Published online Aug 7, 2019. doi: 10.3748/wjg.v25.i29.3972
Peer-review started: April 28, 2019
First decision: May 24, 2019
Revised: June 7, 2019
Accepted: June 25, 2019
Article in press: June 26, 2019
Published online: August 7, 2019
Processing time: 102 Days and 23.1 Hours
Colorectal cancer (CRC) is the third most prevalent malignancy and has the fourth highest global cancer mortality rate. Early diagnosis and prompt medical attention can improve quality of life and the prognosis of CRC patients. Accumulating evidence reveals that long non-coding RNAs (lncRNAs) function as oncogenes or anti-oncogenes, as well as biomarkers in various cancers.
To investigate the levels and molecular mechanism of the lncRNA maternally expressed gene 3 (MEG3) in CRC.
The levels of lncRNA MEG3 in CRC tissue, serum and cell line samples were explored via qRT-PCR. The relationship between MEG3 levels and clinicopathological features in CRC was investigated. The diagnostic and prognostic values of serum MEG3 levels were analyzed with ROC curves and Kaplan‑Meier survival curves, respectively.
Significant decreased levels of MEG3 existed in CRC tissue, cell lines and serum. CRC patients with down-regulated serum MEG3 levels had larger tumor sizes, and advanced clinical stages. The sensitivity and specificity of serum MEG3 levels in CRC detection was 0.667 and 0.875, respectively. Tumor size, T stages, and serum MEG3 levels are indie factors that produce an effect on CRC patients' prognosis. Kaplan‑Meier survival curves suggested that CRC patients with high levels of MEG3 had a remarkably better overall survival rate.
LncRNA MEG3 is down-regulated in CRC, and regulates cell functions by targeting adenosine deaminase’s effect on RNA 1 in CRC.
Core tip: Long non-coding RNA (LncRNA) maternally expressed gene 3 (MEG3) is down-regulated in tissue, cell lines and serum. Colorectal cancer (CRC) patients with down-regulated serum MEG3 levels were had larger tumor sizes, and advanced clinical stages. LncRNA MEG3 functions as a diagnostic and prognostic marker in CRC. LncRNA MEG3 promotes cell proliferation and induced apoptosis in CRC. The effect of adenosine deaminase on RNA 1 may be the target of lncRNA MEG3 in CRC.
- Citation: Wang W, Xie Y, Chen F, Liu X, Zhong LL, Wang HQ, Li QC. LncRNA MEG3 acts a biomarker and regulates cell functions by targeting ADAR1 in colorectal cancer. World J Gastroenterol 2019; 25(29): 3972-3984
- URL: https://www.wjgnet.com/1007-9327/full/v25/i29/3972.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i29.3972
Colorectal cancer (CRC), which is the third most prevalent malignancy and causes the fourth highest cancer mortality rate globally, is a severe disease and significant threat to human health[1]. Worldwide, there are an estimated 1.2 million new cases and 0.6 million deaths from CRC each year[2]. Nevertheless, the clinical methods for screening, diagnosing, and treating CRC are limited. For example, early course screening of CRC, such as fecal occult blood tests and carcinoembryonic antigens, are often affected by other health disorders and factors, resulting in low specificity and sensitivity, as well as inaccurate clinical diagnoses. At malignancy stages, the tumor node metastasis (TNM) staging system can be used to describe the stage of malignancy and assess patient prognosis. However, TNM suffers from difficulties in invasion and specimen collection, limiting the application of this method for the prediction and prognosis of CRC. Previous studies have highlighted the need for better clinical methods, revealing that nearly 90% of patients with early-stage CRC were alive five years following prognosis, while 14% of patients with advanced-stage CRC were alive five years later. However, only 39.6% of CRC cases are diagnosed at early stages[3,4], thus indicating a need for clinical markers with high sensitivity and specificity that can be used in the early detection and prognosis of CRC.
Typically consisting of more than 200 nucleotides, long non-coding RNAs (lncRNAs) are referred to as endogenous cellular RNAs[5]. LncRNAs lack classically-defined open reading frames, and thus have limited or no protein-coding potential[6-8], yet a large number of aberrant lncRNAs are known to be involved in carcinogenesis, dissemination, and metastasis[9,10]. Moreover, investigations into the roles of lncRNAs as oncogenes or anti-oncogene factors, and their potential as serum biomarkers for the detection of various cancers including CRC, have increasingly garnered the attention of experts[11-14]. For example, CRC tissues up-regulate the lncRNA SPINT1-AS1, which is associated with partial clinical features (e.g., regional lymph node metastasis, distant metastasis, and shorter relapse-free survival time), suggesting that SPINT1-AS1 is a prognostic marker for CRC[3].
One of the best-studied lncRNAs called maternally expressed gene 3 (MEG3) was reported to be aberrantly expressed in multiple types of malignancies, such as hemangioma, glioma, cervical cancer, and bladder cancer[15-18]. In addition, MEG3 was recently found to act as an anti-oncogene in CRC, specifically by targeting the clusterin in CRC cells to inhibit cell proliferation and migration[19]. Another study revealed that down-regulation of MEG3 in CRC cells activates sphingosine kinase 1, accelerating cell proliferation and suppressing transforming growth factor β1-mediated apoptosis[11]. However, the potential biomarker applications and the mechanisms underlying the roles of MEG3 in CRC require further investigation. Here, we aimed to determine the levels of MEG3 in CRC tissue, cell lines, and serum, further exploring the roles of MEG3 in cellular processes. We uncovered the diagnostic and prognostic value of MEG3 in CRC.
Forty-two CRC tissue specimens and corresponding normal tissues were collected from the First Affiliated Hospital of China Medical University. Among the 42 patients in this study, none of them received any therapy pre-operation. All patients diagnosed with CRC had been verified via pathological methods, and patients with other tumors or diseases were excluded from our study. Fresh surgical specimens were processed within half an hour, and then submerged in RNAlater reagent (Qiagen) for half an hour. After that, CRC tissue specimens were stored in liquid nitrogen until RNA extraction.
Serum samples were obtained from 126 CRC patients, as well as 48 healthy control individuals. Serum from 35 paired pre- and post-operative CRC individuals was also collected. All venous blood was disposed within 1 h after extraction. Briefly, serum samples were isolated by centrifugation (1200 × g, 10 min) followed by another centrifugation (10000 × g, 10 min) to discard residual cellular debris. All centri-fugations were performed at 4 °C. Similarly, serum samples were stored in liquid nitrogen until RNA extraction.
