Published online Jan 7, 2017. doi: 10.3748/wjg.v23.i1.93
Peer-review started: July 17, 2016
First decision: August 19, 2016
Revised: September 5, 2016
Accepted: September 14, 2016
Article in press: September 14, 2016
Published online: January 7, 2017
Processing time: 175 Days and 19.5 Hours
To determine the functional role of miR-490-5p in mast cell proliferation and apoptosis, and in the mast cell tryptase/PAR-2 signal pathway.
The 3rd generation of lentivirus vector systems containing enhanced green fluorescent protein (EGFP) (Ruisai Inc., Shanghai, China), which acts as a reporter gene was used to construct the mmu-miR-490-5p lentivirus expression vector pEGFP-antagomiR-490-5p, and the lentivirus vector pEGFP-negative was used as a negative control. The stably transfected mast cell line p815 was then constructed. GFP positive cells were successfully transfected cells. We determined the expression of miR-490-5p in p815 mast cells before and after transfection using quantitative real-time PCR (qRT-PCR). In addition, after transduction with the lentivirus vectors, the role of miR-490-5p in mast cell proliferation and apoptosis was investigated using the CCK-8 assay and flow cytometry, respectively. The mRNA levels of tryptase and PAR-2 were detected by qRT-PCR and the protein levels were detected by Western blot.
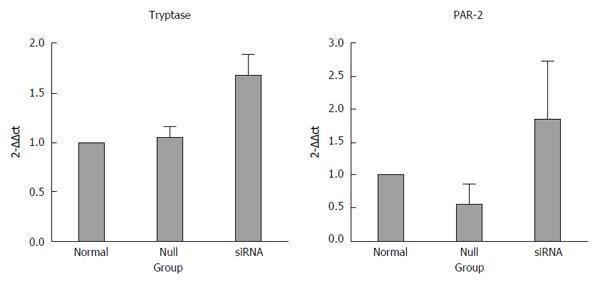
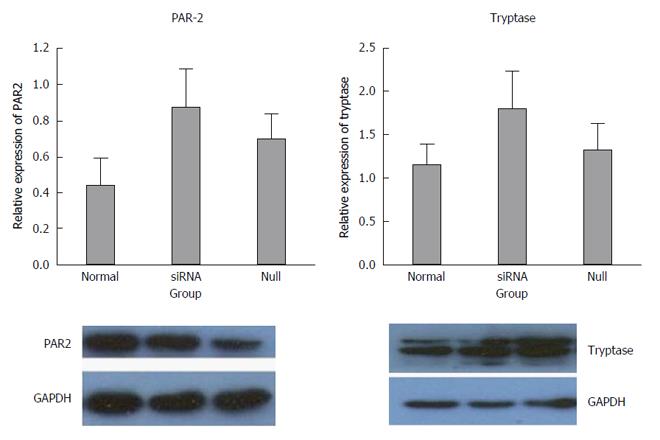
The inhibition of miR-490-5p expression promoted apoptosis and inhibited proliferation of p815 mast cells. The mRNA levels of tryptase and PAR-2 were significantly increased after transfection compared with the control group, tryptase (P = 0.721, normal vs null; P = 0.001, siRNA vs normal; P = 0.002, siRNA vs null) and PAR-2 (P = 0.027, siRNA vs null; P = 0.353, normal vs null; P = 0.105, siRNA vs normal). The protein levels of tryptase and PAR2 were slightly higher in the siRNA group than those in the control group, but the difference was not statistically significant (P > 0.05).
miR-490-5p plays a vital role in the pathogenesis of irritable bowel syndrome by affecting mast cell proliferation and apoptosis; with down-regulation of miR-490-5p, the mRNA level of mast cell tryptase and PAR-2 increased, and the protein level increased, but the difference was not statistically significant.
Core tip: Mast cells and mast cell degranulation play important roles in the pathophysiology of irritable bowel syndrome. miRNA is a class of important endogenous single-stranded non-coding RNA, and plays an essential regulatory role in complex biological systems without protein translation. In the present study, we aimed to determine the role of miR-490-5p in regulating mast cell proliferation and apoptosis, the expression of mast cell tryptase and PAR-2, and thus predict the role of miR-490-5p in the pathogenesis of irritable bowel syndrome.
