Published online Apr 7, 2017. doi: 10.3748/wjg.v23.i13.2318
Peer-review started: November 17, 2016
First decision: December 19, 2016
Revised: January 5, 2017
Accepted: March 2, 2017
Article in press: March 2, 2017
Published online: April 7, 2017
Processing time: 141 Days and 21.6 Hours
To explore the functional role of cullin 4A (CUL4A), a core subunit of E3 ubiquitin ligase, in perihilar cholangiocarcinoma (PHCC).
The expression of CUL4A in PHCC cell lines was evaluated by Western blot and quantitative reverse transcription-polymerase chain reaction. Immunohistochemistry (IHC) was adopted to investigate the relationship between CUL4A expression and clinicopathological characteristics of PHCC. Univariate analysis and multivariate regression analysis were performed to analyze the risk factors related to overall survival (OS) and progression-free survival (PFS) of PHCC patients. Wound healing, Transwell and Matrigel assays were utilized to explore the function of CUL4A in PHCC metastasis. Furthermore, expression of epithelial to mesenchymal transition (EMT) markers was verified in cells with CUL4A knockdown or overexpression. The relationship between CUL4A expression and E-cadherin expression was also analyzed by IHC assay. Finally, the role of ZEB1 in regulating CUL4A mediated PHCC was detected by IHC, Western blot, Transwell and Matrigel assays.
CUL4A overexpression was detected in PHCC cell lines and clinical specimens. Clinicopathological analysis revealed a close correlation between CUL4A overexpression and tumour differentiation, T, N and TNM stages in PHCC. Kaplan-Meier analysis revealed that high CUL4A expression was correlated with poor OS and PFS of PHCC patients. Univariate analysis identified the following four parameters as risk factors related to OS rate of PHCC: T, N, TNM stages and high CUL4A expression; as well as three related to PFS: N stage, TNM stage and high CUL4A expression. Further multivariate logistic regression analysis identified high CUL4A expression as the only independent prognostic factor for PHCC. Moreover, CUL4A silencing in PHCC cell lines dramatically inhibited metastasis and the EMT. Conversely, CUL4A overexpression promoted these processes. Mechanistically, ZEB1 was discovered to regulate the function of CUL4A in promoting the EMT and metastasis.
CUL4A is an independent prognostic factor for PHCC, and it can promote the EMT by regulating ZEB1 expression. CUL4A may be a potential therapeutic target for PHCC.
Core tip: Cullin 4A (CUL4A), a core subunit of E3 ubiquitin ligase, was confirmed to promote the metastasis and the epithelial to mesenchymal transition (EMT) in perihilar cholangiocarcinoma (PHCC) in the present study. High CUL4A expression was revealed to be correlated with poor overall survival (OS) and progression-free survival (PFS) of PHCC patients. CUL4A expression was detected to be the only independent risk factor for OS and PFS in PHCC. Mechanistically, ZEB1 was verified to mediate the function of CUL4A in regulating PHCC metastasis and the EMT.
- Citation: Zhang TJ, Xue D, Zhang CD, Zhang ZD, Liu QR, Wang JQ. Cullin 4A is associated with epithelial to mesenchymal transition and poor prognosis in perihilar cholangiocarcinoma. World J Gastroenterol 2017; 23(13): 2318-2329
- URL: https://www.wjgnet.com/1007-9327/full/v23/i13/2318.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i13.2318
Cholangiocarcinoma (CCA), a primary epithelial cancer originating from the hepatobiliary tract and exhibiting characteristics of cholangiocyte differentiation, is classified into intrahepatic CCA, perihilar CCA (PHCC) and distal CCA based on its anatomical location[1]. During the past 2-3 decades, the incidence and mortality rates of PHCC, accounting for 50% of CCA cases[2], have been steadily increasing[3]. The vast majority of patients diagnosed with PHCC usually present with an advanced disease that develops without an identifiable aetiology. Curative surgical resection, the only effective treatment to achieve a possible cure in PHCC, can be achieved in less than 19%-75% of patients[4]. Even worse, currently available chemotherapy and radiotherapy regimens are usually unresponsive to advanced PHCC. When left untreated, the survival time is only 6 to 12 mo[5]. Therefore, to improve the prognosis of PHCC patients, it is essential to further elucidate the underlying molecular mechanisms of PHCC progression and identify more effective prognostic biomarkers that might also serve as potential targets for PHCC treatment.
Cullin 4A (CUL4A), a core subunit of E3 ubiquitin ligase, encodes a member of the cullin ubiquitin-ligase family, which includes seven members in mammals (Cul1, 2, 3, 4A, 4B, 5, and 7)[6]. CUL4A controls diverse cellular processes, including proliferation, differentiation, apoptosis and metastasis. Additionally, CUL4A has been shown to mediate the ubiquitin-dependent proteolysis of several well-defined tumour suppressor genes, such as p21, p27, DDB2, and p53, indicating that CUL4A may be a potential oncogene[7,8]. Furthermore, CUL4A amplification and overexpression have been found in several human cancers, including primary human breast cancer[9], esophageal squamous cell carcinoma[10], adrenocortical carcinoma[11], childhood medulloblastoma[12], hepatocellular carcinoma[13], and primary malignant pleural mesothelioma[14]. Furthermore, strong CUL4 expression is an independent prognostic factor for survival in node-negative breast cancer, prostate cancer and intrahepatic CCA[15-17]. However, to the best of our knowledge, its functional role in PHCC has not been previously investigated.
Metastasis, the final stage of solid cancer progression, is responsible for the vast majority of cancer-related deaths. However, the exact mechanism for cancer metastasis remains unclear. The biology of most PHCC cases is aggressive, involving rapid invasion and metastases. The epithelial to mesenchymal transition (EMT) is a cellular process during which epithelial cells lose their polarity and cell-cell adhesion, undergo changes in cell shape and cytoskeletal organization and acquire mesenchymal characteristics[18]. It has been confirmed that the EMT is closely involved in increasing cell migratory and invasive properties, inducing stem cell properties, preventing apoptosis and senescence, and resisting chemotherapy and immunotherapy[19-21]. Therefore, the EMT process is now considered a potential target in tumour metastasis prevention and treatment.