Our research was managed under the Ethics Committee of the First Affiliated Hospital of China Medical University. Every participant in our study provided full consent. Table 1 shows the clinical characteristics of the CRC patients.
| Clinical features | Cases | Serum MEG3 expression | P value |
| Age in yr | 0. 956 | ||
| ≥ 60 | 54 | 0.678 ± 0.013 | |
| < 60 | 72 | 0.679± 0.043 | |
| Gender | 0.400 | ||
| Male | 85 | 0.665 ± 0.032 | |
| Female | 41 | 0.696 ± 0.038 | |
| Tumor site | 0.843 | ||
| Colon | 58 | 0.671 ± 0.044 | |
| Rectum | 68 | 0.676 ± 0.032 | |
| Tumor size in cm | 0.001 | ||
| ≥ 5 | 43 | 0.748 ± 0.031 | |
| < 5 | 83 | 0.567 ± 0.039 | |
| pT, TNM | 0.098 | ||
| T1+T2 | 93 | 0.617 ± 0.025 | |
| T3+T4 | 33 | 0.693 ± 0.019 | |
| Lymph node metastasis | 0.236 | ||
| Positive | 71 | 0.717 ± 0.046 | |
| Negative | 55 | 0.662 ± 0.034 | |
| Clinical stage, TNM | |||
| III | 72 | 0.867 ± 0.049 | 0.001 |
| I+II | 54 | 0.562 ± 0.018 | |
| Pathological differentiation | |||
| Well/moderate | 59 | 0.665 ± 0.050 | 0.322 |
| Poor | 67 | 0.710 ± 0.032 |
Human CRC cell lines (HCT-116 and HT29) and normal colorectal mucosa epithelial cells (NCM460) were obtained from the American Type Culture Collection. All cells were cultivated with DMEM (Gibco; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific), 5% antibiotics (penicillin and streptomycin sulfates), and 20mM glutamine. HCT-116, HT29, and NCM460 cells were maintained in an incubator (37 °C, 5% CO2). For cell transfection, a pCDNA3.1 vector containing the MEG3 sequence was purchased from Invitrogen. HCT-116 and HT29 cells were pre-seeded in 6-well plates and cultivated until they reached 50-60% confluency. After that, HCT-116 and HT29 cells were transfected with pCDNA-MEG3 or empty vector using the X-tremeGENE HP DNA transfection reagent (Roche).
For RNA isolation of tissue specimens, TRIzol reagent (Invitrogen) was used according to the manufacturer’s procedures. For RNA isolation of serum samples, the miRNeasy Serum/Plasma Kit (Qiagen) was used. The quantity of RNA in all samples was measured with a NanoDrop 2000c (Thermo Fisher Scientific), and any RNA samples that exhibited an optical density ratio (260/280) of less than 1.8 or over 2.0 were excluded from further experiments. RNA samples were either stored in liquid nitrogen or subsequently used for cDNA synthesis.
cDNA was synthesized via the PrimeScript RT Master Mix (Takara Biotechnology) with 0.1 μg of sample-derived RNA, then used for RT-qPCR to detect and quantify the levels of lncRNA MEG3 in tissues and serum. This assay was conducted on the Roche Lightcycler 480 Real-Time PCR system (Roche Diagnostics) with SYBR-Green PCR master mix (Roche). After normalization to GADPH, changes in lncRNA MEG3 expression were calculated using the 2−ΔCt method. The primers used were as follows: MEG3 forward, 5′-CTGCCCATCTACACCTCACG -3′ and reverse, 5′-CTCTCCGCCGTCTGCGCTAGGGGCT- 3′; GAPDH forward, 5′-AGCCACAT CGCTCAGACAC-3′ and reverse, 5′-GCCCAATACGACCAAATCC-3′.
A CCK-8 kit (US Everbright, lnc.) was used for evaluating cell proliferative capacity. After transfection, we plated CRC cells (HCT-116 and HT29) in a 96-well plate, and measured the optical density (OD) every 24 h according to the manufacturer’s protocol. Before each detection, the CCK-8 kit (10 μL) was added to each well, and an enzyme immunoassay instrument (Bio Rad Laboratories) was used for value readings.
Annexin V (Invitrogen; Thermo Fisher Scientific, Inc.) was used to assess the apoptosis rate of HCT-116 and HT29 cells. After transfection with pCDNA-MEG3 or empty vector, CRC cells (HCT-116 and HT29) were grown to 100% confluency, then collected via centrifugation and washed with phosphate buffered saline. After resuspending in binding buffer, the cells were stained with 5 μL Annexin V. Fifteen min later, apoptotic cells were detected by flow cytometry (EPICS XL 4; Beckman Coulter, Inc.).
After rinsing with pre-cooled phosphate buffered saline, total protein in CRC (HCT-116 and HT29) cells was extracted with RIPA buffer (Thermo Fisher Scientific) and a protease/phosphatase inhibitor cocktail (Roche). Protein concentration was evaluated with a Bio-Rad BCA assay system, and equal amounts of protein were loaded into each lane of a polyacrylamide gel. Proteins were separated via SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore, Billerica), and blocked with 5% skim milk. Membranes were stained with primary antibodies [anti-adenosine deaminase acting on RNA 1 (ADAR1) 1:500, Proteintech; anti-GAPDH 1:1000, Proteintech] for 16 h, then rinsed with TBST (3 × 10 min), and stained with secondary antibody (HRP conjugated goat anti-rabbit 1:1000, Sigma) at room temperature for 60 min. An enhanced chemiluminescence kit (Merck Millipore) and Kodak film (Kodak) was used to detect the blots. ADAR1 protein abundance was normalized to GAPDH.
All reported statistics were visualized with SPSS 21.0 and Graphpad Prism 7 software. Significance values were determined by student’s t-test. Associations between MEG3 levels and clinicopathological features were analyzed with a Chi-square test and Fisher’s exact test, and receiver operating characteristic (ROC) curves were drawn to evaluate the value of serum MEG3 for CRC detection. The Kaplan-Meier method was applied for assessing the value of serum MEG3 in the prognosis of CRC. A P value < 0.05 is indicated as a significant difference.
