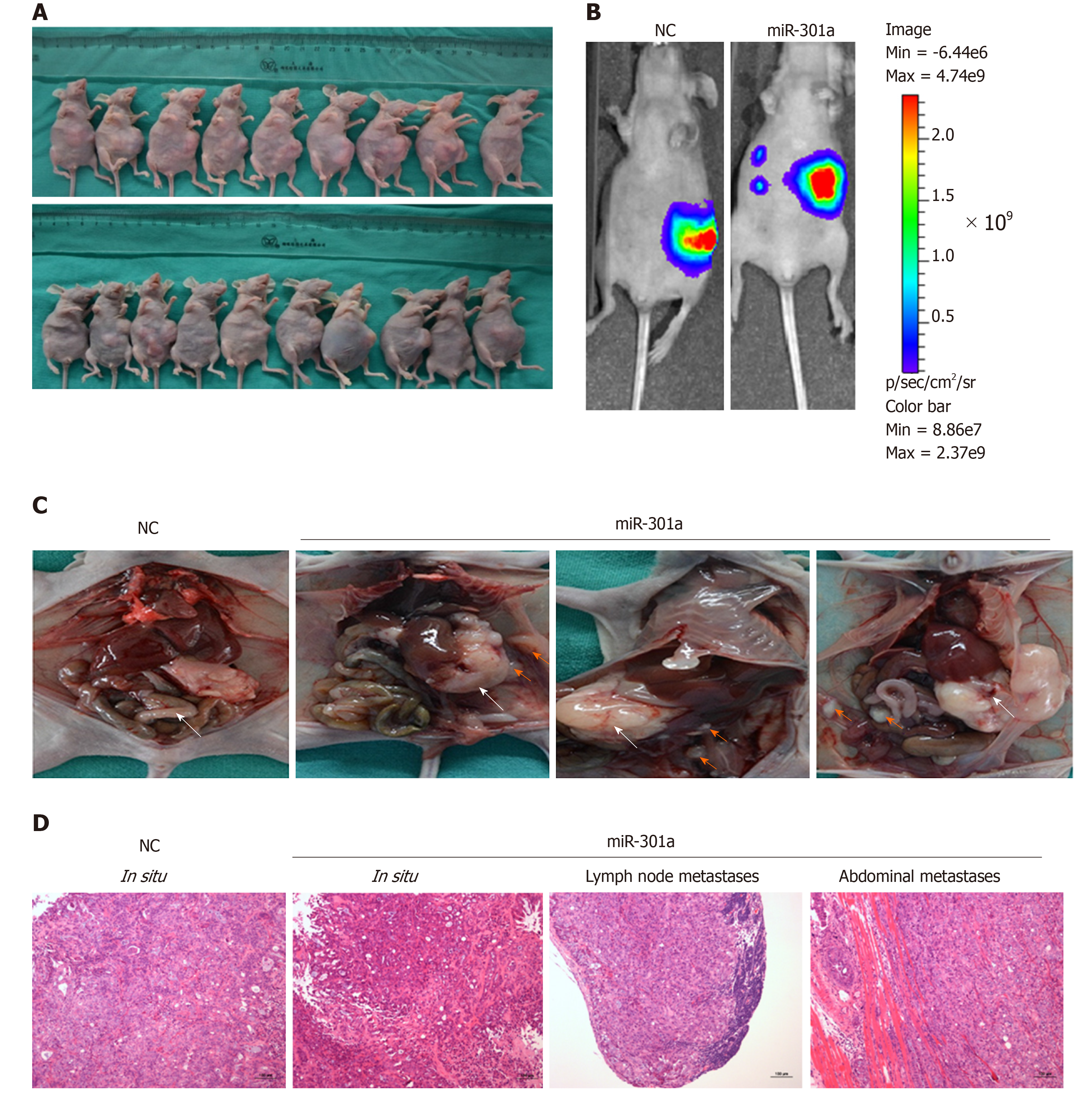
The above experiments confirmed in vitro that miR-301a participated in hypoxia-induced EMT and metastasis of PC cells by targeting the TP63 gene. Next, we explored the biological function of miR-301a in vivo. First, we constructed a CFPAC-1-LUC cell line stably expressing miR-301a and established nude mouse models with the orthotopic implantation of PC cells (Figure 6A). The results showed that metastasis more easily occurred in the miR-301a overexpression group (Figure 6B). The statistical results showed that no significant difference in the weight of the primary tumor was evident between the miR-301a overexpression group and the control group (1.96 ± 0.93 vs 2.37 ± 0.87, P = 0.36), and no differences were evident in the incidences of hepatic and pulmonary metastasis (Table 1). The incidence of ascites in the miR-301a overexpression group was increased (6/10 vs 3/9), although the result was not statistically significant (P = 0.483). More importantly, the incidences of abdominal metastasis and lymph node metastasis were increased after miR-301a overexpression (Table 1 and Figure 6C). Consistently, HE staining showed that the miR-301a overexpression group displayed obvious lymph node metastasis and distant metastasis of the PC cells, while the miR-NC group displayed localized growth (Figure 6D). Collectively, these results demonstrated that ectopic miR-301a expression was capable of promoting tumor metastasis in vivo.