With respect to these notions, the present study investigated the expression of CUL4A and its clinical significance in PHCC. The role and mechanisms by which CUL4A promotes the EMT and PHCC cell motility were also explored. The results not only further elucidated the metastasis mechanism of PHCC but also identified that CUL4A may serve as a potential target for PHCC treatment.
Primary PHCC tissues (n = 78), 12 of which had matched adjacent normal bile duct tissues, were obtained from PHCC patients who underwent curative surgery at the Department of General Surgery, The People’s Hospital of Binzhou, China, between 2003 and 2010. Informed consent was obtained from each patient, and the study protocol was approved by the Clinical Research Ethics Committee of The People’s Hospital of Binzhou. The diagnosis of PHCC was confirmed by routine pathology. Pathologic tumour-node-metastasis staging was classified according to the 7th staging classification of American Joint Committee on Cancer criteria. Patient latest follow-up was terminated in May 2016. Overall survival (OS) was defined as the interval between the date of surgery and death or when censored at the latest date. Progression-free survival (PFS) was defined as the time from the date of surgery to the date of disease relapse/progression or the date of death or when censored at the latest date. Patients died from causes other than PHCC were censored.
Immunohistochemistry (IHC) analysis of CUL4A, ZEB1 and E-Cadherin expression in clinical samples was performed as previously described[22]. Scoring was performed by two pathologists who were blinded to the pathology and clinical features in The People’s Hospital of Binzhou. The scoring system includes the extent and intensity of staining. Briefly, the intensity was assigned a score of 0, 1, 2, or 3, representing negative, weak, moderate, or strong expression, respectively. Whereas, the extent was assigned a score of 0, 1, 2, or 3, representing negative, < 10%, 10%-50%, and > 50% of cells stained. The overall quantitation of the IHC score was obtained by multiplying the average intensity and score of five different high-power fields (× 400 magnification).
The human normal biliary epithelial cell line, HIBEpiC, and human PHCC cell lines, QBC939 and FRH0201, were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). HIBEpiC was grown in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. QBC939 and FRH0201 were cultured in RPMI-1640 medium, supplemented with 10% FBS and 1% penicillin/streptomycin in an atmosphere of 5% CO2 at 37 °C.
TRIzol reagent (15596-026, Invitrogen) was used to extract total RNA according to the manufacturer’s instructions, and 5 μg of total RNA was used for cDNA synthesis. Assays were performed in triplicate on an ABI PRISM 7900HT sequence detection system according to standard protocols as recommended by the manufacturer. Melting curve analysis was conducted to distinguish specific products from non-specific products and primer dimers. The GAPDH gene was amplified as an internal control in each sample.
Total cellular proteins were isolated using RIPA buffer (Thermo Fisher Scientific, United States) according to the manufacturers’ protocols. Aliquots (12 μg) of total protein were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis. The separated proteins were transferred to polyvinylidene difluoride membranes (Millipore). The membranes were blocked with 5% bovine serum albumin for 2 h and incubated with each primary antibody overnight at 4 °C. The signals from the primary antibody were amplified by HRP conjugated anti-mouse IgG or anti-rabbit IgG, and the bands were visualized with a FluorChem E system (Protein Simple, United States).
The shRNA sequences and cDNA of wild-type CUL4A gene were cloned into pLenti-vectors. The shRNA sequences of CUL4A were 5′-GATCCCCGGTTTATCCACGGTAAAGATTCAAGAGATCTTTACCGTGGATAAACCTTTTTGGAAA-3′ (forward) and 5′-AGCTTTTCCAAAAAGGTTTATCCACGGTAAAGATCTCTTGAATCTTTACCGTGGATAAACCGGG-3′ (reverse). Then, pLenti-shCUL4A and pLenti vectors were transfected into QBC939 cells and pLenti-CUL4A vector and pLenti vector were transfected into FRH0201 cells via lentiviral infection. For the lentiviral infection, lentiviral plasmid vectors including pLenti-vector, pLenti-shCUL4A and pLenti-CUL4A were co-transfected with packaging vectors and the viral particles were produced by 293T cells. Then, lentiviral stocks were concentrated using ultracentrifugation. Subsequently, PHCC cells (QBC939 and FRH0201 cells) were incubated with the infection medium mixed with polybrene (4 μg/mL) at a multiplicity of infection (MOI) of 20. Finally, puromycin (1-3 μg/mL) was used to select resistant cells. The QBC939 cell line with stable knockdown of CUL4A (QBC939-pLenti-shCUL4A), its corresponding control cell line (QBC939-pLenti-Vector), the FRH0201 cell line with stable CUL4A overexpression (FRH0201-pLenti-CUL4A) and its corresponding control cell line (FRH0201-pLenti-Vector) were established. The expression of CUL4A in these cell lines was examined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
siNC, siZEB1.1 and siZEB1.2 were purchased from GENEWIZ (Suzhou, China). Transfection was performed with Lipofectamine 2000 (Invitrogen, United States) according to the manufacturer’s instructions.
Cells were cultured in medium that was deprived of serum and grown to confluence in a 6-well plate. Then, a wound was made in the middle of a culture plate with a 200 μL sterile pipette tip. The wound healing process was photographed using phase-contrast microscopy at 0, 24 and 48 h. The quantification was performed by measuring the uncovered areas compared with the controls.
A Boyden chamber (8 μm pore size, BD Biosciences, United States) was used to test cell motility. In the Matrigel invasion assay, the Boyden chambers were coated with Matrigel (40 lg/well; BD Biosciences, Bedford, MA, United States) and incubated for at least 1 h at 37 °C before the cells were seeded. Cells (5 × 104) were suspended in 200 μL of serum-free medium and plated in the upper chamber, whereas 600 μL of medium with 10% FBS was added to the lower well. After 24 h of incubation, the cells were removed from the upper side of the chamber, and cells attached to the lower surface of the membrane were fixed with methanol for 30 min, and then stained with 10% Giemsa solution (Solarbo, United States). The statistical results of cell numbers per image field were obtained from three independent experiments that were averaged from five image fields.
To select the clinically essential cut-off scores for CUL4A, receiver operating characteristic (ROC) curve analysis was performed. Previous studies have verified the reproducibility of ROC curve analysis in determining a biologically or clinically relevant cutoff score[23,24]. The OS and PFS data were dichotomized according to death due to PHCC or censored (lost to follow-up, alive or death from other causes) for evaluation by the ROC curve analysis. Based on the cut-off score, the patients were categorized into the high CUL4A expression and low CUL4A expression groups. Tumours labelled with high CUL4A expression were those with scores equal to or higher than the cut-off value, while low CUL4A expression indicated those with scores below the cut-off value.