- Citation: Ren HX, Zhang FC, Luo HS, Zhang G, Liang LX. Role of mast cell-miR-490-5p in irritable bowel syndrome. World J Gastroenterol 2017; 23(1): 93-102
- URL: https://www.wjgnet.com/1007-9327/full/v23/i1/93.htm
- DOI: https://dx.doi.org/10.3748/wjg.v23.i1.93
Irritable bowel syndrome (IBS) is one of the most common functional gastrointestinal (GI) disorders. The cause of IBS is unknown and seems to be multifactorial[1-4]. Several mechanisms including visceral hypersensitivity, motility disorder, infection, and psychiatric factors, have been suggested as possible etiologic links to the development of IBS[5,6]. In addition, there is increasing evidence of a genetic contribution in IBS such as gene polymorphism[7-9] and dysregulated microRNA (miRNA) expression[10-14]. Mast cells and mast cell degranulation function as a bridge in the neuro-immuno-endocrine system in IBS[15]. Tryptase is one of the most important proteins in mast cell degranulation and it exerts its effects mainly through the tryptase-PAR-2 signal pathway[16]. However, considering the complexity of the expression and function of tryptase, there are obvious limitations in the single analysis of the signal pathway. Multiple factors are involved in the regulation of the above signaling pathway. Among these factors, intracellular concentration of Ca2+, calmodulin, DAG/PKC and Rho GTPase have been noted to affect mast cell degranulation[17,18]. miRNA is a an important endogenous single-stranded non-coding RNA, which plays an essential regulatory role in complex biological systems without protein translation. Mature miRNA down-regulates gene expression by binding to the 3′-UTR region of the target gene and causes translational inhibition or mRNA degradation ultimately moderating the protein expression level. To date, multiple dysregulated microRNA expression has been reported in IBS. For example, miRNA-29a affects intestinal membrane permeability through its regulation of the glutamate-ammonia ligase gene and miRNA-510 plays an important role in the regulation of 5-HT3E expression[19,20]. Previously, we detected an elevation of miR-490-5p in diarrhea predominant IBS (IBS-D) patients using high-throughput microarray, but miR-490-5p has not been investigated in IBS. TargetScan was used to predict the target gene of miR-490-5p, and we noted the following: CABP5: encoding calcium binding protein5; CAMK1D: encoding calcium/calmodulin-dependent protein kinase ID; CASP3: encoding caspase 3, apoptosis-related cysteine peptidase; TNFSF18: encoding tumor necrosis factor (ligand) superfamily, member 18; BOP1: encoding proliferation-associated protein. Of these, both calcium and calmodulin-dependent protein kinase were highly correlated with mast cell degranulation[17,18]. Caspase, apoptosis-related cysteine peptidase and proliferation associated protein have been reported to be involved in cell proliferation and apoptosis in other diseases[21]. However, there are no studies available concerning the relationship between these factors and IBS. By comprehensively analyzing the results of gene microarray and bioinformatics, we focused our studies on the expression and functional role of miR-490-5p in mast cells and the mast cell/tryptase/PAR-2 signaling pathway, and then to predict whether and how miR-490-5p was involved in the pathogenesis of IBS. The results of this study may provide a theoretical basis for further study of biomarkers for the diagnosis and new treatment of IBS.
The p815 mast cell line was purchased from Shanghai Cell Biochemical Institute, China Academy of Science (Shanghai, China). This cell line is a suitable transfection host. P815 cells phagocytose latex beads, but not zymosan or BCG. They do not function in antibody-dependent cell-mediated cytotoxicity. Growth of these cells is not inhibited by dextran sulfate, lipopolysaccharide (LPS) or tuberculin purified protein derivative (PPD). The cells were also found to be negative for the ectromelia virus (mousepox).
Mast cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS), phosphate-buffered saline (PBS), trypsin, penicillin/streptomycin combination, miR-490-5p-siRNA vector, siRNA negative lentiviral vector, blasticidin, RIPA lysate [50 mmol/L Tris (pH 7.4), 150 mmol/L NaCl, 5 mmol/L EDTA, 1% Triton X-100 (v/v)], protease inhibitor cocktail, 1 mmol/L sodium vanadate and 10 mmol/L NaF. DNA marker, agarose, EB substitute, Trizol and cDNA reverse transcription kit were obtained from Takara. Polymerase chain reaction (PCR) primers were synthesized by Sangon Biotechnology (Shanghai, China). SuperReal PreMix Plus was purchased from Tiangen Biotechnology (Beijing, China). Mouse anti-GAPDH monoclonal antibody for western blot was obtained from Cell Signaling Technology (Danvers, MA, United States). Anti-PAR-2 monoclonal antibody for western blot was obtained from Abcam (Shanghai, China). Antitryptase clonal antibody for western blot was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, United States).
Based on the results of a previous high-throughput microarray and bioinformatics analysis, TargetScan analysis (http://www.targetscan.org) was used to predict the target gene of the miRNA. A GO enrichment analysis and a KEGG pathway analysis of the target gene were subsequently performed. Based on the results of the bioinformatics analysis, we searched miRDB and miRBase to query the specific features of target genes of interest. Our selection criteria for the target miRNA were as follows: (1) it is abnormally expressed in IBS-D patients[22]; (2) the biological function of the target gene should be related to mast cell degranulation, proliferation and apoptosis or the signal transduction pathway or visceral sensitivity or neurotransmitter-release associated protein or intracellular material transportation-associated protein, as the products of these target genes may correlate highly with the pathogenesis of IBS[23]; and (3) it exists both in human and mouse mast cells. Finally, we choose miR-490-5p as our target miRNA.