| MiR-301a | NC | P value | |
| Mouse weight (g) | 20.90 ± 2.42 | 20.07 ± 1.58 | 0.42 |
| Tumor weight (g) | 1.96 ± 0.93 | 2.37 ± 0.87 | 0.36 |
| Incidence of ascites | 6/10 | 3/9 | 0.483 |
| Abdominal metastasis | 9/10 | 2/9 | 0.012 |
| Liver metastasis | 2/10 | 2/9 | 1.000 |
| Pulmonary metastasis | 2/10 | 1/9 | 1.000 |
| Lymph node metastases | 5/10 | 0/10 | 0.039 |
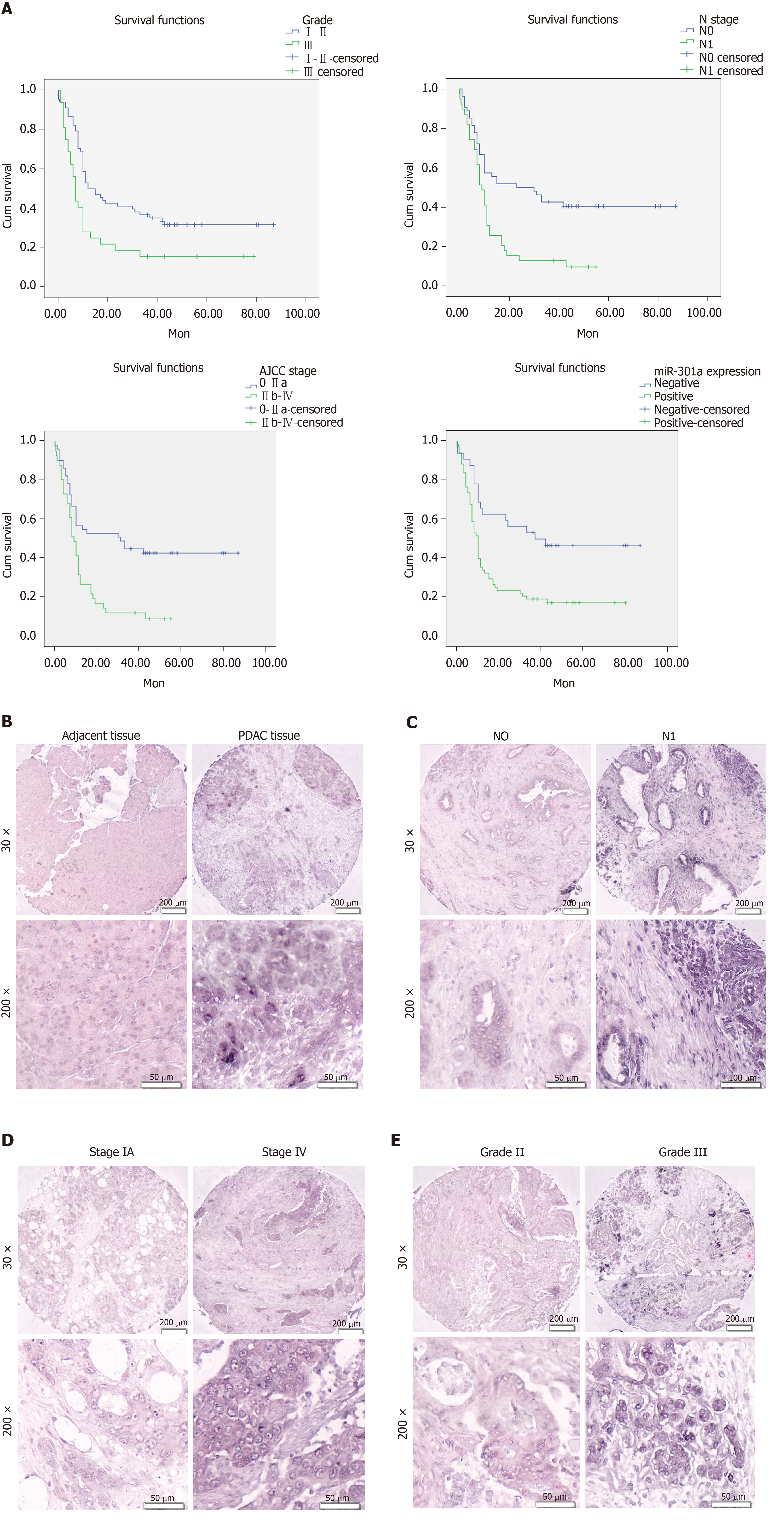
To investigate the relationship between miR-301a expression and the clinicopathological features and prognosis of PDAC patients, we detected the expression of miR-301a in PC and adjacent tissues by in situ hybridization. We observed miR-301a-positive staining in the cytoplasm of the tumor cells and no staining in the nuclei or cell membrane. Statistical analysis showed that tumor tissues from 68 of 100 cases were miR-301a positive, while the remaining 32 tissues were miR-301a negative. However, 38 cases comprising 80 paracancerous tissues were miR-301a positive, and 42 cases were miR-301a negative. MiR-301a expression was higher in the PC tissues than in the paracancerous tissues (P = 0.005) (Table 2 and Figure 7B). As expected, miR-301a expression was higher in the cancerous tissues with lymph node metastasis than in those without (P = 0.001) (Table 2 and Figure 7C). In addition, increasing miR-301a expression correlated with a decreased level of tumor differentiation (Table 2 and Figure 7E) and an advanced stage of PDAC (Table 2 and Figure 7D) but showed no significant difference in terms of the patient’s age, gender, tumor size, tumor location, or T stage (P > 0.05) (Table 2). These findings strongly indicate that miR-301a expression plays a critical role in the development and progression PC and that miR-301a is a valuable biomarker for this disease.
| MiR-301a expression | |||||
| Parameters | n | Negative | Positive | P value | |
| Tissueb | 0.005 | ||||
| Nontumor | 80 | 42 | 38 | ||
| Tumor | 100 | 32 | 68 | ↑ | |
| Age, yr | 0.064 | ||||
| ≤ 60 | 49 | 20 | 29 | ||