Statistical analyses were performed using SPSS 19.0 software (SPSS, Inc., Chicago, IL, United States). Data are shown as median ± SD. ROC analysis was utilized to get the optimal cutoff scores of CUL4A expression for survival analysis. Differences among variables were assessed by two-tailed Student’s t-test or one-way analysis of variance. Linear regression was tested by using the Spearman rank correlation. The Kaplan-Meier method was used to estimate survival, and the survival difference was compared using the log-rank test. P < 0.05 was considered to be statistically significant.
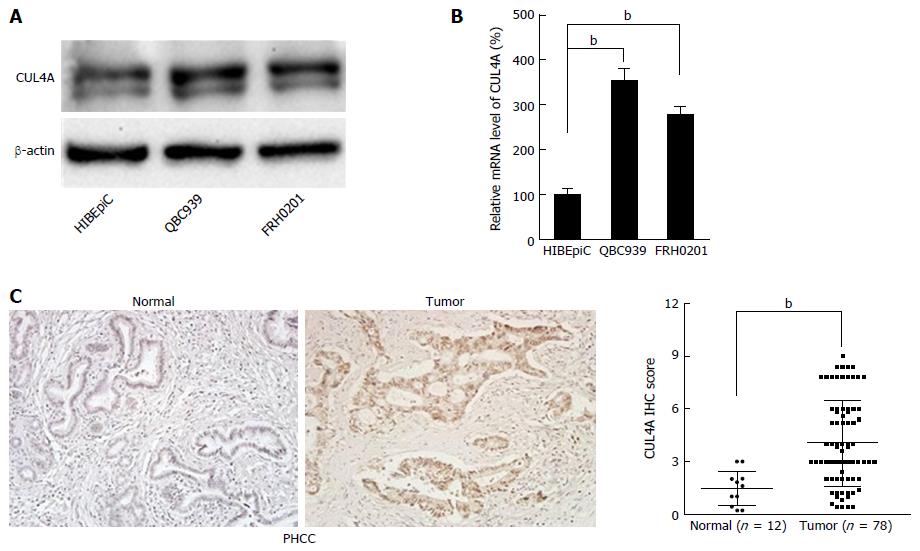
To explore the role of CUL4A in PHCC development, we evaluated the expression of CUL4A in various human PHCC cell lines. As shown in Figure 1A, elevated CUL4A expression was observed in PHCC cell lines compared to a human normal biliary epithelial cell line (HIBEpiC). This result was further verified by qRT-PCR in the studied cell lines (Figure 1B). To determine the clinical significance of CUL4A expression in PHCC, the relationship between CUL4A expression and clinicopathological features was analysed. Validation with IHC also determined that an increased CUL4A IHC score could be found in primary PHCC tumours relative to adjacent non-tumoural intrahepatic bile ducts (4.04 ± 2.45 vs 1.47 ± 0.96, P = 0.001) (Figure 1C). Additionally, statistical analyses revealed a strong correlation between the expression of CUL4A and tumour differentiation (P < 0.001), T stage, N stage (P < 0.001) and TNM stage (P < 0.001) (Table 1). PHCC patients with higher CUL4A expression had a higher tendency to have poorer differentiation and a higher incidence of advanced T, N and TNM stages. There were no statistically significant connections between CUL4A expression and other clinicopathological parameters, such as age, gender, tumour size, and M stage (P > 0.05, Table 1). These results indicated that CUL4A overexpression is positively correlated with PHCC progression.
| Parameter | n | CUL4A IHC score | P value |
| Age (yr) | 0.902 | ||
| < 65 | 54 | 4.07 ± 2.46 | |
| ≥ 65 | 24 | 3.99 ± 2.47 | |
| Gender | 0.606 | ||
| Male | 45 | 3.92 ± 2.34 | |
| Female | 33 | 4.21 ± 2.62 | |
| Tumour size (cm) | 0.330 | ||
| < 3 | 36 | 3.75 ± 2.55 | |
| ≥ 3 | 42 | 4.30 ± 2.36 | |
| Differentiation grade | < 0.001 | ||
| Well + moderate | 66 | 3.56 ± 2.30 | |
| Poor + undifferentiated | 12 | 6.68 ± 1.24 | |
| T stage | 0.001 | ||
| T1 + T2 | 63 | 3.62 ± 2.34 | |
| T3 + T4 | 15 | 5.81 ± 2.12 | |
| N stage | < 0.001 | ||
| N0 | 64 | 3.35 ± 2.08 | |
| N1 + N2 | 14 | 7.21 ± 1.13 | |
| M stage | 0.980 | ||
| M0 | 76 | 4.04 ± 2.46 | |
| M1 | 2 | 4.00 ± 2.83 | |
| TNM stage | < 0.001 | ||
| I + II | 54 | 3.14 ± 2.03 | |
| III + IV | 24 | 6.08 ± 2.07 |
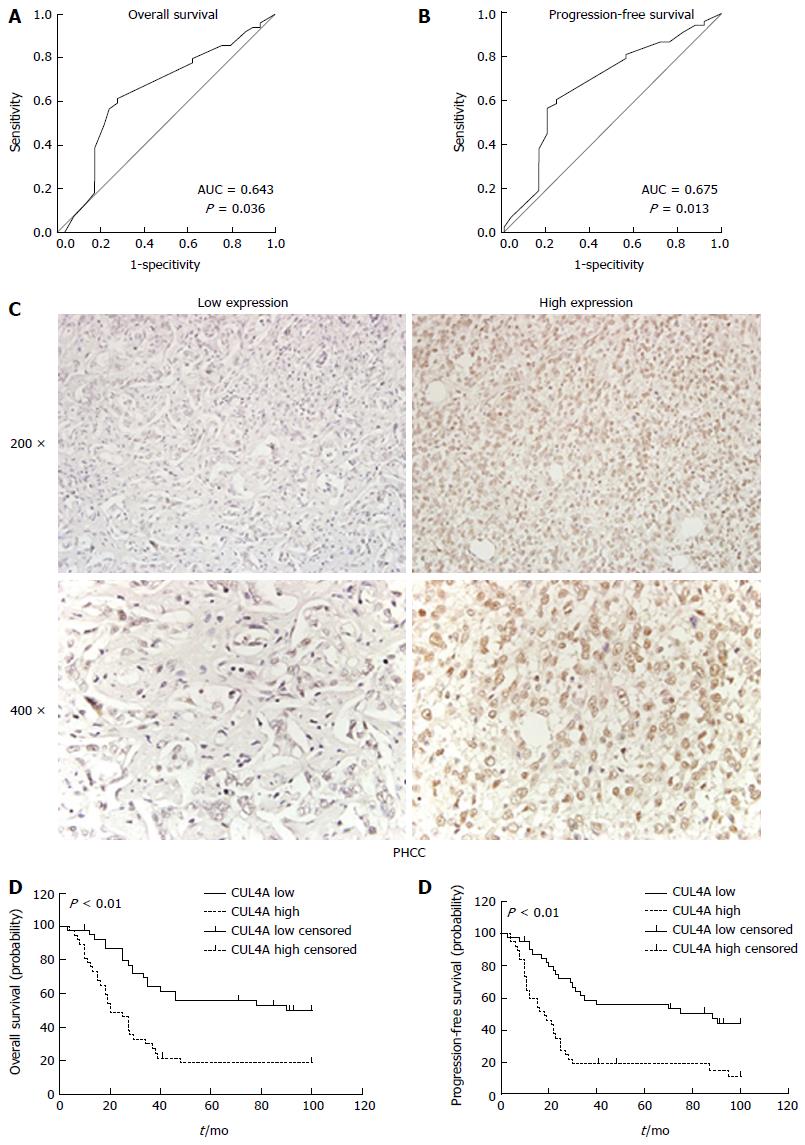