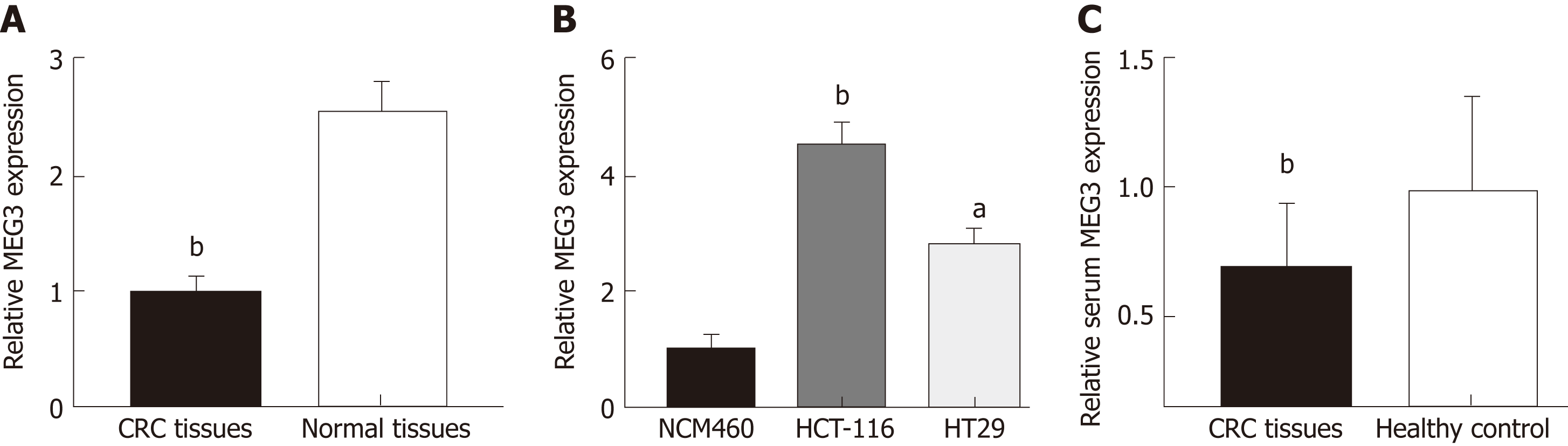
MEG3 levels in CRC tissue, serum, and cell lines were first detected by qRT-PCR. Neither age nor gender appeared to affect MEG3 expression when comparing between the CRC group and healthy group (Table 2). As expected, lncRNA MEG3 expression was significantly decreased in CRC tissues versus corresponding colorectal tissue (P < 0.01; Figure 1A). Results from cell lines agreed with these findings, and revealed significant down-regulation of MEG3 in CRC cell lines compared to NCM460 cells (P < 0.01 for HCT-116, P < 0.05 for HT29; Figure 1B). Moreover, significant down-regulation of MEG3 existed in serum samples of CRC patients versus the NC (P < 0.05; Figure 1C).
| Tissue samples, n = 42 | Serum samples | ||
| Colorectal cancer, n = 126 | Healthy controls, n = 48 | ||
| Age | |||
| mean ± SD | 56.8 ± 6.5 | 56.9 ± 5.9 | 56.3 ± 6.4 |
| Median (range) | 57 (44-73) | 58 (38-74) | 58 (38-73) |
| Gender | |||
| Male | 27 | 85 | 33 |
| Female | 15 | 41 | 15 |
| Tumor site | |||
| Colon | 17 | 58 | |
| Rectum | 25 | 68 | |
| Tumor size in cm | |||
| ≥ 5 | 12 | 43 | |
| < 5 | 30 | 83 | |
| pT, TNM | |||
| T1+T2 | 33 | 93 | |
| T3+T4 | 9 | 33 | |
| Lymph node metastasis | |||
| Positive | 18 | 71 | |
| Negative | 22 | 55 | |
| Clinical stage, TNM | |||
| III | 13 | 72 | |
| I+II | 29 | 54 | |
| Pathological differentiation | 26 | 59 | |
| Well/moderate Poor | 16 | 67 | |
| high vs low | |||
| Lymph node metastasis, positive vs negative | 0.988 | ||
| 0.225 | |||
| Clinical stage of | 0.440 | ||
| TNM, III vs I + II | 2.789 | ||
| Pathological differentiation, well/moderate vs poor | |||
| Serum lncRNA | |||
| MEG3 expression, | |||
We next investigated the relationship between MEG3 levels and clinical pathological features of CRC patients. In our study, the 126 CRC patients were divided into two equal-sized groups of high and low MEG3 expression, with a cut-off point at the median MEG3 level (63 high serum MEG3 level, and 63 low serum MEG3 level). The comparisons displayed in Table 1 reveal that no statistically significant associations were uncovered between the serum MEG3 concentration and age (P = 0.956), gender (P = 0.400), tumor site (P = 0.843), T stage (P = 0.098), lymph node metastasis (P = 0.236) or pathological differentiation (P = 0.322). However, serum MEG3 levels clearly correlated with tumor size (P = 0.001) and clinical stage (P = 0.001). Thus, our results demonstrate that CRC patients with down-regulated serum MEG3 levels are more prone to developing larger tumors and reaching advanced clinical stages.
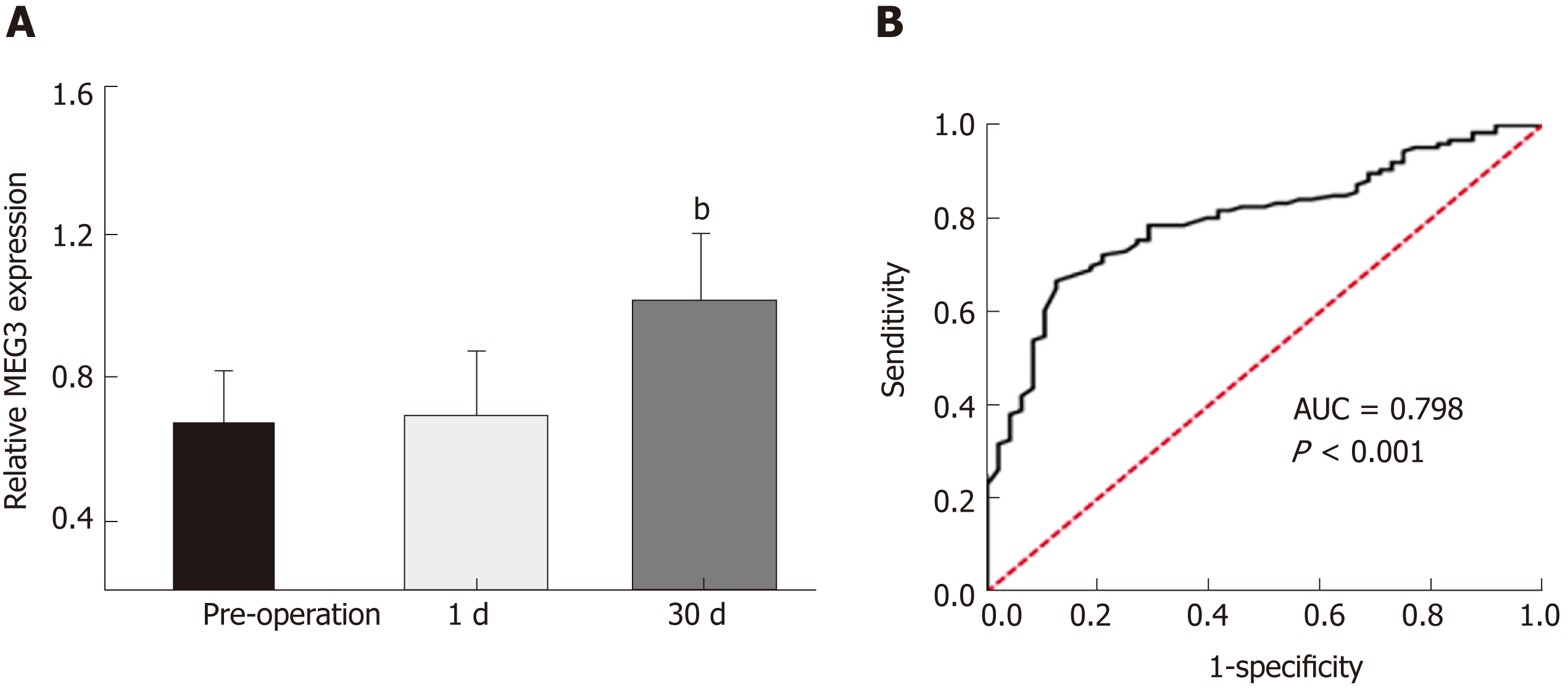
In order to assess the value of measuring MEG3 expression in diagnosing CRC, the ROC method was applied. As shown in Figure 2A, no significant difference was observed between the serum MEG3 levels in samples 24 h post-operation and serum MEG3 levels in the preoperative samples (P > 0.05). The opposite result was obtained in the samples 1 mo after surgery, which revealed that the serum MEG3 levels were significantly elevated compared to the preoperative samples (P < 0.01). A ROC curve was applied to the results, which demonstrated that the sensitivity and specificity of serum MEG3 levels in CRC detection was 0.667 and 0.875, respectively [area under the curve (AUC) 0.798; 95% confidence interval (CI) 0.730-0.866, P < 0.001; Figure 2B]. Overall, lncRNA MEG3 is a reliable marker for CRC diagnosis.