| > 60 | 51 | 12 | 39 | ||
| Sex | 0.338 | ||||
| Male | 63 | 18 | 45 | ||
| Female | 37 | 14 | 23 | ||
| Tumor size (cm) | 0.794 | ||||
| ≤ 5 | 60 | 19 | 41 | ||
| > 5 | 38 | 13 | 25 | ||
| Missing data | 2 | ||||
| Tumor location | 0.378 | ||||
| Head | 62 | 21 | 41 | ||
| Body or tail | 32 | 8 | 24 | ||
| Missing data | 6 | ||||
| T stage | 0.363 | ||||
| T1 | 4 | 2 | 2 | ||
| T2 | 74 | 26 | 48 | ||
| T3 | 20 | 4 | 16 | ||
| Missing data | 2 | ||||
| N stagec | 0.001 | ||||
| N0 | 54 | 28 | 26 | ||
| N1 | 39 | 5 | 34 | ↑ | |
| Missing data | 7 | ||||
| Tumor gradea | 0.016 | ||||
| I-II | 68 | 27 | 41 | ||
| III | 32 | 5 | 27 | ↑ | |
| AJCC stageb | 0.007 | ||||
| 0-IIa | 51 | 22 | 29 | ||
| IIb- IV | 41 | 7 | 34 | ↑ | |
| Missing data | 8 | ||||
Because the miR-301a expression level had an intimate association with pathological characteristics, we inferred that miR-301a overexpression promoted PDAC progression and predicted a poor patient prognosis. A Kaplan–Meier analysis of the different prognostic parameters from 100 PC patients indicated that the overall survival time was significantly lower for patients with worse tumor differentiation, advanced stage, positive lymph node metastasis, and miR-301a positivity (Figure 7A).
In this study, we determined the critical roles of miR-301a in hypoxia-induced EMT in PC cells. We found that miR-301a was upregulated in a HIF-2α-dependent manner and that miR-301a could significantly promote the hypoxia-induced EMT process by targeting TP63. Furthermore, the ectopic expression of miR-301a facilitated PDAC distant metastasis and lymph nodes metastasis in vivo. In addition, miR-301a overexpression was indicative of aggressive clinicopathological behaviors and poor prognosis. Collectively, our novel clinical and mechanistic data strongly suggest that miR-301a overexpression contributes to hypoxia-induced EMT in PC cells and that miR-301a may serve as a new prognostic biomarker and candidate miRNA for tumor diagnosis and treatment.