To better assess the expression of CUL4A in PHCC, we employed ROC curve analysis to define an optimal cut-off value for high CUL4A expression, based on the results of IHC evaluation. The ROC curves for the OS and PFS clearly illustrate the point on the curve closest to (0.0, 1.0), which maximizes both the sensitivity and specificity for the outcome (Figure 2A). Tumours with scores equal to or higher than the obtained cut-off value were considered to have high CUL4A expression, resulting in the highest number of tumours that were correctly classified as having or not having the positive clinical outcome. As a result, the cut-off score of the OS for high CUL4A expression is 3.30, and the cut-off score of the PFS for high CUL4A expression is 3.90. To increase the sensitivity of the cut-off score, one was defined as high CUL4A expression when the IHC score was not lower than 3.30 (Figure 2C). According to the obtained IHC score cutoff, the 78 PHCC patients were classified into two groups, the high CUL4A group (n = 38) and low CUL4A group (n = 40).
To evaluate the prognostic potential of CUL4A expression in PHCC, Kaplan-Meier analysis was performed, which showed that high CUL4A expression was associated with a poorer OS rate of PHCC patients (P < 0.001; Figure 2D and E). Moreover, high CULA4 expression was correlated with a lower PFS in PHCC patients (P < 0.001; Figure 3B). Univariate analysis identified the following four prognostic factors for OS: T stage (HR = 2.181, 95%CI: 1.129-4.213, P = 0.020), N stage (HR = 3.120, 95%CI: 1.613-6.036, P < 0.001), TNM stage (HR = 3.001, 95%CI: 1.671-5.387, P < 0.001) and CUL4A expression level (HR = 2.823, 95%CI: 1.577-5.053, P < 0.001); as well as three prognostic factors for FPS: N stage (HR = 2.917, 95%CI: 1.515-5.619, P = 0.001), TNM stage (HR = 2.729, 95%CI: 1.538-4.842, P = 0.001) and CUL4A expression level (HR = 2.964, 95%CI: 1.691-5.196, P < 0.001). Other clinicopathological features, such as gender and age, were not statistically significant prognostic factors (Table 2). Furthermore, multivariate analysis identified a high CUL4A expression level as the only independent prognostic factor for OS (HR = 2.117, 95%CI: 1.086-4.125, P = 0.028) and PFS (HR = 2.248, 95%CI: 1.240-4.446, P = 0.009) (Table 2).Taken together, these results suggested that CUL4A could serve as a potential predictive factor for recurrence and poor survival in PHCC patients.
| Parameter | Overall survival | Progression-free survival | ||||
| HR | 95%CI | P value | HR | 95%CI | P value | |
| Univariate analysis | ||||||
| Age: ≥ 65 yr vs < 65 yr | 1.218 | 0.676-2.193 | 0.512 | 1.097 | 0.616-1.954 | 0.753 |
| Gender: male vs female | 1.003 | 0.569-1.767 | 0.993 | 0.931 | 0.538-1.610 | 0.798 |