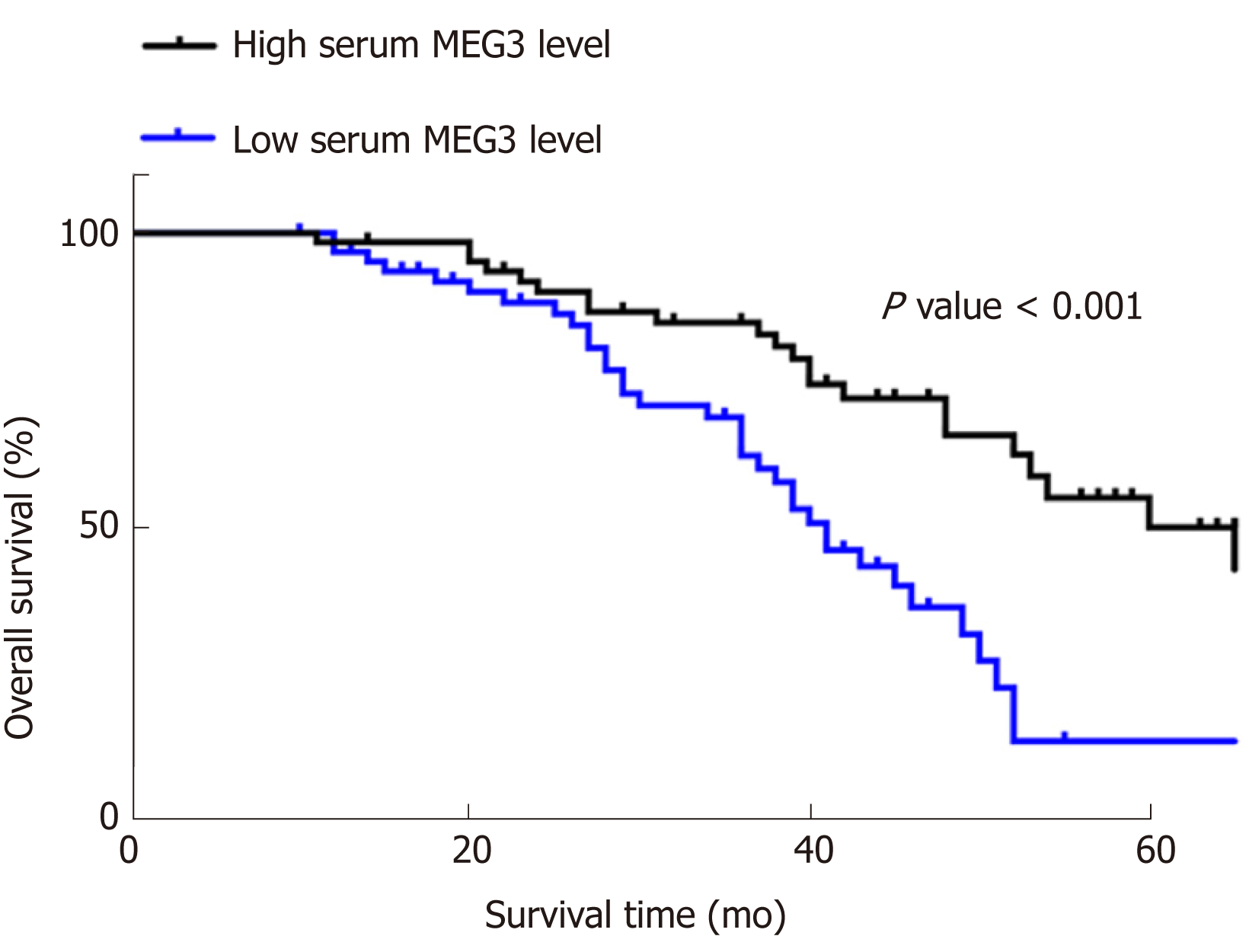
Kaplan‑Meier survival curves and Cox proportional hazard regression analyses were applied to assess the prognostic value of lncRNA MEG3 in CRC. Data from univariate analysis showed that tumor size [hazard ratio (HR) = 0.576, 95%CI = 0.342-0.968, P = 0.037], T stage (HR = 0.572, 95%CI = 0.122-0.414, P = 0.044), clinical stage (HR = 0.225, 95%CI = 0.122-0.414, P = 0.001), pathological differentiation (HR = 0.440, 95%CI = 0.258-0.752, P = 0.003) and serum MEG3 levels (HR = 2.789, 95%CI = 1.615-4.816, P = 0.001) are indeed factors that affect a CRC patient’s prognosis. In multivariate analysis, tumor size (HR = 0.436, 95%CI = 0.236-0.806, P = 0.008), T stage (HR = 0.039, 95%CI = 0.012-0.124, P = 0.001), and serum MEG3 levels (HR = 0.173, 95%CI = 0.063-0.480, P = 0.002) were identified as factors affecting CRC patient prognosis (Table 3). Kaplan‑Meier survival curves (Figure 3) were plotted, and suggested that CRC patients with high levels of MEG3 had remarkably better overall survival (OS) rates (P < 0.001). These data demonstrate that lncRNA MEG3 is a reliable marker for CRC prognosis.