MiRNAs play important roles in the occurrence and development of many types of tumors by participating in cell proliferation, invasion, metastasis, chemoresistance, and other biological processes. Among these miRNAs, miR-301 is overexpressed in various malignancies. A number of studies have confirmed that miR-301 functions as an oncogene in tumor progression. Shi et al[38] have found that miR-301 is a negative prognostic factor for invasive ductal breast cancer. The authors have also demonstrated that miR-301 promotes cell proliferation, clone formation, invasion, migration, and tamoxifen resistance by targeting FOXF2, BBC3, PTEN, and COL2A1[38]. In addition, Yue et al[39] have found that miR-301a is overexpressed in glioma tissues and that miR-301a expression correlates with tumor differentiation. More importantly, patients with high miR-301a expression have a significantly poorer prognosis[39]. Another study has shown that miR-301a promotes NF-κB activation by inhibiting NKrf, a negative regulator of NF-κB[31]. NF-κB activation is correlated with tumor invasion-related clinicopathological features, such as lymph node metastasis and high pathological grade[40]. In our previous study, we found that miR-301a overexpression enhanced PDAC cell colony formation, invasion, and migration in vitro and tumorigenicity in vivo[35].
In this study, we further explored the role and mechanism of miR-301a in hypoxia-induced EMT in PC. We found that hypoxia was capable of inducing EMT in PC cells and significantly upregulated miR-301a expression. Moreover, we observed that elevated miR-301a expression could contribute to the acquisition of the EMT phenotype via the upregulation of mesenchymal cell markers and downregulation of epithelial cell markers, whereas miR-301a deletion could contribute to the acquisition of the mesenchymal-epithelial transition phenotype. Consistent with our results, a recent study revealed that miR-301a overexpression downregulated E-cadherin expression in prostate cancer cells, thereby promoting tumor cell EMT and metastasis. Therefore, we speculate that miR-301a upregulation is a key factor in tumor cell EMT.
Given the important role of miR-301a in hypoxia-induced EMT in PC, we further examined the underlying mechanisms responsible for miR-301a overexpression in malignances. The phenomenon whereby miR-301a is upregulated in hypoxia is also evident in prostate cancer, but the underlying mechanisms have not been reported. Our study demonstrates that miR-301a is a downstream target of HIF-2α, an important transcription factor that can control the adaptive responses of cells to hypoxia. The data are as follows: miR-301a is greatly decreased after HIF-2α knockdown, and dual luciferase reporter gene assays show that HIF-2α indeed promotes the activity of the miR-301a promoter region. Notably, the HIF-1α protein levels in miR-301a-overexpressing cells are significantly increased compared with those in the control cells, while the HIF-1α protein levels in the miR-301a-deleted cells are significantly lower than the levels in the control group, suggesting that altered miR-301a expression may affect HIF-1α protein expression. HIF-1α participates in several biological processes in cancer, including EMT and metastasis, by regulating target genes, such as HDAC1[7], TWIST[41,42], and ZEB1[10].
To identify the target genes of miR-301a in PC, we used the TargetScan and microRNAs.org online tools to find potential miR-301a target genes. We have selected two genes, namely, TP63 and PTEN, based on the following criteria: The target gene must be associated with two algorithm overlaps and must be associated with the EMT process. PTEN has been reported as a target gene for miR-301 in breast cancer[38]. Although a previous study by our group showed that miR-301a bound to the PTEN 3'UTR region and inhibited PTEN protein expression, PTEN protein expression was not significantly altered under hypoxia in subsequent studies. However, the levels of TP63, a tumor suppressor gene, were significantly reduced in hypoxia-cultured PC cell lines, and miR-301a was confirmed to bind the 3'UTR region of TP63 to reduce TP63 protein levels. Based on these findings, we conclude that TP63 is a novel target gene of miR-301a.
TP63 is a member of the p53 tumor suppressor gene family and transcribes from two different promoters to produce two subtypes, namely, TAp63 and ΔNp63[43]. Previous studies have shown that TAp63 inhibits EMT in tumor cells, whereas ΔNp63 promotes the development of EMT[37,44]. Thus, we hypothesized that miR-301a played a role in EMT in PC cells, possibly by inhibiting TAp63. To verify this possibility, we upregulated TAp63 in the miR-301a-overexpressing cell line. As expected, TAp63 overexpression resulted in the upregulation of epithelial marker E-cadherin and the downregulation of mesenchymal marker vimentin, indicating that PC cells underwent mesenchymal-epithelial transition. Our results suggested that TAp63 was a key regulator in the process of miR-301a-induced PC cell EMT.