| Differentiation: well + moderate vs poor + undifferentiated | 0.432 | 0.171-1.092 | 0.076 | 0.461 | 0.196-1.083 | 0.075 |
| Tumour size: ≥ 3 cm vs < 3 cm | 1.049 | 0.598-1.839 | 0.869 | 1.008 | 0.586-1.733 | 0.977 |
| T stage: (T3 + T4) vs (T1 + T2) | 2.181 | 1.129-4.213 | 0.020 | 1.871 | 0.977-3.583 | 0.059 |
| N stage: N1 + N2 vs N0 | 3.120 | 1.613-6.036 | 0.001 | 2.917 | 1.515-5.619 | 0.001 |
| M stage: M1 vs M0 | 4.761 | 0.54-38.170 | 0.142 | 2.766 | 0.363-21.092 | 0.326 |
| TNM stage: (III + IV) vs (I + II) | 3.001 | 1.671-5.387 | < 0.001 | 2.729 | 1.538-4.842 | 0.001 |
| CUL4A: high vs low | 2.823 | 1.577-5.053 | < 0.001 | 2.964 | 1.691-5.196 | < 0.001 |
| Multivariate analysis | ||||||
| T stage: (T3 + T4) vs (T1 + T2) | 0.811 | 0.271-2.424 | 0.708 | - | - | - |
| N stage: N1 + N2 vs N0 | 0.969 | 0.305-3.081 | 0.957 | 1.106 | 0.428-2.857 | 0.836 |
| TNM stage: (III + IV) vs (I + II) | 2.483 | 0.632-9.752 | 0.193 | 1.699 | 0.735-3.928 | 0.215 |
| CUL4A: high vs low | 2.117 | 1.086-4.125 | 0.028 | 2.248 | 1.240-4.446 | 0.009 |
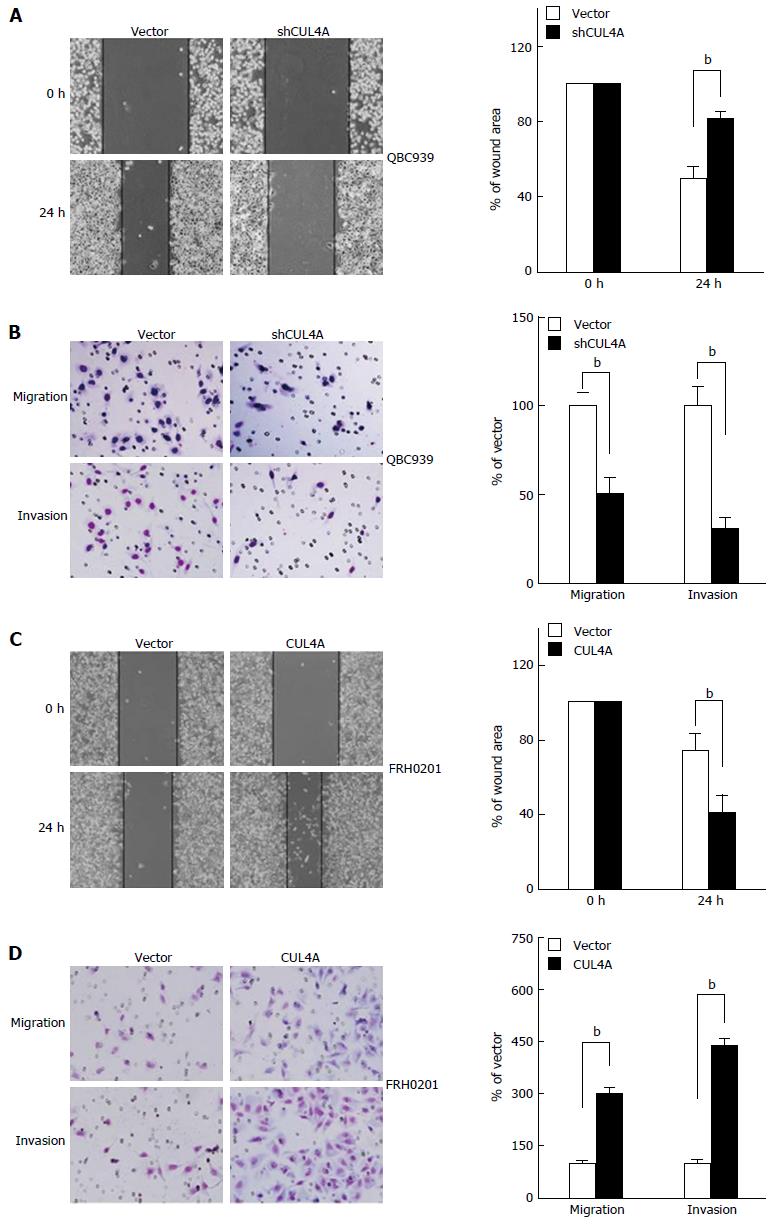
To further investigate the functional role of CUL4A in PHCC cell metastasis, a wound healing assay was first performed. As shown in Figure 3A, CUL4A depletion dramatically suppressed the migration of QBC939 cells. This result was also confirmed in a Transwell assay (Figure 3B, up). Furthermore, the invasiveness of QBC939 cells with CUL4A depletion was markedly reduced in a Matrigel assay (Figure 3B, down). Consistently, CUL4A ectopic expression significantly increased the migration and invasion of FRH0201 cells (Figure 3C and D). Collectively, CUL4A could promote the migration and invasion of PHCC cells.
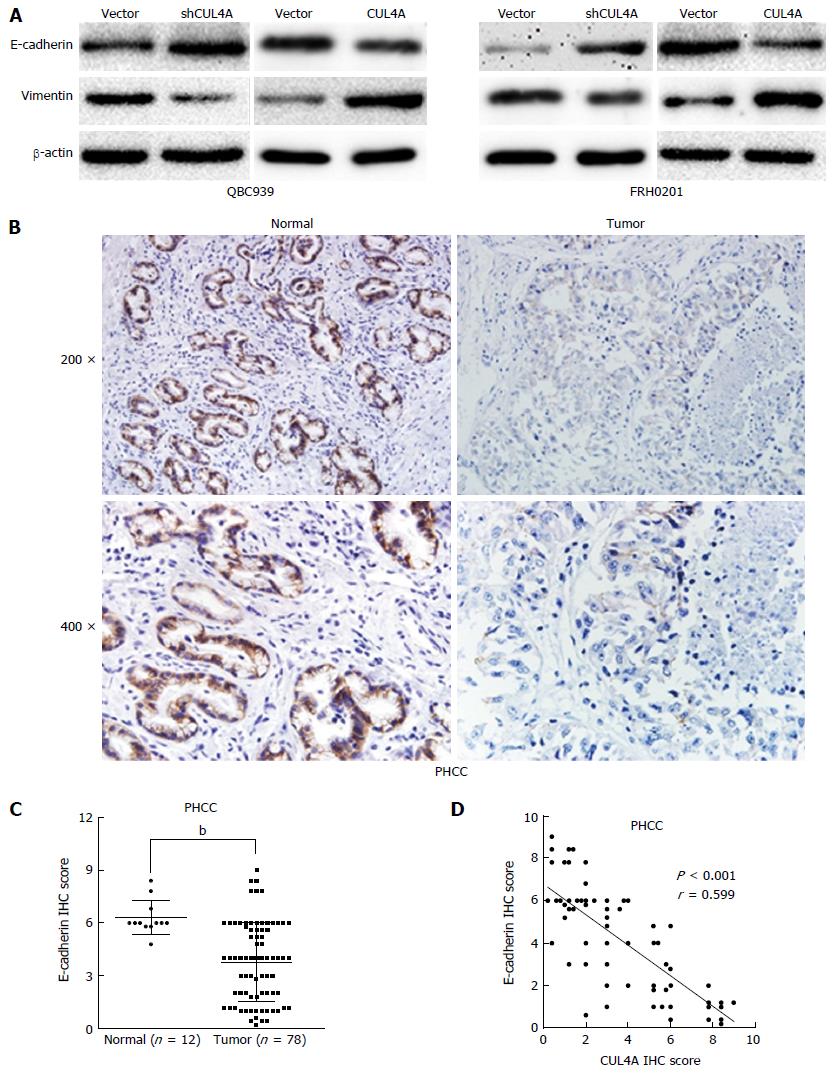
Mounting evidence has confirmed that the EMT endows tumour cells with migratory and invasive properties[20,25]. To explore whether CUL4A could promote the EMT process, we evaluated EMT markers in PHCC cells with CUL4A depletion and control cells. CUL4A depletion in QBC939 cells and FRH0201 cells enhanced the expression of E-cadherin, a hall mark of the EMT, in Western blot assays, which is accompanied by decreased expression of vimentin (Figure 4A). Consistently, CUL4A overexpression in QBC939 cells and FRH0201 cells dramatically increased the expression of E-cadherin) and decreased that of vimentin) in Western blot assays (Figure 4A). Moreover, the expression of E-cadherin in PHCC tissues was also investigated by IHC, which verified high E-cadherin expression in tumour adjacent tissues relative to primary tumour tissues (Figure 4B and C). In addition, an negative correlation between CUL4A expression and E-cadherin expression was observed in PHCC tissues in IHC assays (Figure 4D). Together, these results indicated that CUL4A could promote metastasis through, at least partially, the induction of the EMT in PHCC.