| Variables | Univariate analysis | Multivariate analysis | ||||
| HR | 95%CI | P value | HR | 95%CI | P value | |
| Age in yr, ≥ 60 vs < 60 | 0.806 | 0.481-1.352 | 0.414 | - - - | - - - | - - - |
| Gender, male vs female | 1.265 | 0.757-2.178 | 0.396 | - - - | - - - | - - - |
| Tumor site, colon vs rectum | 0.681 | 0.405-1.143 | 0.146 | - - - | - - - | - - - |
| Tumor size in cm, ≥ 5 vs < 5 | 0.576 | 0.342-0.968 | 0.037 | 0.436 | 0.236-0.806 | 0.008 |
| pT of TNM, T1 + T2 vs T3 + T4 | 0.572 | 0.332-0.986 | 0.044 | 0.942 | 0.477-1.861 | 0.863 |
| Lymph node metastasis, positive vs negative | 0.988 | 0.591-1.651 | 0.964 | - - - | - - - | - - - |
| Clinical stage of TNM, III vs I + II | 0.225 | 0.122-0.414 | 0.001 | 0.039 | 0.012-0.124 | 0.001 |
| Pathological differentiation, well/moderate vs poor | 0.440 | 0.258-0.752 | 0.003 | 0.641 | 0.324-1.267 | 0.201 |
| Serum lncRNA MEG3 expression, high vs low | 2.789 | 1.615-4.816 | 0.001 | 0.173 | 0.063-0.480 | 0.002 |
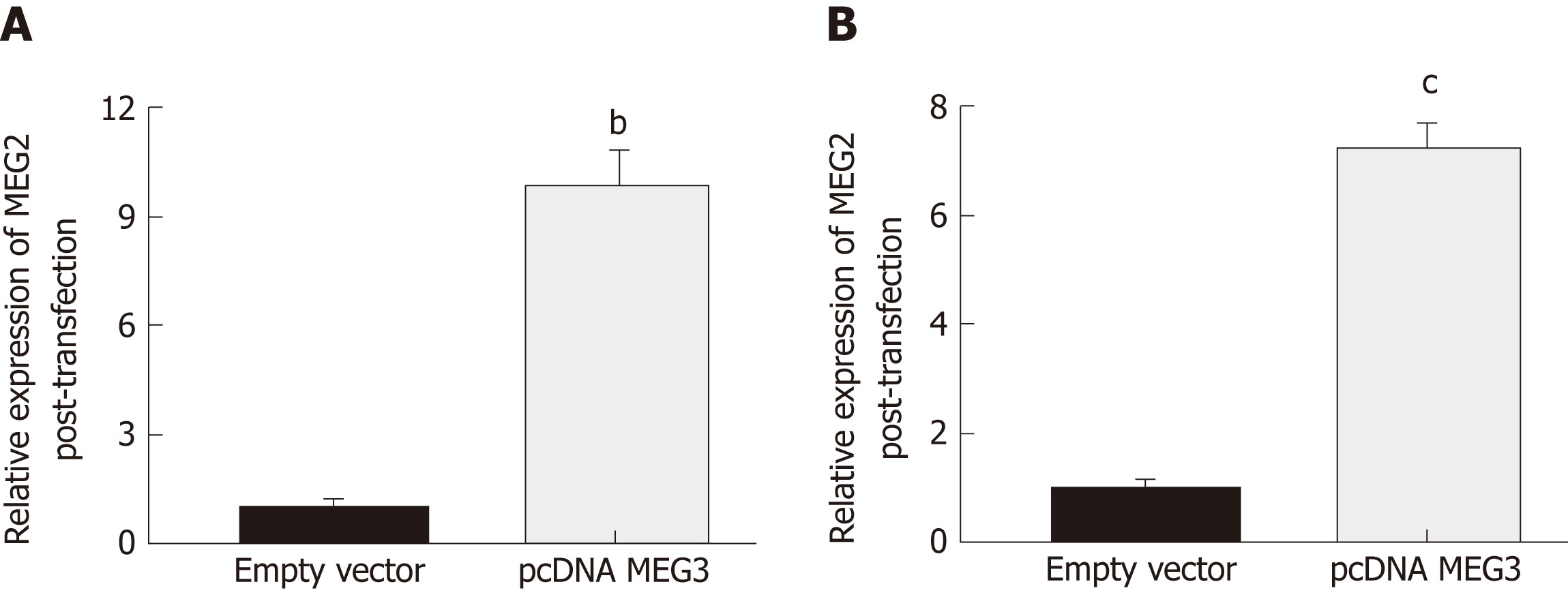
Changes in lncRNA MEG3 expression in two different CRC cell lines (HCT-116 and HT29) after transfection with pcDNA MEG3 were measured with RT-qPCR. The amount of lncRNA MEG3 present in transfected HCT-116 cells was 9.87 times higher than the control group (P < 0.01; Figure 4A), and MEG3 expression in similarly transfected HT-29 cells was increased 7.32 times compared to cells transfected with an empty vector (P < 0.001; Figure 4B).
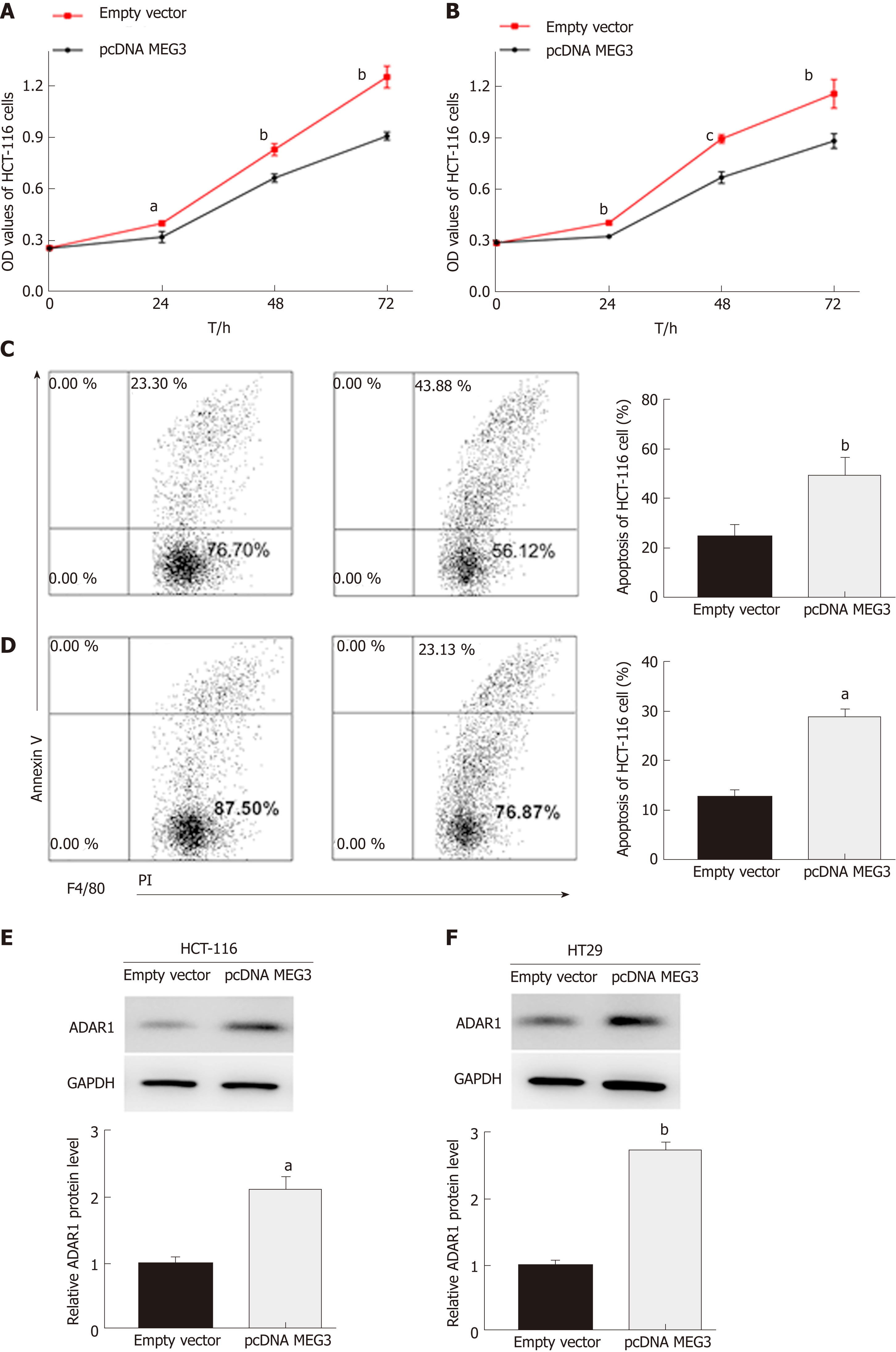
In our study, CCK-8 and flow cytometry assays were conducted to evaluate cell proliferation and apoptosis, respectively. As expected, the CCK-8 assay revealed that up-regulation of MEG3 could suppress cell proliferation both in HCT-116 (P < 0.05; Figure 5A) and HT29 (P < 0.01; Figure 5B) cell lines compared with the control group. Flow cytometry showed that elevated expression of MEG3 induced a significant amount of cell apoptosis compared with the control group [(HCT-116, P < 0.01; Figure 5C) (HT-29, P < 0.05; Figure 5D)].
Western blotting was conducted on cells that were transfected to overexpress MEG3. From these blots, we were able to elucidate that ADAR1 protein expression in both HCT-116 (P < 0.01; Figure 5E) and HT29 (P < 0.01; Figure 5F) cells was elevated when MEG3 was expressed at higher levels. MEG3 levels were up-regulated in HCT-116 cells.
The carcinogenesis and progression of CRC is an extremely complex process involving multiple steps of cellular reprogramming. In recent years, despite many efforts to improve clinical treatments, the prognosis of CRC patients has not significantly improved. Early detection of CRC is one of the main prerequisites for satisfactory therapy, and can help significantly improve a patient’s chances for survival. Thus, it is an issue of particular significance and urgency to explore and understand the mechanism behind the carcinogenesis and progression of CRC, so as to unearth relevant biomarkers for diagnosis and prognosis, as well as enable the accurate monitoring of CRC progression.