The 3rd generation of a lentivirus vector system containing an expression vector and 3 packaging auxiliary plasmids was used to construct the miR-490-5p recombinant silencing vector. The expression vector contained the basic components of HIV 5 ‘LTR and 3’ LTR as well as other auxiliary components, CMV promoter, blasticidin markers and enhanced green fluorescent protein (EGFP). Packaging auxiliary plasmid contained pGag-Pol, PRev and pVSVG. According to the mouse mmu-miR-490-5p (MIMAT0017261) gene sequence in the miBase database (5’-CCAUGGAUCUCCAGGUGGGU-3’), a pair of oligonucleotide chains containing the miR-490-5p sequence and the reverse complementary miR-145 sequence, which can form an shRNA precursor sequence containing a stem-loop structure after annealing, were designed. (Top strand: 5’-TGCTGACCCACCTGGAGATCCATGGGTTTTGGCCACT GACTGACCCATGGATCCAGGTGGGT-3’ Bottom strand: 5’-CCTGACCCACCTGGATCCATGGGTCAGT CAGTGGCCAAAACCCATGGATCTCCAGGTGGGTC-3’). The shRNA was then connected with the linearized miRNA vector, the product was used to transform Escherichia coli DH5α, and the recombinant plasmids were extracted and analyzed by sequencing, and named pcDNA6.2-EGFP-mmu-490-5p, which was then used as a template to amplify the following primers:
(Lenti-Asc1-F: 5’-TACTGGCGCGCCGCCACCATGGTG AGCAAGGGCGAGGA-3’;
Lenti-Pme1-R: 5’-ACTAGTTTAAACTGCGGCCAGA TCTGGGC-3’).
The pLV-shRNA lentiviral expression vector was generated via T4 DNA ligase, and an unrelated negative control sequence was established in the same manner. The above protocol was completed by Shanghai RS Biotechnology Co., Ltd (Shanghai, China).
HEK-293T cells (5 × 104; ATCC, Maryland, United States) were seeded in a 6-well cell culture plate in DMEM and incubated at 37 °C with 5% CO2 for 24 h. When the cells reached 50%-70% confluence, pLV-shRNA plasmids and auxiliary plasmids were co-transfected into them using Lipo3000. After 48 h of transfection, the cell supernatant was collected for titer determination. Gradient dilution of the supernatant was performed using PBS at ratios of 10-1-10-6. Three wells were used for each gradient, and 50 μL of lentiviral diluent was placed in each well for infection. After 48 h of infection, we recorded the number of infected fluorescent cells at the dilution gradient where the ratio of green fluorescent protein (GFP)+ cells was approximately 20%, and mean values were calculated. Lentiviral titers were calculated according to the following formula (BT = TU/mL): TU/μL = (P × N/100 × V) × 1/DF (P = number of GFP+ cells, N = 105, V = volume of lentiviral diluent = 50 μL, DF = dilution factor).
Growing mast cells were seeded in 24-well plates (30000 cells per well). After adherence and the cell density reached 50%, the cells were transduced with recombinant lentivirus vectors at a multiplicity of infection (MOI) of 60. The cells were maintained at 37 °C in a humidified atmosphere of 5% CO2 for 24 h, and the medium was then changed. After 48 h of transfection, the transfection efficiency was assessed by inverted fluorescence microscopy. Thereafter, positively transfected cells were selected by adding blasticidin to the medium at the minimal effective dose of 6 μg/mL (the minimal effective dose was defined as the dose at which most of the cells died within 7 d). The efficiency of GFP expression was detected by flow cytometry. When the expression efficiency of GFP was more than 85%, the dose of blasticidin was replaced by half of the minimal effective dose.
Total RNA was extracted both from normal cells and stably transfected cells using Trizol (Roche, Mannheim, Germany) according to the manufacturer’s instructions. Complementary DNA (cDNA) templates were synthesized using the PrimeScript™ RT reagent Kit (Takara, Dalian, China) after the concentration and purity of total RNA were measured by NanoDrop. The expression level of miR-490-5p was measured by qRT-PCR. The primers used in RT-PCR are listed in Table 1.
