Published online Jun 28, 2016. doi: 10.3748/wjg.v22.i24.5589
Peer-review started: March 11, 2016
First decision: March 31, 2016
Revised: April 25, 2016
Accepted: May 21, 2016
Article in press: May 23, 2016
Published online: June 28, 2016
Processing time: 102 Days and 6.7 Hours
AIM: To determine the expression of miR-422a in colorectal cancer (CRC) tissues and to further explore the prognostic value and function of miR-422a in CRC carcinogenesis.
METHODS: miR-422a expression was analyzed in 102 CRC tissues and paired normal mucosa adjacent to carcinoma by quantitative real-time PCR. The relationship of miR-422a expression with clinicopathological parameters was also analyzed. Kaplan-Meier analysis and Cox multivariate analysis were performed to estimate the potential role of miR-422a. Cell proliferation, migration, and invasion were used for in vitro functional analysis of miR-422a.
RESULTS: The levels of miR-422a were dramatically reduced in CRC tissues compared with normal mucosa (P < 0.05), and significantly correlated with local invasion (P = 0.004) and lymph node metastasis (P < 0.001). Kaplan-Meier survival and Cox regression multivariate analyses revealed that miR-422a expression (HR = 0.568, P = 0.015) and clinical TNM stage (HR = 2.942, P = 0.003) were independent prognostic factors for overall survival in CRC patients. Furthermore, in vitro experiments showed that overexpression of miR-422a inhibited the proliferation, migration, and invasion of SW480 and HT-29 cells.
CONCLUSION: Down-regulation of miR-422a may serve as an independent prognosis factor in CRC. MiR-422a functions as a tumor suppressor and regulates progression of CRC.
Core tip: In the present study, we found that miR-422a was dramatically reduced in colorectal cancer (CRC) tissues, and significantly correlated with local invasion and lymph node metastasis. miR-422a expression and clinical TNM stage were independent prognostic factors for overall survival in CRC patients. Furthermore, in vitro experiments showed that overexpression of miR-422a inhibited the proliferation, migration, and invasion of SW480 and HT-29 cells. These results indicated that down-regulation of miR-422a might serve as an independent prognosis factor in CRC, and miR-422a functions as a tumor suppressor and regulates progression of CRC.
- Citation: Zheng GX, Qu AL, Yang YM, Zhang X, Zhang SC, Wang CX. miR-422a is an independent prognostic factor and functions as a potential tumor suppressor in colorectal cancer. World J Gastroenterol 2016; 22(24): 5589-5597
- URL: https://www.wjgnet.com/1007-9327/full/v22/i24/5589.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i24.5589
Colorectal cancer (CRC) is one of the most common malignant tumors with increasing incidence and mortality over the past several decades[1]. Despite the significant progress made in diagnostic methods and improved treatment strategies, the prognosis of CRC patients remains poor, especially in those with advanced CRC[2,3]. CRC carcinogenesis is associated with multiple alterations in oncogenes and tumor suppressor genes. A growing number of studies have revealed that microRNAs (miRNAs) might regulate up to 30% of human genes and play a pivotal role in various cellular processes including proliferation, differentiation, apoptosis, migration, and invasion[4-8]. While the functional mechanisms of miRNAs remain largely unknown in the pathogenesis of CRC, dysregulated expression of miRNAs can serve as potential biomarkers for the diagnosis and prognosis of cancer[9-11].
Several studies have determined the importance of miR-422a in human diseases such as cancer, multiple sclerosis, and postmenopausal osteoporosis. Gougelet et al[12] reported that miR-422a could inhibit signaling pathways regulating tumor cell proliferation in osteosarcoma. A study by Mao et al[13] demonstrated that miR-422a targeted key mismatch repair protein (MutLα) by suppressing MLH1 expression, resulting in genome instability and tumorigenesis. Faltejskova et al[14] reported that dysregulation of miR-378, miR-375, miR-422a, miR-215 and miR-135b in CRC patients played an important role in CRC pathogenesis. However, further study is needed to confirm whether miR-422a is an independent predictive factor in patients with CRC. Moreover, the functional mechanism by which miR-422a regulates CRC progression and whether it can serve as a prognostic biomarker in CRC remain largely unknown.
Previously, we have identified a serum 4-miRNA panel that included miR-19a-3p, miR-92a-3p, miR-223-3p, and miR-422a. This miRNA panel served as biomarkers for early diagnosis of colorectal adenocarcinoma[15,16]. In this study, we showed that the expression of miR-422a is dysregulated in CRC tissues. The correlation between miR-422a and certain clinical characteristics, as well as its potential as a prognostic marker for CRC, was also investigated. The effects of miR-422a on CRC cell proliferation, invasion, and migration were also assessed.
All written informed consent was obtained from every participant for use of tissue samples. This project was approved by the Clinical Research Ethics Committee of Qilu Hospital of Shandong University. All CRC patients were recruited from the Department of General Surgery, Qilu Hospital of Shandong University between November 2004 and December 2013. The diagnosis of CRC was confirmed by histopathology or histobiopsy. A total of 102 CRC tissues were collected and the adjacent normal mucosa tissues were used as controls since CRC is a malignant epithelial tumor and originates from glandular epithelium of the colorectal mucosa. All tissues were immediately frozen in liquid nitrogen and stored at -80 °C until RNA extraction. The median follow-up period for patients enrolled in this study was 63 mo (range, 14-78 mo).