Previous studies have revealed that lncRNAs exert significant effects on numerous biological processes, such as neoplastic angiopoiesis, cell migration, and drug resistance[20,21]. An estimated 1 × 105 lncRNAs have been identified thus far, but it is difficult to determine the precise number due to many factors, such as varying tissues and stages[22]. Despite extensive research conducted on the role of lncRNAs, fewer than 2% of lncRNA species have been ascribed to a particular biological role[23]. Moreover, how lncRNAs exert their functions at the molecular level, and how lncRNAs are targeted to specific genomic sites remains elusive[24]. In addition to the important functions of lncRNA in tissues and cells, the function of lncRNAs in serum has also interested many researchers, who agree that lncRNAs in serum have great potential for clinical use in diagnosing tumors and predicting patient prognosis.
Previous studies have demonstrated that dysregulated lncRNA MEG3 was widespread in malignancies. It was found that MEG3 levels were reduced in the tissues of prostate cancer patients, and that the MEG3 inhibitory role in various cellular functions (invasion, proliferation and migration) relied on regulating the miR-9-5p/QKI-5 axis[25]. Decreased levels of MEG3 were found in bladder cancer tissues, and increased levels of MEG3 could hinder the ability of BC cell migration and invasion. Furthermore, bladder cancer cells with up-regulated MEG3 were sensitized to cisplatin, which is a drug used for bladder cancer chemotherapy[26]. Likewise, MEG3 levels were significantly reduced in liver cancer tissues, and hepatoma cell proliferation and invasion could be promoted by down-regulation of MEG3[27]. In gastric cancer, MEG3 was lowly expressed in tumor tissue, and up-regulation of MEG3 suppressed cancer cell proliferation, metastasis, and p53 levels[28]. MEG3 also plays a fatal role in kidney cancer, chronic myeloid leukemia, thyroid carcinoma, and endometrial carcinoma[29-32]. In our study, MEG3 was significantly reduced in CRC tumor tissue, serum, and cell lines. Up-regulation of MEG3 by transfection inhibited CRC cellular proliferation and induced apoptosis.
ADAR1, a significant member of the ADAR protein family, has been reported to participate in multiple biological functions, such as cell proliferation and apoptosis[33,34]. There are two major isoforms of ADAR1: An interferon-inducible ADAR1 p150 that contains both the Za and Zb Z-DNA-binding domains and a constitutive ADAR1 p110 that lacks the N-terminal Za Z-DNA-binding domain[35]. ADAR1 has emerged as a biomarker in numerous solid tumors, including gastric cancer and esophageal cancer[36,37]. In our study, StarBase 3.0 (http://starbase .sysu.edu.cn/index.php) was used, and ADAR1 was found to be co-expressed with MEG3 in CRC. Thus, we assume that ADAR1 is a potential target regulated by MEG3 in CRC cells. Western blotting was then performed to test our hypothesis. As expected, the cells overexpressing MEG3 exhibited increased ADAR1 expression, thus implicating ADAR1 as a target of MEG3 in CRC cells. These results agree with the StarBase data.
Previous studies demonstrated that MEG3 acts as a diagnostic biomarker in malignancies. In bladder cancer, the AUC of the three lncRNA panel (MEG3, SNHG16 and MALAT1) in detecting bladder cancer was 0.828, and the diagnostic performances of the lncRNA panel for Ta, T1, and T2-T4 were 0.778, 0.805, and 0.880, respectively. This thus identified a three lncRNA panel for use in diagnosing bladder cancer[38]. In our study, MEG3 could discriminate between CRC patients and healthy controls with a sensitivity and specificity of 0.667 and 0.875, respectively, thus demonstrating that lncRNA MEG3 was a reliable marker for CRC diagnosis.