| mmu-miR-490-5p | Forward | TGGCGGCCATGGATCTCCAG |
| RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCCAC | |
| Reverse | ATCCAGTGCAGGGTCCGAGG | |
| U6 | Forward | CGCTTCACGAATTTGCGTGTGTCAT |
| Reverse | GCTTCGGCAGCACATATACTAAAAT |
The cell counting kit-8 (CCK-8) assay was used to examine cell proliferation. A standard curve was constructed, and the cells were then washed three times with PBS, resuspended in PBS, counted, and diluted 1:2 geometrically. The cells were seeded in 96-well plates at different final concentrations (3 × 104, 6 × 104, 12 × 104, 24 × 104, 48 × 104 per mL) in assay medium and incubated at 37 °C in a 5% CO2 incubator for approximately 2-4 h. Next, 10 μL of CCK-8 was added to each well, and the cells were incubated at 37 °C. At different time points (1 h, 2 h, 3 h, 4 h), the absorbance at 490 nm was read on a GF-M3000 microplate reader (Gaomicaihong Analysis Instrument Company, Shandong, China). The absorbance was used as the Y-axis, and the cell number was used as the X-axis to draw a standard curve and the optimal number of cells and the time of detection were determined. In addition, using the standard curve, we assessed whether CCK-8 was suitable for determining the level of proliferation of mast cells. In the second step, we washed, resuspended, and counted the cells as described above. The cells were then seeded in 96-well plates at a final concentration of 3 × 104 per mL in assay medium and incubated at 37 °C in a 5% CO2 incubator. At different time points (24 h, 48 h, 72 h), 10 μL of CCK-8 was added to each well, and the cells were incubated at 37 °C for another 4 h. The absorbance at 490 nm was read on a microplate reader.
Cell apoptosis was detected by flow cytometry (FCM) and analyzed by CellQuest software (Becton Dickinson, Bedford, MA, United States). The transfected and normal cells were collected, washed three times with cool PBS, and then resuspended in PBS. 1 × 106 cells were then processed for labeling with Annexin V/7AAD according to the PE Annexin V apoptosis detection kit (BD Biosciences, Franklin Lakes, NJ, United States).
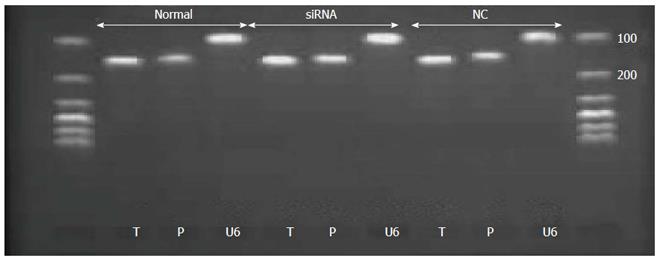
Real-time PCR was used to detect the mRNA level of tryptase and PAR-2 in p815 mast cells after transfection. Total RNA was extracted from both normal cells and stably transfected cells using Trizol. Total RNA was transcribed using the reverse transcription kit after measuring the concentration of total RNA with Nano drop. Total RNA was quantified to 1 μg. Information regarding the primers of tryptase, PAR-2 and the endogenous control U6 are listed in Table 2. The PCR samples were prepared in a volume of 20 μL, containing 2 μL of cDNA diluted 1:5 with PCR grade water, 10.4 μL of SYBR Green Supermix, and 300 nmol/L each of the forward and reverse primers. The PCR conditions consisted of preliminary denaturation at 95 °C for 15 min, followed by 40 cycles at 95 °C for 10 s, and 60 °C for 32 s. Melting curve analysis was performed to confirm the specificity and the integrity of the PCR products by the presence of a single peak. Products were subjected to agarose gel electrophoresis. Expression levels of mRNA were quantified by calculating threshold cycle values compared with the U6 endogenous control using 2-DDCt.
| Gene | Forward/reverse primer | PCR product, bp | Gene bank No. |
| Tryptase | GCCTCTCCCACCTCCTTATC/GGTATTTCCAGCACACAGCA | 145 | NM_010781.3 |
| PAR-2 | AGTTCTCTGCGTCCATCCTC/GGGTGTTTCTTCTTCGTTCG | 139 | NM_007974.4 |
| U6 | CGCTTCACGAATTTGCGTGTGTCAT/ATCCAGTGCAGGGTCCGAGG | 106 | NM-0012042741 |
Total protein was extracted from p815 mast cells and lysed in RIPA lysate [50 mmol/L Tris (pH 7.4), 150 mmol/L NaCl, 5 mmol/L EDTA, 1% Triton X-100 (v/v), 1 × protease inhibitor cocktail, 1 mmol/L sodium vanadate, and 10 mmol/L NaF]. Equal amounts of total protein were separated by SDS-PAGE with a 12% resolving layer and a 4% stacking layer and the proteins were transferred to PVDF membranes at 300 mA for 1.5 h. After blocking in 5% nonfat milk diluted with TBS-T for 1 h at room temperature with shaking, the membranes were incubated with primary antibodies (working dilution of tryptase 1:300; PAR-2 1:400) with gentle shaking overnight at 4 °C, washed with TBS-T (3 times for 5 min each time) and then incubated with species-appropriate secondary antibodies for 2 h at 37 °C. After washing in TBS-T (3 times for 5 min each time), the membranes were incubated using the ECL Developer Kit in a dark room. The efficiency of protein loading and transfer was assessed by reprobing the membranes with an anti-GAPDH antibody. The density of each band was analyzed using Image J software. Expression of the target protein band was compared with that of the corresponding control band.