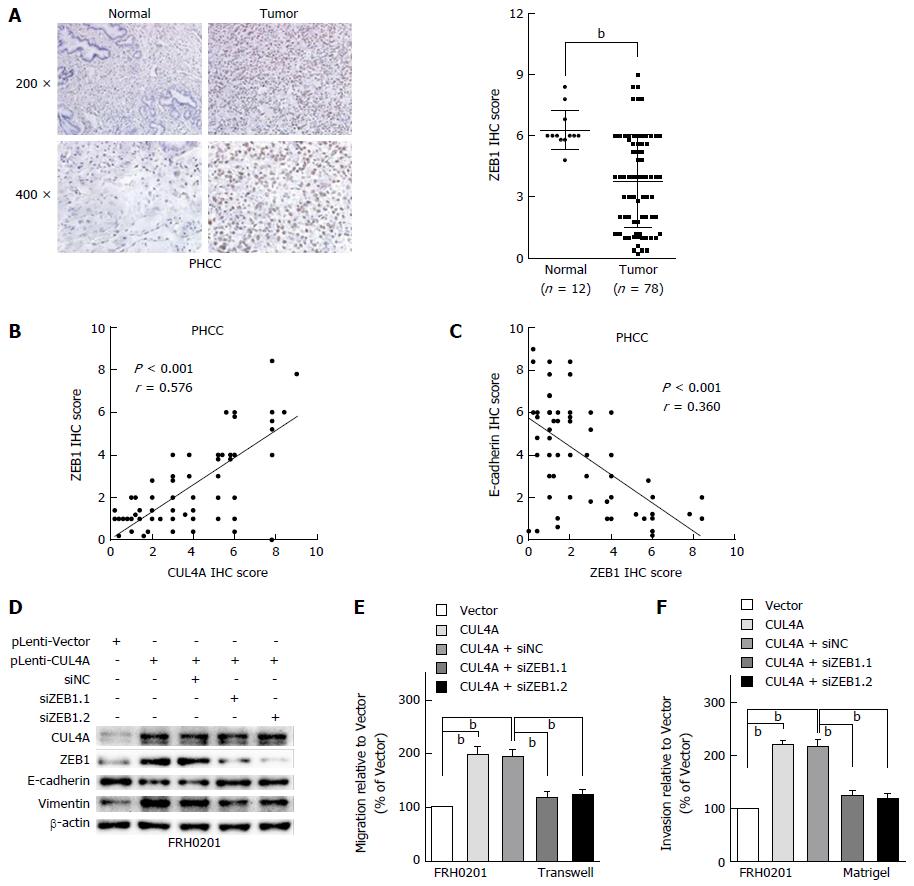
Various transcriptional factors are involved in regulating the EMT process[20]. A previous study revealed that ZEB1 is the transcriptional factor in the CUL4A-regulated EMT in breast cancer[26]. The present study further investigated the functional role of ZEB1 in the CUL4A related-EMT in PHCC. To investigate the role of ZEB1 in the CUL4A-related EMT and metastasis, we analysed the phenotypic changes in CUL4A overexpressing FRH0201 cells with ZEB1 knockdown. First, ZEB1 expression was analysed by IHC assay, which revealed that ZEB1 was overexpressed in primary tumour tissues compared with tumour adjacent tissues (Figure 5A). Furthermore, the positive correlation between ZEB1 expression and CUL4A expression was confirmed (Figure 5B). Additionally, it was verified that ZEB1 expression was negatively correlated with E-cadherin expression (Figure 5C). In in vitro assays, ZEB1 deficiency obviously reversed the decreased expression of E-cadherin and increased the expression of vimentin induced by CUL4A overexpression (Figure 5D). Furthermore, the increased migratory and invasive capacities induced by CUL4A overexpression were also inhibited by ZEB1 knockdown in the FRH0201 cell line (Figure 5E and F). Thus, these findings suggest that ZEB1 mediates the CUL4A-induced EMT, migration and invasion in PHCC.
CUL4, one of three founding cullins conserved from yeast to humans, uses a large β-propeller protein, DDB1, as a linker to interact with a subset of WD40 proteins that serve as substrate receptors, forming as many as 90 E3 complexes in mammals[27]. In addition to the function of CUL4A-DDB1 ligases in ubiquitinating several important proteins[28], many studies have revealed its role in tumour development and progression. In the current study, we delineated, for the first time, that the CUL4A gene is amplified and overexpressed in PHCC cell lines and tissues. The association study of CUL4A and clinicopathologic characteristics revealed that CUL4A expression was significantly correlated with tumour differentiation and the T, N and TNM stages of PHCC. Furthermore, an essential finding of this study was that PHCC patients with high CUL4A expression had a worse prognosis than those with low CUL4A expression. Multivariable Cox’s regression analysis indicated that CUL4A was the only independent prognostic factor for OS and PFS. Functional analysis with CUL4A suppression in PHCC cell lines showed a marked reduction in cell invasion and migration. In agreement with this, ectopic expression of CUL4A in FRH0201 cells promoted cell migratory and invasive abilities. Therefore, our results indicated that CUL4A may serve as a potential prognostic marker for PHCC and an important oncogene in PHCC metastasis.