More importantly, TAp63 is capable of suppressing tumor metastasis through different mechanisms, such as the regulation of HIF-1α signaling[45]. TAp63 can upregulate the expression of Sharp-1/Dec-2[46]. The combination of the Sharp-1 and HIF-1α proteins promotes the ubiquitin-independent proteasomal degradation of the HIF-1α protein. Therefore, TAp63 indirectly regulates HIF-1α through Sharp-1[46]. In this study, we found HIF-1α accumulation in miR-301a-overexpressing cells. The possible mechanism of this observation is that TAp63 downregulation by miR-301a relieves the Sharp-1-mediated degradation of HIF-α proteins. Consequently, the accumulation of HIF-1α in PC cells induces EMT in multiple ways, such as the transcriptional activation of various transcription factors, including VEGF, Snail, and Twist. In addition, TAp63 may also directly inhibit a variety of EMT-related transcription factors, including Snail, Slug, and Twist, to regulate tumor cell EMT and metastasis.
In summary, we have found that miR-301a expression in PC cells is increased during the process of hypoxia-induced EMT. MiR-301a overexpression facilitates hypoxia-induced EMT and HIF-1α expression, while miR-301a knockout inhibits hypoxia-induced EMT and HIF-1α expression. The increased expression of miR-301a may be transcriptionally regulated by HIF-2α. In addition, we have identified a new target gene of miR-301a, namely, TP63, and confirmed that TP63 is involved in EMT and metastasis of PC. Therefore, the results of this study showed that miR-301a may be a new molecular marker for the prognosis of patients with PC.
Pancreatic cancer (PC) continues to be a leading cause of cancer-related death worldwide. PC metastasis involves a complex set of events, including epithelial-mesenchymal transition (EMT), that increase tumor cell invasiveness. Recent evidence has shown that hypoxia is a major EMT regulator in PC cells and facilitates metastasis; however, the mechanisms remain elusive.
This study aimed to investigate the key regulators and signaling pathways in hypoxia-induced EMT in PC cells. This study may enrich the mechanism of PC metastasis and provide a target for the treatment of PC.
The objectives of this research was to explore the role and the mechanism of miR-301a in hypoxia-induced EMT in PC cells. Realizing these objectives will provide strong evidence that miR-301a can be used as a new molecular marker for the prognosis of patients with PC.
Real-time PCR and Western blot analysis were used to detect the expression of miR-301a and EMT markers in PDAC cells cultured in hypoxic and normoxic conditions. Western blot analysis was used to detect the expression of EMT markers in PDAC cells with miR-301a overexpression and knockout. Wound healing assay and Transwell assay were used to detect the migration capabilities of PDAC cells with miR-301a overexpression and knockout. Luciferase assay was used to detect the miR-301a promoter and the 3’ untranslated region of TP63. Orthotopic PC mouse models were used to study the role of miR-301a in metastasis of PDAC cells in vivo. In situ hybridization assay was used to detect the expression of miR-301a in PDAC patient samples.
MiR-301a was increased in a HIF-2α dependent manner in the process of hypoxia-induced EMT in PC cells. MiR-301a promoted EMT of PC cells by inhibiting the expression of TP63. Furthermore, miR-301a upregulation facilitated PDAC distant metastasis and lymph node metastasis in vivo. Additionally, miR-301a overexpression was indicative of aggressive clinicopathological behaviors and poor prognosis. These results are helpful to enrich the metastasis mechanism of PC and provide targets for clinical treatment. How to develop an effective drug to inhibit miRNAs in PC patient is an urgent problem to be solved.
MiR-301a regulated by HIF-2α plays an important role in the process of hypoxia-induced EMT in PC cells. In addition, TP63 as a new target gene of miR-301a, is involved in the EMT and metastasis of PC. Therefore, the results of this study showed that miR-301a may be a new therapeutic target for patients with PC.
A prospective study is expected to confirm the role of miR-301a in PDAC patients with advanced metastases. It is worth further studying whether dysfunctions of the miR-301a by effective drugs could prevent PDAC metastasis.