The experimental data are represented by the mean ± SD. SPSS 20.0 was used for data analysis. One-way analysis of variance was used for analysis. Spearman analysis was used for correlation analysis. A P value less than 0.05 was considered statistically significant.
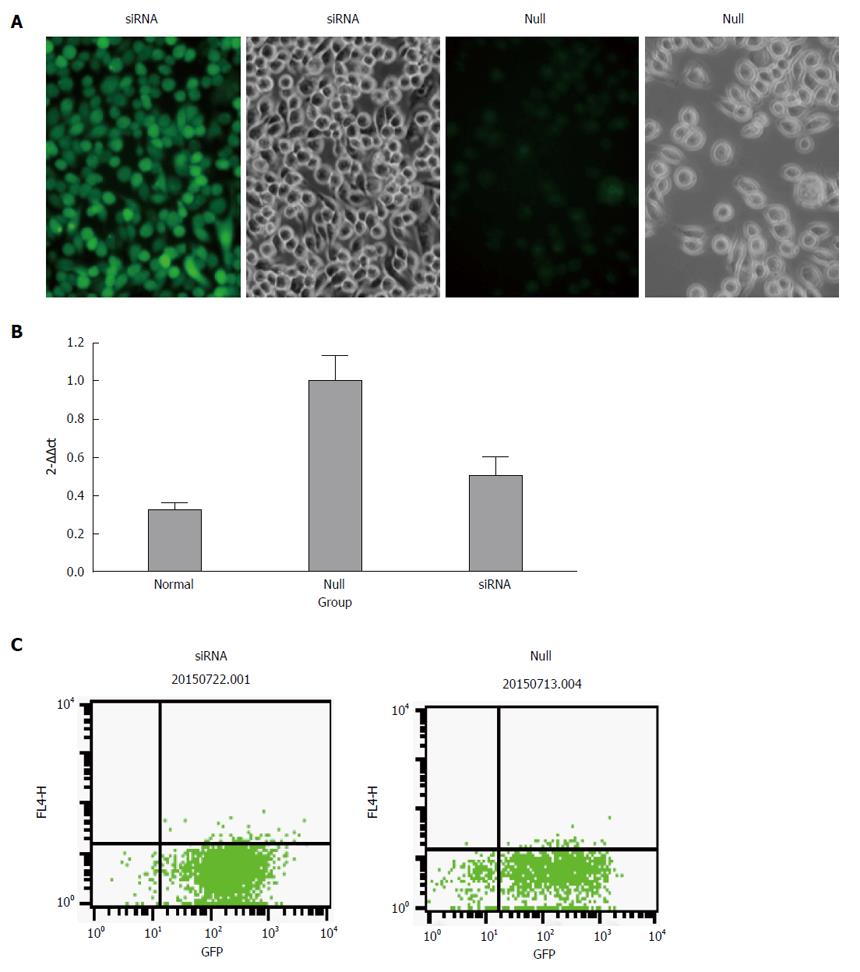
The transfection rate was measured by flow cytometry and the results showed that the efficiency was more than 85% in both the siRNA group and null group. We performed qRT-PCR to confirm the expression of miR-490-5p in p815 mast cells after silencing. As shown in Figure 1, the level of miR-490-5p in the siRNA group was significantly decreased compared with the empty vector group (P < 0.05).