Previous studies demonstrated that lncRNA MEG3 acts as a prognostic biomarker in malignancies. Bioinformatics analysis revealed that high MEG3 levels were a suitable prognostic factor for patients with lung cancer, especially in younger patients (≤ 60 years old), indicating MEG3 as a promising prognostic factor in lung cancer[39]. In breast cancer, MEG3 levels are closely related to the TNM stage differentiation grade, and lymph node metastasis. MEG3 levels were an insusceptible undesirable factor of prognosis [5-year OS and 5-year progression-free survival (PFS)] in BC patients. Breast cancer patients that had MEG3 levels would experience a poor prognosis (poor OS and PFS)[40]. In osteosarcoma, MEG3 levels were particularly lower in tumor tissues, and correlated with both clinical stage and metastasis. The results of Kaplan-Meier analysis suggested that patients with high MEG3 levels generally live longer. These data therefore demonstrate that MEG3 acts as a prognostic biomarker in osteosarcoma[38]. In our study, we found that MEG3 expression was indeed a factor affecting the prognosis of CRC patients, and that patients with high levels of MEG3 had a remarkably higher OS rate, which demonstrated that lncRNA MEG3 was a good marker for the prognosis of CRC. In conclusion, lncRNA MEG3 is down-regulated in CRC, and regulates cell function by targeting ADAR1 in CRC.
Among common types of gastrointestinal malignancies, colorectal cancer (CRC) has seen a dramatic increase in annual global incidence rate. Many recent studies have demonstrated the molecular mechanisms involved in the transcriptional regulation in CRC, and shown that long non-coding RNAs (lncRNAs) play an irreplaceable role in the initiation and progression of CRC, such as maintaining cell growth, evasion of apoptosis, promotion of invasion and metastasis, stemness maintenance and EMT.
To identify more biomarkers for the diagnosis and treatment of CRC.
To investigate the underlying mechanisms of lncRNA maternally expressed gene 3 (MEG3) in CRC.
LncRNA MEG3 expression was observed by qRT-PCR assays on CRC tissue, cell lines and serum. Clinicopathological characteristics were collected, arranged and combined with expression analysis of CRC to evaluate the functions of lncRNA MEG3. Cell function assays were performed to explore the functions of MEG3 in CRC cell lines. Moreover, western blots were performed to explore the targeted regulation by MEG3 in CRC cell lines.
We found that levels of LncRNA MEG3 decreased in CRC tissues, cell lines and serum, and exhibited a significant negative relation with tumor size, TNM stage, and lymph node metastasis. Cell experiments showed that MEG3 levels declined during CR cell line proliferation and invasion, and that ADAR1 may be the target regulated by lncRNA MEG3 in CRC cells. Importantly, CRC patients with higher lncRNA MEG3 levels have a better overall survival rate.
Our study demonstrated that lncRNA MEG3 can significantly inhibit cell growth, migration and invasion of gastric cancer. Furthermore, it can work through ADAR1. Therefore, our study provides some molecular mechanism and two new biomarkers for CRC.
In the future, research may reveal the important role of lncRNA MEG3 that enhances the sensitivity of CRC detection, and further develop its application for anti-cancer treatments. The identification of the lncRNA MEG3/ADAR1 molecular axis may further explain the underlying mechanism.
| 1. | Liu HN, Liu TT, Wu H, Chen YJ, Tseng YJ, Yao C, Weng SQ, Dong L, Shen XZ. Serum microRNA signatures and metabolomics have high diagnostic value in colorectal cancer using two novel methods. Cancer Sci. 2018;109:1185-1194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 2. | Conev NV, Donev IS, Konsoulova-Kirova AA, Chervenkov TG, Kashlov JK, Ivanov KD. Serum expression levels of miR-17, miR-21, and miR-92 as potential biomarkers for recurrence after adjuvant chemotherapy in colon cancer patients. Biosci Trends. 2015;9:393-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 3. | Liao G, Zhang X, Li Z, Lan S, Huang M, Huang W. Renal-type clear cell carcinoma of prostate: A case report and review of literature. Indian J Pathol Microbiol. 2018;61:431-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 4. | Nikolaou S, Qiu S, Fiorentino F, Rasheed S, Tekkis P, Kontovounisios C. Systematic review of blood diagnostic markers in colorectal cancer. Tech Coloproctol. 2018;22:481-498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 100] [Article Influence: 12.5] [Reference Citation Analysis (4)] |
| 5. | Zhao W, Song M, Zhang J, Kuerban M, Wang H. Combined identification of long non-coding RNA CCAT1 and HOTAIR in serum as an effective screening for colorectal carcinoma. Int J Clin Exp Pathol. 2015;8:14131-14140. [PubMed] |
| 6. | Tong YS, Wang XW, Zhou XL, Liu ZH, Yang TX, Shi WH, Xie HW, Lv J, Wu QQ, Cao XF. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 221] [Cited by in RCA: 273] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 7. | van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150:1170-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 205] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 8. | Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4004] [Cited by in RCA: 3982] [Article Influence: 234.2] [Reference Citation Analysis (0)] |
| 9. | Zhang L, Zhou Y, Huang T, Cheng AS, Yu J, Kang W, To KF. The Interplay of LncRNA-H19 and Its Binding Partners in Physiological Process and Gastric Carcinogenesis. Int J Mol Sci. 2017;18:pii: E450. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 10. | Li J, Meng H, Bai Y, Wang K. Regulation of lncRNA and Its Role in Cancer Metastasis. Oncol Res. 2016;23:205-217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 246] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 11. | Kong H, Wu Y, Zhu M, Zhai C, Qian J, Gao X, Wang S, Hou Y, Lu S, Zhu H. Long non-coding RNAs: Novel prognostic biomarkers for liver metastases in patients with early stage colorectal cancer. Oncotarget. 2016;7:50428-50436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 12. | Chen S, Wu DD, Sang XB, Wang LL, Zong ZH, Sun KX, Liu BL, Zhao Y. The lncRNA HULC functions as an oncogene by targeting ATG7 and ITGB1 in epithelial ovarian carcinoma. Cell Death Dis. 2017;8:e3118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 100] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 13. | Wang J, Yang K, Yuan W, Gao Z. Determination of Serum Exosomal H19 as a Noninvasive Biomarker for Bladder Cancer Diagnosis and Prognosis. Med Sci Monit. 2018;24:9307-9316. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 98] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 14. | Zhang S, Du L, Wang L, Jiang X, Zhan Y, Li J, Yan K, Duan W, Zhao Y, Wang L, Wang Y, Shi Y, Wang C. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med. 2019;23:1396-1405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 106] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 15. | Chen X, Qu J. Long non-coding RNA MEG3 suppresses survival, migration, and invasion of cervical cancer. Onco Targets Ther. 2018;11:4999-5007. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 16. | Dai Y, Wan Y, Qiu M, Wang S, Pan C, Wang Y, Ou J. lncRNA MEG3 Suppresses the Tumorigenesis of Hemangioma by Sponging miR-494 and Regulating PTEN/ PI3K/AKT Pathway. Cell Physiol Biochem. 