We would like to thank Professor Qian Zhao and the Key Laboratory of Cell Differentiation and Apoptosis of National Ministry of Education.
| 1. | Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371:1039-1049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1500] [Cited by in RCA: 1743] [Article Influence: 145.3] [Reference Citation Analysis (0)] |
| 2. | Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, Hruban RH. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63:318-348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 572] [Cited by in RCA: 686] [Article Influence: 52.8] [Reference Citation Analysis (0)] |
| 3. | Diepenbruck M, Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr Opin Cell Biol. 2016;43:7-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 390] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 4. | Li H, Wang X, Wen C, Huo Z, Wang W, Zhan Q, Cheng D, Chen H, Deng X, Peng C, Shen B. Long noncoding RNA NORAD, a novel competing endogenous RNA, enhances the hypoxia-induced epithelial-mesenchymal transition to promote metastasis in pancreatic cancer. Mol Cancer. 2017;16:169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 203] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 5. | Azimi I, Beilby H, Davis FM, Marcial DL, Kenny PA, Thompson EW, Roberts-Thomson SJ, Monteith GR. Altered purinergic receptor-Ca²⁺ signaling associated with hypoxia-induced epithelial-mesenchymal transition in breast cancer cells. Mol Oncol. 2016;10:166-178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 6. | Ibrahim AA, Schmithals C, Kowarz E, Köberle V, Kakoschky B, Pleli T, Kollmar O, Nitsch S, Waidmann O, Finkelmeier F, Zeuzem S, Korf HW, Schmid T, Weigert A, Kronenberger B, Marschalek R, Piiper A. Hypoxia Causes Downregulation of Dicer in Hepatocellular Carcinoma, Which Is Required for Upregulation of Hypoxia-Inducible Factor 1α and Epithelial-Mesenchymal Transition. Clin Cancer Res. 2017;23:3896-3905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 7. | Lin CW, Wang LK, Wang SP, Chang YL, Wu YY, Chen HY, Hsiao TH, Lai WY, Lu HH, Chang YH, Yang SC, Lin MW, Chen CY, Hong TM, Yang PC. Daxx inhibits hypoxia-induced lung cancer cell metastasis by suppressing the HIF-1α/HDAC1/Slug axis. Nat Commun. 2016;7:13867. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 8. | Mak P, Leav I, Pursell B, Bae D, Yang X, Taglienti CA, Gouvin LM, Sharma VM, Mercurio AM. ERbeta impedes prostate cancer EMT by destabilizing HIF-1alpha and inhibiting VEGF-mediated snail nuclear localization: implications for Gleason grading. Cancer Cell. 2010;17:319-332. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 316] [Cited by in RCA: 316] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 9. | Yang MH, Wu MZ, Chiou SH, Chen PM, Chang SY, Liu CJ, Teng SC, Wu KJ. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 973] [Cited by in RCA: 1092] [Article Influence: 60.7] [Reference Citation Analysis (0)] |
| 10. | Joseph JV, Conroy S, Pavlov K, Sontakke P, Tomar T, Eggens-Meijer E, Balasubramaniyan V, Wagemakers M, den Dunnen WF, Kruyt FA. Hypoxia enhances migration and invasion in glioblastoma by promoting a mesenchymal shift mediated by the HIF1α-ZEB1 axis. Cancer Lett. 2015;359:107-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 249] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 11. | Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 808] [Cited by in RCA: 1088] [Article Influence: 98.9] [Reference Citation Analysis (0)] |
| 12. | Robins H, Li Y, Padgett RW. Incorporating structure to predict microRNA targets. Proc Natl Acad Sci U S A. 2005;102:4006-4009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 179] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 13. | Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, Singh B, Franklin JL, Wang J, Hu H, Wei T, Yang M, Yeatman TJ, Lee E, Saito-Diaz K, Hinger S, Patton JG, Chung CH, Emmrich S, Klusmann JH, Fan D, Coffey RJ. lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat Med. 2017;23:1331-1341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 343] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 14. | Zhuang J, Shen L, Yang L, Huang X, Lu Q, Cui Y, Zheng X, Zhao X, Zhang D, Huang R, Guo H, Yan J. TGFβ1 Promotes Gemcitabine Resistance through Regulating the LncRNA-LET/NF90/miR-145 Signaling Axis in Bladder Cancer. Theranostics. 2017;7:3053-3067. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 132] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 15. | Chang HY, Ye SP, Pan SL, Kuo TT, Liu BC, Chen YL, Huang TC. Overexpression of miR-194 Reverses HMGA2-driven Signatures in Colorectal Cancer. Theranostics. 