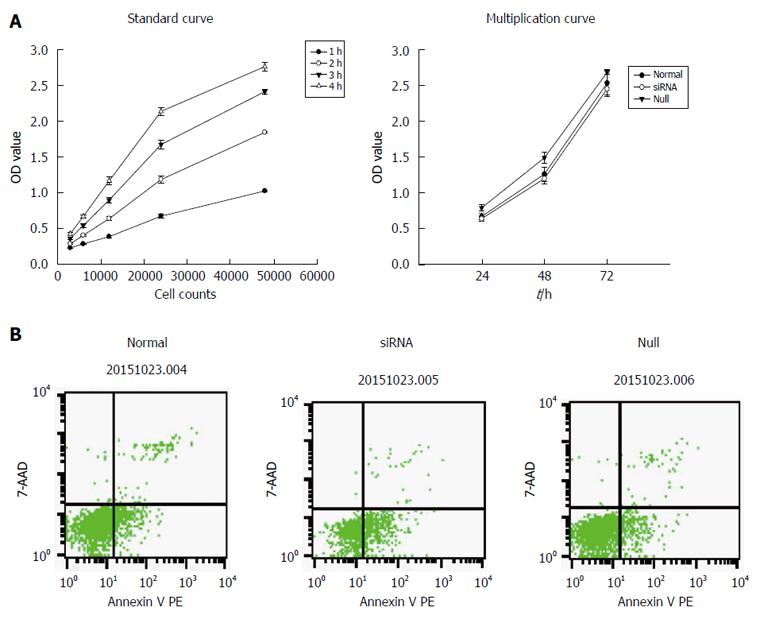
From the results of the standard curve, we found that when the number of cells varied between 3000 and 48000, there was a linear correlation between the number of cells and OD value. Therefore, CCK-8 was suitable for determining the level of proliferation of mast cells. In the proliferation test, the results showed that inhibition of miR-490-5P expression significantly reduced cell viability compared with the control group (P < 0.05). In the apoptosis test, the rate of apoptosis was significantly higher following the inhibition of miR-490-5p expression than in the control group (P < 0.05), as shown in Figure 2.
The mRNA levels of tryptase (P = 0.721 normal vs null, P = 0.001 siRNA vs normal, P = 0.002 siRNA vs null) and PAR-2 (P = 0.027 siRNA vs null, P = 0.353 normal vs null, P = 0.105 siRNA vs normal) were higher in the siRNA group than in the normal and null groups. In addition, the difference between the groups was statistically significant, and all the real-time PCR products from the three groups migrated as expected (Figures 3 and 4).
The expression results of the siRNA group were compared with normal and null controls. The density of each band was compared with the corresponding control band and normalized to the GAPDH gene. Elevated expression of tryptase and PAR-2 was found in the siRNA group compared with both the normal and null group, but there were no significant differences between the siRNA group and the controls (P > 0.05) (Figure 5).
In this study, we found that miR-490-5p promotes mast cell proliferation and resistance to apoptosis. Inhibition of miR-490-5p expression significantly increased the mRNA level of mast cell tryptase and PAR-2, but the protein level of tryptase and PAR-2 increased slightly, and a statistically significant difference between the siRNA and the control groups was not observed.
miRNAs are an abundant class of 20-22-nt non-coding single-stranded RNA and play significant roles in various physiological and pathological processes[24,25]. Many miRNAs have been shown to be associated with IBS. We previously detected an elevation of miR-490-5p in IBS-D patients. miR-490-5p is a member of the miR-490 family. The expression and functional role of miR-490-5p has also been reported in other diseases. It is involved in cell proliferation, apoptosis and the regulation of signaling pathways in different ways. In the study by Shiqi Li, miR-490-5p was found to be a novel tumor suppressor of bladder cancer cell proliferation by targeting c-Fos[26,27]. In renal cancer, miR-490-5p was confirmed to directly bind to the 3’UTR of PIK3CA mRNA and reduce the expression of PIK3CA at both the mRNA and protein level, which further inhibited the PIK3/Akt signaling pathway[28]. While, the role of miRNA-490-5p in the development and progression of IBS has not been reported. Its target genes include CABP5: encoding calcium binding protein 5; CAMK1D: encoding calcium/calmodulin-dependent protein kinase ID; CASP3: encoding caspase 3, apoptosis-related cysteine peptidase; TNFSF18: encoding tumor necrosis factor (ligand) superfamily, member 18; BOP1: encoding proliferation-associated protein. Of these, both calcium and calmodulin-dependent protein kinase were highly correlated with mast cell degranulation[17,18]. Caspase, apoptosis-related cysteine peptidase and proliferation associated protein have been reported to be involved in cell proliferation and apoptosis. In the present study, we found that inhibition of miR-490-5p significantly reduced mast cell proliferation and promoted apoptosis. Furthermore, the level of miR-490-5p was highly correlated with the mRNA level of mast cell tryptase and PAR-2. However, why the increase in PAR-2 and tryptase mRNA did not lead to a significant elevation in PAR-protein and tryptase-protein remains unknown. Possible reasons are as follows: Firstly, some other factors may be involved in the post-transcriptional regulation or the protein translation process. Secondly, the role of miR-490-5p is mainly up-regulating the function of tryptase/PAR-2 rather than affecting their expression. Thirdly, mast cells are merely normal cells in the resting state, and lack activating factors; however, in IBS-D patients, mast cells are activated. This is the first time that the role of miR-490-5p in IBS has been demonstrated, and information on the biological role of miRNA in IBS which could be used for reference was limited. Previous studies have reported that the number of intestinal mucosal mast cells increased in IBS patients and the mast cell tryptase/PAR-2 signal pathway played an important role in the pathogenesis of IBS. Thus, any factors associated with the above process are important. In summary, although problems occurred in our study, which require further investigation, we can conclude that mast cell miR-490-5p may participate in the occurrence and development of IBS by regulating the proliferation and apoptosis of mast cells; however, its effect on the mast cell tryptase/PAR-2 signal pathway is complex. The limitations in this study were that mir-490-5p was highly correlated with mast cell proliferation and apoptosis, but the specific target gene through which miR-490-5p plays a crucial role in IBS remains unknown. Furthermore, the elevation of tryptase and PAR-2 mRNA did not lead to an increase in protein level, which should be explored. The next step is to screen out the target gene of miR-490-5p involved in the regulation of the above biological process using bioinformatics analysis, selectively knockout the target gene, and then determine the level of mast cell tryptase and PAR-2. This may provide a new target for the treatment of IBS.