Accumulating evidence has indicated that the EMT mediates tumour progression including local invasion, dissemination from the primary tumour, intravasation into blood circulation, and metastasis. The EMT is a complex process requiring extensive changes in cell adhesion and morphology as well as activation of signaling pathways. These considerations indicate that EMT induction is likely to be a centrally important mechanism for the progression of carcinomas to a metastatic stage and the maintenance of malignancy[29]. EMT inhibition may be a potential strategy for cancer treatment. In the present study, CUL4A depletion caused rapid regression of EMT features, which is characterized by decreased cell motility and invasive phenotypes. Conversely, gain of CUL4A promoted a global acquisition of mesenchymal characteristics and accelerated metastatic progression. In addition, loss of E-cadherin is considered a fundamental event in the EMT[20]. E-cadherin is expressed in epithelial cells, and its expression is decreased during the EMT in embryonic development, tissue fibrosis, and cancer[30]. In this report, loss of E-cadherin was accompanied by CUL4A overexpression in PHCC cells and specimens, indicating that CUL4A may represent an upstream molecule that can induce the EMT. Vimentin is another commonly used EMT-associated marker that has been clarified in tumour samples to evaluate mesenchymal specific features[21]. In the present study, Western blot assay discovered that the expression of vimentin was up-regulated in CUL4A overexpressing cells, however, the expression of vimentin was decreased in cells with CUL4A knockdown. These results suggested that CUL4A could promote EMT in PHCC.
Among the transcriptional factors regulating EMT, ZEB1 is an essential E-cadherin repressor which can directly bind to and repress the activity of E-cadherin promoter[20]. A previous study revealed that CUL4A could promote ZEB1 transcription by modulating histone H3K4me3 at the promoter of ZEB1 in breast cancer[26]. Mechanistically, this report also showed that CUL4A could regulate EMT via ZEB1 in PHCC cell lines and tissues. Furthermore, ZEB1 interference dramatically reduced the metastasis and EMT caused by CUL4A overexpression in FRH0201 cells. Interestingly, previous studies revealed that depletion of ZEB1, either chemically or by RNAi, resulted in a partial epithelial metaplasia and drug sensitivity in mesenchymal-like cells[31,32]. Considering the positive correlation between CUL4A and ZEB1 in PHCC that was demonstrated in this study, inhibition of CUL4A may be a potential therapeutic strategy for PHCC that can increase the chemotherapy sensitivity and effectiveness through downregulating the expression of ZEB1, especially for patients with CUL4A overexpression.
In conclusion, the present study indicates that CUL4A is overexpressed in PHCC tumour tissues than in normal intrahepatic bile ducts, and it is obviously correlated with the poor prognosis of PHCC patients. Moreover, CUL4A plays a critical role in PHCC metastasis by facilitating PHCC cell motility and cell invasion as well as consequent induction of EMT via, at least partially, up-regulating transcriptional regulation factor ZEB1. CUL4A may serve as a valuable prognostic biomarker and a potential therapeutic target for PHCC.
During the past 2-3 decades, the incidence and mortality rates of perihilar cholangiocarcinoma (PHCC), accounting for 50% of cholangiocarcinoma cases, have been steadily increasing. The vast majority of patients diagnosed with PHCC are usually presented with an advanced disease that develops without an identifiable etiology. Curative surgical resection, the only effective treatment to achieve possible cure in PHCC, can be achieved in less than 19%-75% of patients. What is worse, currently available chemotherapy and radiotherapy regimens are usually unresponsive to advanced PHCC. Left untreated, survival is only 6 to 12 mo.
Cullin 4A (CUL4A), a core subunit of E3 ubiquitin ligase, has been suggested to play an essential role in cellular transformation, however, its role in PHCC has not been identified.
The present study indicates that CUL4A is overexpressed in PHCC tumour tissues than in normal intrahepatic bile ducts and is obviously correlated with a poor prognosis of PHCC patients. Moreover, CUL4A plays a critical role in PHCC metastasis by facilitating PHCC cell motility and cell invasion as well as consequent induction of epithelial to mesenchymal transition (EMT) via up-regulating transcriptional regulation factor ZEB1.
CUL4A may serve as a potential prognostic marker for evaluating the overall survival and progression-free survival of patients with PHCC. Besides, CUL4A may also be a therapeutic target for PHCC.
EMT is a cellular process during which epithelial cells lose their polarity and cell-cell adhesion, undergo changes in cell shape and in cytoskeletal organization and acquire mesenchymal characteristics. It has been confirmed that EMT is closely involved in increasing cell migratory and invasive properties, inducing stem cell properties, preventing apoptosis and senescence, and resisting to chemotherapy and immunotherapy.
The authors have put together an excellent paper that is a mix of solid basic science with a clinical correlation.