2017;7:3889-3900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 35] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 16. | Lv C, Li F, Li X, Tian Y, Zhang Y, Sheng X, Song Y, Meng Q, Yuan S, Luan L, Andl T, Feng X, Jiao B, Xu M, Plikus MV, Dai X, Lengner C, Cui W, Ren F, Shuai J, Millar SE, Yu Z. MiR-31 promotes mammary stem cell expansion and breast tumorigenesis by suppressing Wnt signaling antagonists. Nat Commun. 2017;8:1036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 101] [Cited by in RCA: 148] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 17. | Zhang WC, Chin TM, Yang H, Nga ME, Lunny DP, Lim EK, Sun LL, Pang YH, Leow YN, Malusay SR, Lim PX, Lee JZ, Tan BJ, Shyh-Chang N, Lim EH, Lim WT, Tan DS, Tan EH, Tai BC, Soo RA, Tam WL, Lim B. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun. 2016;7:11702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 160] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 18. | Jiang X, Hu C, Arnovitz S, Bugno J, Yu M, Zuo Z, Chen P, Huang H, Ulrich B, Gurbuxani S, Weng H, Strong J, Wang Y, Li Y, Salat J, Li S, Elkahloun AG, Yang Y, Neilly MB, Larson RA, Le Beau MM, Herold T, Bohlander SK, Liu PP, Zhang J, Li Z, He C, Jin J, Hong S, Chen J. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat Commun. 2016;7:11452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 119] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 19. | Ye G, Huang K, Yu J, Zhao L, Zhu X, Yang Q, Li W, Jiang Y, Zhuang B, Liu H, Shen Z, Wang D, Yan L, Zhang L, Zhou H, Hu Y, Deng H, Liu H, Li G, Qi X. MicroRNA-647 Targets SRF-MYH9 Axis to Suppress Invasion and Metastasis of Gastric Cancer. Theranostics. 2017;7:3338-3353. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 84] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 20. | Kang M, Shi J, Peng N, He S. MicroRNA-211 promotes non-small-cell lung cancer proliferation and invasion by targeting MxA. Onco Targets Ther. 2017;10:5667-5675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Yang Y, Ahn YH, Gibbons DL, Zang Y, Lin W, Thilaganathan N, Alvarez CA, Moreira DC, Creighton CJ, Gregory PA, Goodall GJ, Kurie JM. The Notch ligand Jagged2 promotes lung adenocarcinoma metastasis through a miR-200-dependent pathway in mice. J Clin Invest. 2011;121:1373-1385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 176] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 22. | Davalos V, Moutinho C, Villanueva A, Boque R, Silva P, Carneiro F, Esteller M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2012;31:2062-2074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 269] [Cited by in RCA: 294] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 23. | Ahn YH, Gibbons DL, Chakravarti D, Creighton CJ, Rizvi ZH, Adams HP, Pertsemlidis A, Gregory PA, Wright JA, Goodall GJ, Flores ER, Kurie JM. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J Clin Invest. 2012;122:3170-3183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 135] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 24. | Celià-Terrassa T, Liu DD, Choudhury A, Hang X, Wei Y, Zamalloa J, Alfaro-Aco R, Chakrabarti R, Jiang YZ, Koh BI, Smith HA, DeCoste C, Li JJ, Shao ZM, Kang Y. Normal and cancerous mammary stem cells evade interferon-induced constraint through the miR-199a-LCOR axis. Nat Cell Biol. 2017;19:711-723. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 91] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 25. | Chen SY, Teng SC, Cheng TH, Wu KJ. miR-1236 regulates hypoxia-induced epithelial-mesenchymal transition and cell migration/invasion through repressing SENP1 and HDAC3. Cancer Lett. 2016;378:59-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 26. | He M, Zhan M, Chen W, Xu S, Long M, Shen H, Shi Y, Liu Q, Mohan M, Wang J. MiR-143-5p Deficiency Triggers EMT and Metastasis by Targeting HIF-1α in Gallbladder Cancer. Cell Physiol Biochem. 2017;42:2078-2092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 27. | Li H, Rokavec M, Jiang L, Horst D, Hermeking H. Antagonistic Effects of p53 and HIF1A on microRNA-34a Regulation of PPP1R11 and STAT3 and Hypoxia-induced Epithelial to Mesenchymal Transition in Colorectal Cancer Cells. Gastroenterology. 