The authors thank Dr. Wei Jiao, Dr. Fei Liu, Dr. Jiao Lan and Rui-Ping Xiao from Scientific Research Center of the People’s Hospital of Guangxi Zhuang Autonomous Region for their valuable technological assistance on our project.
Previous studies have reported that mast cells and mast cell degranulation are involved in the pathogenesis of irritable bowel syndrome (IBS). Tryptase is an important component of mast cell degranulation which leads to visceral hypersensitivity in IBS patients by activation of PAR-2. The authors previous studies showed that IBS patients demonstrated dysregulation of miR-490-5p whose target gene was highly correlated with visceral sensitivity or neurotransmitter release associated protein, or intracellular material transportation associated protein. Thus, we predicted that miR-490-5p may be indirectly involved in the regulation of the mast cell-tryptase-PAR-2 signaling pathway.
A growing number of studies have reported that different miRNAs are involved in the regulation of proliferation and differentiation of digestive tract epithelial cells and smooth muscle cells. Recent research reported that miRNA plays an important role in the differentiation, proliferation and functional regulation of bone marrow mast cells in the inflammatory response. Another study reported that miR-29a was highly correlated with increased intestinal permeability in IBS-D patients. Thus, the authors can conclude that the regulation of miRNA may be closely related to the pathogenesis of IBS.
The authors proved, for the first time, that miR-490-5p may be directly or indirectly involved in mast cell proliferation and apoptosis and the mast cell/ tryptase/PAR-2 signaling pathway may play an important role in IBS-D.
miRNA-490-5p may be a new biomarker for the diagnosis of IBS and a new target for therapy.
RNA interference (RNAi), a fundamental biological process by which cells regulate gene expression, acts through complementary base-pairing with target mRNA and retrieves cellular RNases which in turn degrade mRNA transcripts. RNAi is now routinely used to evaluate gene function both in vitro and in vivo and many innovative screens reported the use of RNAi to investigate potential drug targets.
The author supported that miR-490-5p may directly or indirectly involved in the pathogenesis of IBS. It was the first research focusing on the miR-490-5p and IBS and it really could improve our understanding of IBS to some extent.
| 1. | Liang WJ, Zhang G, Luo HS, Liang LX, Huang D, Zhang FC. Tryptase and Protease-Activated Receptor 2 Expression Levels in Irritable Bowel Syndrome. Gut Liver. 2016;10:382-390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 37] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 2. | La JH, Kim TW, Sung TS, Kim HJ, Kim JY, Yang IS. Role of mucosal mast cells in visceral hypersensitivity in a rat model of irritable bowel syndrome. J Vet Sci. 2004;5:319-324. [PubMed] |
| 3. | Park JH. Dysregulated MicroRNA Expression in Irritable Bowel Syndrome. J Neurogastroenterol Motil. 2016;22:166-167. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 4. | Cenac N, Andrews CN, Holzhausen M, Chapman K, Cottrell G, Andrade-Gordon P, Steinhoff M, Barbara G, Beck P, Bunnett NW. Role for protease activity in visceral pain in irritable bowel syndrome. J Clin Invest. 2007;117:636-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 432] [Cited by in RCA: 466] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 5. | Chey WD, Kurlander J, Eswaran S. Irritable bowel syndrome: a clinical review. JAMA. 2015;313:949-958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 629] [Cited by in RCA: 784] [Article Influence: 71.3] [Reference Citation Analysis (0)] |
| 6. | Lacy BE. Diagnosis and treatment of diarrhea-predominant irritable bowel syndrome. Int J Gen Med. 2016;9:7-17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 7. | Zhang ZF, Duan ZJ, Wang LX, Yang D, Zhao G, Zhang L. The serotonin transporter gene polymorphism (5-HTTLPR) and irritable bowel syndrome: a meta-analysis of 25 studies. BMC Gastroenterol. 2014;14:23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 8. | Qin SY, Jiang HX, Lu DH, Zhou Y. Association of interleukin-10 polymorphisms with risk of irritable bowel syndrome: a meta-analysis. World J Gastroenterol. 2013;19:9472-9480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 14] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 9. | Bashashati M, Rezaei N, Bashashati H, Shafieyoun A, Daryani NE, Sharkey KA, Storr M. Cytokine gene polymorphisms are associated with irritable bowel syndrome: a systematic review and meta-analysis. Neurogastroenterol Motil. 