2018;51:2872-2886. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 17. | Gong X, Huang M. Long non-coding RNA MEG3 promotes the proliferation of glioma cells through targeting Wnt/β-catenin signal pathway. Cancer Gene Ther. 2017;24:381-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 43] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 18. | Liu G, Zhao X, Zhou J, Cheng X, Ye Z, Ji Z. Long non-coding RNA MEG3 suppresses the development of bladder urothelial carcinoma by regulating miR-96 and TPM1. Cancer Biol Ther. 2018;1-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 44] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 19. | Zhu Y, Chen P, Gao Y, Ta N, Zhang Y, Cai J, Zhao Y, Liu S, Zheng J. MEG3 Activated by Vitamin D Inhibits Colorectal Cancer Cells Proliferation and Migration via Regulating Clusterin. EBioMedicine. 2018;30:148-157. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 63] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 20. | Liu Z, Wu C, Xie N, Wang P. Long non-coding RNA MEG3 inhibits the proliferation and metastasis of oral squamous cell carcinoma by regulating the WNT/β-catenin signaling pathway. Oncol Lett. 2017;14:4053-4058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 63] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 21. | Xu DH, Chi GN, Zhao CH, Li DY. Long noncoding RNA MEG3 inhibits proliferation and migration but induces autophagy by regulation of Sirt7 and PI3K/AKT/mTOR pathway in glioma cells. J Cell Biochem. 2018;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 22. | Sherpa C, Rausch JW, Le Grice SF. Structural characterization of maternally expressed gene 3 RNA reveals conserved motifs and potential sites of interaction with polycomb repressive complex 2. Nucleic Acids Res. 2018;46:10432-10447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 38] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | Hon CC, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJ, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J, Lizio M, Kawaji H, Kasukawa T, Itoh M, Burroughs AM, Noma S, Djebali S, Alam T, Medvedeva YA, Testa AC, Lipovich L, Yip CW, Abugessaisa I, Mendez M, Hasegawa A, Tang D, Lassmann T, Heutink P, Babina M, Wells CA, Kojima S, Nakamura Y, Suzuki H, Daub CO, de Hoon MJ, Arner E, Hayashizaki Y, Carninci P, Forrest AR. An atlas of human long non-coding RNAs with accurate 5' ends. Nature. 2017;543:199-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 673] [Cited by in RCA: 756] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 24. | Blank-Giwojna A, Postepska-Igielska A, Grummt I. lncRNA KHPS1 Activates a Poised Enhancer by Triplex-Dependent Recruitment of Epigenomic Regulators. Cell Rep. 2019;26:2904-2915.e4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 82] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 25. | Wu M, Huang Y, Chen T, Wang W, Yang S, Ye Z, Xi X. LncRNA MEG3 inhibits the progression of prostate cancer by modulating miR-9-5p/QKI-5 axis. J Cell Mol Med. 2019;23:29-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 26. | Feng SQ, Zhang XY, Fan HT, Sun QJ, Zhang M. Up-regulation of LncRNA MEG3 inhibits cell migration and invasion and enhances cisplatin chemosensitivity in bladder cancer cells. Neoplasma. 2018;65:925-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 47] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 27. | Sun Y, Cao FL, Qu LL, Wang ZM, Liu XY. MEG3 promotes liver cancer by activating PI3K/AKT pathway through regulating AP1G1. Eur Rev Med Pharmacol Sci. 2019;23:1459-1467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 28. | Wei GH, Wang X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:3850-3856. [PubMed] |
| 29. | He H, Dai J, Zhuo R, Zhao J, Wang H, Sun F, Zhu Y, Xu D. Study on the mechanism behind lncRNA MEG3 affecting clear cell renal cell carcinoma by regulating miR-7/RASL11B signaling. J Cell Physiol. 2018;233:9503-9515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 30. | Li Z, Yang L, Liu X, Nie Z, Luo J. Long noncoding RNA MEG3 inhibits proliferation of chronic myeloid leukemia cells by sponging microRNA21. Biomed Pharmacother. 2018;104:181-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 31. | Liu Y, Yue P, Zhou T, Zhang F, Wang H, Chen X. LncRNA MEG3 enhances 131I sensitivity in thyroid carcinoma via sponging miR-182. Biomed Pharmacother. 2018;105:1232-1239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 32. | Sun KX, Wu DD, Chen S, Zhao Y, Zong ZH. LncRNA MEG3 inhibit endometrial carcinoma tumorigenesis and progression through PI3K pathway. Apoptosis. 2017;22:1543-1552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 33. | Nemlich Y, Greenberg E, Ortenberg R, Besser MJ, Barshack I, Jacob-Hirsch J, Jacoby E, Eyal E, Rivkin L, Prieto VG, Chakravarti N, Duncan LM, Kallenberg DM, Galun E, Bennett DC, Amariglio N, Bar-Eli M, Schachter J, Rechavi G, Markel G. MicroRNA-mediated loss of ADAR1 in metastatic melanoma promotes tumor growth. J Clin Invest. 2013;123:2703-2718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 148] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 34. | Nakano M, Fukami T, Gotoh S, Nakajima M. A-to-I RNA Editing Up-regulates Human Dihydrofolate Reductase in Breast Cancer. J Biol Chem. 2017;292:4873-4884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 100] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 35. | Shevchenko G, Morris KV. All I's on the RADAR: Role of ADAR in gene regulation. FEBS Lett. 2018;592:2860-2873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 35] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 36. | Takeda S, Shigeyasu K, Okugawa Y, Yoshida K, Mori Y, Yano S, Noma K, Umeda Y, Kondo Y, Kishimoto H, Teraishi F, Nagasaka T, Tazawa H, Kagawa S, Fujiwara T, Goel A. Activation of AZIN1 RNA editing is a novel mechanism that promotes invasive potential of cancer-associated fibroblasts in colorectal cancer. Cancer Lett. 2019;444:127-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 53] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 37. | Qiao JJ, Chan TH, Qin YR, Chen L. ADAR1: A promising new biomarker for esophageal squamous cell carcinoma? Expert Rev Anticancer Ther. 2014;14:865-868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 38. | Tian ZZ, Guo XJ, Zhao YM, Fang Y. Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma. Int J Clin Exp Pathol. 2015;8:15138-15142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 39. | Zhang Z, Liu T, Wang K, Qu X, Pang Z, Liu S, Liu Q, Du J. Down-regulation of long non-coding RNA MEG3 indicates an unfavorable prognosis in non-small cell lung cancer: Evidence from the GEO database. Gene. 2017;630:49-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 26] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 40. | Zhang JJ, Guo SH, Jia BQ. Down-regulation of long non-coding RNA MEG3 serves as an unfavorable risk factor for survival of patients with breast cancer. Eur Rev Med Pharmacol Sci. 2016;20:5143-5147. [PubMed] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See:
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Katuchova J, Tanabe S S-Editor: Yan JP L-Editor: Filipodia E-Editor: Ma YJ