2017;153:505-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 121] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 28. | Wang X, Yu M, Zhao K, He M, Ge W, Sun Y, Wang Y, Sun H, Hu Y. Upregulation of MiR-205 under hypoxia promotes epithelial-mesenchymal transition by targeting ASPP2. Cell Death Dis. 2016;7:e2517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 29. | Dou C, Liu Z, Xu M, Jia Y, Wang Y, Li Q, Yang W, Zheng X, Tu K, Liu Q. miR-187-3p inhibits the metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting S100A4. Cancer Lett. 2016;381:380-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 91] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 30. | Zhou P, Jiang W, Wu L, Chang R, Wu K, Wang Z. miR-301a is a candidate oncogene that targets the homeobox gene Gax in human hepatocellular carcinoma. Dig Dis Sci. 2012;57:1171-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 60] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 31. | Lu Z, Li Y, Takwi A, Li B, Zhang J, Conklin DJ, Young KH, Martin R, Li Y. miR-301a as an NF-κB activator in pancreatic cancer cells. EMBO J. 2011;30:57-67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 198] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 32. | Kawano M, Tanaka K, Itonaga I, Iwasaki T, Tsumura H. MicroRNA-301a promotes cell proliferation via PTEN targeting in Ewing's sarcoma cells. Int J Oncol. 2016;48:1531-1540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | Wang M, Li C, Yu B, Su L, Li J, Ju J, Yu Y, Gu Q, Zhu Z, Liu B. Overexpressed miR-301a promotes cell proliferation and invasion by targeting RUNX3 in gastric cancer. J Gastroenterol. 2013;48:1023-1033. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 34. | Cui L, Li Y, Lv X, Li J, Wang X, Lei Z, Li X. Expression of MicroRNA-301a and its Functional Roles in Malignant Melanoma. Cell Physiol Biochem. 2016;40:230-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 35. | Xia X, Zhang K, Cen G, Jiang T, Cao J, Huang K, Huang C, Zhao Q, Qiu Z. MicroRNA-301a-3p promotes pancreatic cancer progression via negative regulation of SMAD4. Oncotarget. 2015;6:21046-21063. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 36. | Barbieri CE, Pietenpol JA. p63 and epithelial biology. Exp Cell Res. 2006;312:695-706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 96] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 37. | Zhang Y, Yan W, Chen X. P63 regulates tubular formation via epithelial-to-mesenchymal transition. Oncogene. 2014;33:1548-1557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 38. | Shi W, Gerster K, Alajez NM, Tsang J, Waldron L, Pintilie M, Hui AB, Sykes J, P'ng C, Miller N, McCready D, Fyles A, Liu FF. MicroRNA-301 mediates proliferation and invasion in human breast cancer. Cancer Res. 2011;71:2926-2937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 206] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 39. | Yue X, Cao D, Lan F, Pan Q, Xia T, Yu H. MiR-301a is activated by the Wnt/β-catenin pathway and promotes glioma cell invasion by suppressing SEPT7. Neuro Oncol. 2016;18:1288-1296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 40. | Cao HJ, Fang Y, Zhang X, Chen WJ, Zhou WP, Wang H, Wang LB, Wu JM. Tumor metastasis and the reciprocal regulation of heparanase gene expression by nuclear factor kappa B in human gastric carcinoma tissue. World J Gastroenterol. 2005;11:903-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 16] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 41. | Chen S, Chen JZ, Zhang JQ, Chen HX, Yan ML, Huang L, Tian YF, Chen YL, Wang YD. Hypoxia induces TWIST-activated epithelial-mesenchymal transition and proliferation of pancreatic cancer cells in vitro and in nude mice. Cancer Lett. 2016;383:73-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 76] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 42. | Cho KH, Jeong KJ, Shin SC, Kang J, Park CG, Lee HY. STAT3 mediates TGF-β1-induced TWIST1 expression and prostate cancer invasion. Cancer Lett. 2013;336:167-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 89] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 43. | Gonfloni S, Caputo V, Iannizzotto V. P63 in health and cancer. Int J Dev Biol. 2015;59:87-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 44. | Oh JE, Kim RH, Shin KH, Park NH, Kang MK. DeltaNp63α protein triggers epithelial-mesenchymal transition and confers stem cell properties in normal human keratinocytes. J Biol Chem. 2011;286:38757-38767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 51] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 45. | Amelio I, Melino G. The p53 family and the hypoxia-inducible factors (HIFs): determinants of cancer progression. Trends Biochem Sci. 2015;40:425-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 123] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 46. | Montagner M, Enzo E, Forcato M, Zanconato F, Parenti A, Rampazzo E, Basso G, Leo G, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature. 2012;487:380-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 212] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Unsolicited Manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Tomizawa M S-Editor: Ma YJ L-Editor: Wang TQ E-Editor: Ma YJ