2012;24:1102-e566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 10. | Mansour MA, Sabbah NA, Mansour SA, Ibrahim AM. MicroRNA-199b expression level and coliform count in irritable bowel syndrome. IUBMB Life. 2016;68:335-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Liao XJ, Mao WM, Wang Q, Yang GG, Wu WJ, Shao SX. MicroRNA-24 inhibits serotonin reuptake transporter expression and aggravates irritable bowel syndrome. Biochem Biophys Res Commun. 2016;469:288-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 12. | Zhou Q, Costinean S, Croce CM, Brasier AR, Merwat S, Larson SA, Basra S, Verne GN. MicroRNA 29 targets nuclear factor-κB-repressing factor and Claudin 1 to increase intestinal permeability. Gastroenterology. 2015;148:158-169.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 147] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 13. | Andersen HH, Duroux M, Gazerani P. MicroRNAs as modulators and biomarkers of inflammatory and neuropathic pain conditions. Neurobiol Dis. 2014;71:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 141] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 14. | Fourie NH, Peace RM, Abey SK, Sherwin LB, Rahim-Williams B, Smyser PA, Wiley JW, Henderson WA. Elevated circulating miR-150 and miR-342-3p in patients with irritable bowel syndrome. Exp Mol Pathol. 2014;96:422-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 15. | Wouters MM, Vicario M, Santos J. The role of mast cells in functional GI disorders. Gut. 2016;65:155-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 242] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 16. | Mayer EA, Bradesi S, Chang L, Spiegel BM, Bueller JA, Naliboff BD. Functional GI disorders: from animal models to drug development. Gut. 2008;57:384-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 120] [Article Influence: 6.7] [Reference Citation Analysis (1)] |
| 17. | Chakravarty N, Nielsen EH. Histamine release from saponin-permeabilized rat mast cells by calcium. Agents Actions. 1986;18:65-67. [PubMed] |
| 18. | Aridor M, Sagi-Eisenberg R. Neomycin is a potent secretagogue of mast cells that directly activates a GTP-binding protein involved in exocytosis. J Cell Biol. 1990;111:2885-2891. [PubMed] |
| 19. | Kapeller J, Houghton LA, Mönnikes H, Walstab J, Möller D, Bönisch H, Burwinkel B, Autschbach F, Funke B, Lasitschka F. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet. 2008;17:2967-2977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 149] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 20. | Zhou Q, Souba WW, Croce CM, Verne GN. MicroRNA-29a regulates intestinal membrane permeability in patients with irritable bowel syndrome. Gut. 2010;59:775-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 229] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 21. | Bi X, Cao Y, Chen R, Liu C, Chen J, Min D. MicroRNA-184 Promotes Proliferation and Inhibits Apoptosis in HaCaT Cells: An In Vitro Study. Med Sci Monit. 2016;22:3056-3061. [PubMed] |
| 22. | Guo XW. The microRNAs profiles in the colonic mucosa of IBS patients. Guangxi Yike Daxue Xuebao. 2014;. |
| 23. | Deiteren A, de Wit A, van der Linden L, De Man JG, Pelckmans PA, De Winter BY. Irritable bowel syndrome and visceral hypersensitivity: risk factors and pathophysiological mechanisms. Acta Gastroenterol Belg. 2016;79:29-38. [PubMed] |
| 24. | Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3661] [Cited by in RCA: 4033] [Article Influence: 224.1] [Reference Citation Analysis (0)] |
| 25. | Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9:219-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1034] [Cited by in RCA: 1076] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 26. | Li S, Xu X, Xu X, Hu Z, Wu J, Zhu Y, Chen H, Mao Y, Lin Y, Luo J. MicroRNA-490-5p inhibits proliferation of bladder cancer by targeting c-Fos. Biochem Biophys Res Commun. 2013;441:976-981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 27. | Lan G, Yang L, Xie X, Peng L, Wang Y. MicroRNA-490-5p is a novel tumor suppressor targeting c-FOS in human bladder cancer. Arch Med Sci. 2015;11:561-569. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 28. | Chen K, Zeng J, Tang K, Xiao H, Hu J, Huang C, Yao W, Yu G, Xiao W, Guan W. miR-490-5p suppresses tumour growth in renal cell carcinoma through targeting PIK3CA. Biol Cell. 2016;108:41-50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 43] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Raju J, Zhao HT S- Editor: Qi Y L- Editor: Webster JR E- Editor: Wang CH