| 1. | Razumilava N, Gores GJ. Cholangiocarcinoma. Lancet. 2014;383:2168-2179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1072] [Cited by in RCA: 1460] [Article Influence: 121.7] [Reference Citation Analysis (4)] |
| 2. | DeOliveira ML, Cunningham SC, Cameron JL, Kamangar F, Winter JM, Lillemoe KD, Choti MA, Yeo CJ, Schulick RD. Cholangiocarcinoma: thirty-one-year experience with 564 patients at a single institution. Ann Surg. 2007;245:755-762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 882] [Cited by in RCA: 1041] [Article Influence: 54.8] [Reference Citation Analysis (1)] |
| 3. | Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215-1229. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1070] [Cited by in RCA: 1012] [Article Influence: 77.8] [Reference Citation Analysis (6)] |
| 4. | Kang MJ, Jang JY, Chang J, Shin YC, Lee D, Kim HB, Kim SW. Actual Long-Term Survival Outcome of 403 Consecutive Patients with Hilar Cholangiocarcinoma. World J Surg. 2016;40:2451-2459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 48] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 5. | Brito AF, Abrantes AM, Encarnação JC, Tralhão JG, Botelho MF. Cholangiocarcinoma: from molecular biology to treatment. Med Oncol. 2015;32:245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 6. | Sugasawa K. The CUL4 enigma: culling DNA repair factors. Mol Cell. 2009;34:403-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 7. | Xin H, Herrmann A, Reckamp K, Zhang W, Pal S, Hedvat M, Zhang C, Liang W, Scuto A, Weng S. Antiangiogenic and antimetastatic activity of JAK inhibitor AZD1480. Cancer Res. 2011;71:6601-6610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 8. | Li J, Wang QE, Zhu Q, El-Mahdy MA, Wani G, Praetorius-Ibba M, Wani AA. DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity. Cancer Res. 2006;66:8590-8597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 81] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 9. | Chen LC, Manjeshwar S, Lu Y, Moore D, Ljung BM, Kuo WL, Dairkee SH, Wernick M, Collins C, Smith HS. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 1998;58:3677-3683. [PubMed] |
| 10. | Shinomiya T, Mori T, Ariyama Y, Sakabe T, Fukuda Y, Murakami Y, Nakamura Y, Inazawa J. Comparative genomic hybridization of squamous cell carcinoma of the esophagus: the possible involvement of the DPI gene in the 13q34 amplicon. Genes Chromosomes Cancer. 1999;24:337-344. [PubMed] |
| 11. | Bartek J, Lukas J. Cell cycle. Order from destruction. Science. 2001;294:66-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Bonnet M, Loosveld M, Montpellier B, Navarro JM, Quilichini B, Picard C, Di Cristofaro J, Bagnis C, Fossat C, Hernandez L. Posttranscriptional deregulation of MYC via PTEN constitutes a major alternative pathway of MYC activation in T-cell acute lymphoblastic leukemia. Blood. 2011;117:6650-6659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 13. | Yasui K, Arii S, Zhao C, Imoto I, Ueda M, Nagai H, Emi M, Inazawa J. TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology. 2002;35:1476-1484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 142] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 14. | Hung MS, Mao JH, Xu Z, Yang CT, Yu JS, Harvard C, Lin YC, Bravo DT, Jablons DM, You L. Cul4A is an oncogene in malignant pleural mesothelioma. J Cell Mol Med. 2011;15:350-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 60] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Ren S, Xu C, Cui Z, Yu Y, Xu W, Wang F, Lu J, Wei M, Lu X, Gao X. Oncogenic CUL4A determines the response to thalidomide treatment in prostate cancer. J Mol Med (Berl). 2012;90:1121-1132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 16. | Liu TT, You HL, Weng SW, Wei YC, Eng HL, Huang WT. Recurrent Amplification at 13q34 Targets at CUL4A, IRS2, and TFDP1 As an Independent Adverse Prognosticator in Intrahepatic Cholangiocarcinoma. PLoS One. 2015;10:e0145388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Schindl M, Gnant M, Schoppmann SF, Horvat R, Birner P. Overexpression of the human homologue for Caenorhabditis elegans cul-4 gene is associated with poor outcome in node-negative breast cancer. Anticancer Res. 2007;27:949-952. [PubMed] |
| 18. | Tiwari N, Gheldof A, Tatari M, Christofori G. EMT as the ultimate survival mechanism of cancer cells. Semin Cancer Biol. 2012;22:194-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 331] [Cited by in RCA: 408] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 19. | Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer. 2009;9:265-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2456] [Cited by in RCA: 2600] [Article Influence: 152.9] [Reference Citation Analysis (0)] |
| 20. | Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871-890. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6805] [Cited by in RCA: 7910] [Article Influence: 465.3] [Reference Citation Analysis (1)] |
| 21. | Iwatsuki M, Mimori K, Yokobori T, Ishi H, Beppu T, Nakamori S, Baba H, Mori M. Epithelial-mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010;101:293-299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 574] [Cited by in RCA: 599] [Article Influence: 37.4] [Reference Citation Analysis (0)] |
| 22. | Ning S, Guo S, Xie J, Xu Y, Lu X, Chen Y. TROP2 correlates with microvessel density and poor prognosis in hilar cholangiocarcinoma. J Gastrointest Surg. 2013;17:360-368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 23. | Zlobec I, Steele R, Terracciano L, Jass JR, Lugli A. Selecting immunohistochemical cut-off scores for novel biomarkers of progression and survival in colorectal cancer. J Clin Pathol. 2007;60:1112-1116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 195] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 24. | Lind PA, Wennberg B, Gagliardi G, Rosfors S, Blom-Goldman U, Lideståhl A, Svane G. ROC curves and evaluation of radiation-induced pulmonary toxicity in breast cancer. Int J Radiat Oncol Biol Phys. 2006;64:765-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 70] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 25. | Tsuji T, Ibaragi S, Hu GF. Epithelial-mesenchymal transition and cell cooperativity in metastasis. Cancer Res. 2009;69:7135-7139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 325] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 26. | Wang Y, Wen M, Kwon Y, Xu Y, Liu Y, Zhang P, He X, Wang Q, Huang Y, Jen KY. CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression. Cancer Res. 2014;74:520-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 176] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 27. | Jackson S, Xiong Y. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci. 2009;34:562-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 342] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 28. | Waning DL, Li B, Jia N, Naaldijk Y, Goebel WS, HogenEsch H, Chun KT. Cul4A is required for hematopoietic cell viability and its deficiency leads to apoptosis. Blood. 2008;112:320-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 29. | Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420-1428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6575] [Cited by in RCA: 8113] [Article Influence: 477.2] [Reference Citation Analysis (0)] |
| 30. | Hay ED, Zuk A. Transformations between epithelium and mesenchyme: normal, pathological, and experimentally induced. Am J Kidney Dis. 1995;26:678-690. [PubMed] |
| 31. | Li Y, VandenBoom TG, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704-6712. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 552] [Cited by in RCA: 560] [Article Influence: 32.9] [Reference Citation Analysis (0)] |
| 32. | Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489-500. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 705] [Cited by in RCA: 702] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Bramhall S, Gentilini A S- Editor: Gong ZM L- Editor: Wang TQ E- Editor: Zhang FF