SW480 and HT29 cells were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The two cell lines were cultured in DMEM medium supplemented with 10% fetal bovine serum (Gibco, Carlsbad, CA, United States) at 37 °C in an incubator containing 5% CO2.
CRC cells were seeded at 2 × 105 cells/well in 6-well plates until 30%-50% confluency and then were transfected with miR-422a mimics or negative control mimics (RiboBio, Guangzhou, China) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s introduction. The transfected cells were normalized and used in subsequent assays. The transfection efficiency was determined by RT-qPCR to verify the success of transfection.
Total RNA from cell lines, CRC tissues and normal mucosa was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, United States). The cDNA was reverse-transcribed at 65 °C for 5 min in a 12 μL reaction system including 1 μg RNA, 1 μL reverse transcription primer (RiboBio, Guangzhou, China), 1 μL U6 reverse transcription primer and 1 μL dNTP. Then, 4 μL buffer, 2 μL DTT and 1 μL RNase inhibitor were added and incubated at 37 °C for 2 min followed by adding 1 μL MMLV and incubating at 37 °C for 50 min and 70 °C for 5 min. The PCR reaction was performed as follows: 95 °C for 1 min, and 45 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 45 s. U6 was used as a reference gene and each test was performed in triplicate in this study. RT-qPCR reactions were carried out on ABI Prism 7500 System (Applied Biosystems, Foster City, CA, United States).
miR-422a mimics and control mimics were transfected into SW480 and HT29 cells, then cultured for another 24, 48, 72, or 96 h. Cell proliferation rates was measured with CCK-8 reagent (Beyotime, Hangzhou, China) following the manufacturer’s protocol. A microplate photometer (Multiskan FC, Thermo scientific, Shanghai, China) was used to determine the optical density at 450 nm.
Transwell migration assay was performed using transwell chambers (Corning Costar, United States). After transfection for 24 h, cells were transferred into the upper chamber. RPMI 1640 medium containing 10% FBS functioning as a chemoattractant was added to matched lower chamber. SW480 and HT29 cells unable to migrate were removed from the upper surface of the transwell membrane after incubation for 48 h. Cells that were able to invade on the lower membrane surface were fixed in methanol, stained with 0.1% crystal violet, and counted using an inverted microscope (Olympus, Tokyo, Japan). For invasion assay, the inserts were pre-coated with matrix gel (BD Biosciences, Franklin Lakes, NJ, United States).
Data were analyzed using SPSS 17.0 software (IBM Corporation, Armonk, NY, United States). The expression of miR-422a between groups was compared using Mann-Whitney U test. Kaplan-Meier method was used for overall survival analysis. The Cox regression model was used for univariate and multivariate analyses to estimate the prognostic factors.
We detected the expression of miR-422a in 102 pairs of CRC tissues and normal mucosa adjacent to carcinoma using RT-qPCR. The results indicated that miR-422a levels were significantly lower in CRC tissues compared with normal mucosa (P < 0.05) (Figure 1A). Moreover, 65.7% (67 of 102) of CRC tissues had at least 2-fold down-regulated expression of miR-422a compared with normal mucosa (Figure 1B). The levels of miR-422a in stages I and II samples were significantly higher than those in stage III samples (P < 0.05). There was no significant difference in miR-422a expression between stages I and II samples (P > 0.05) (Figure 1C).
The association between miR-422a level and clinicopathological parameters of CRC patients is showed in Table 1. Our data showed that the level of miR-422a significantly correlated with local invasion (P = 0.004), lymph node metastasis (P = 0.002), and TNM stage (P < 0.001). However, there were no significant correlations between miR-422a level and gender, age, tumor location, differentiation or tumor size (P > 0.05).
| Clinical parameter | Number of cases (n = 102) | miR-422a expression, median (min-max) | P value |
| Gender | 0.704 | ||
| Male | 63 | 1.446 (0.074-130.774) | |
| Female | 39 | 1.617 (0.167-33.543) | |
| Age (yr) | 0.328 | ||
| ≤ median | 49 | 1.637 (0.088-130.774) | |
| > median | 53 | 1.715 (0.074-21.871) | |
| Tumor location | 0.092 | ||
| Colon | 58 | 1.798 (0.088-130.774) | |
| Rectum | 44 | 1.887 (0.074-33.543) | |
| Differentiation | 0.150 | ||
| Well | 12 | 1.798 (0.074-130.744) | |
| Moderate | 67 | 1.922 (0.166-21.871) | |
| Poor | 23 | 2.088 (0.088-20.406) | |
| Tumor size | 0.527 | ||
| ≤ 5 cm | 64 | 2.127 (0.074-130.744) | |
| > 5 cm | 38 | 2.353 (0.167-21.871) | |
| Local invasion | 0.004 | ||
| T1–T2 | 55 | 3.257 (0.206-130.744) | |
| T3–T4 | 47 | 1.617 (0.074-20.406) | |
| Lymph node metastasis | 0.002 | ||
| No | 70 | 2.863 (0.359-130.744) | |
| Yes | 32 | 1.445 (0.074-20.406) | |
| TNM stage | < 0.001 | ||
| I | 23 | 3.877 (0.265-15.999) | |
| II | 42 | 3.253 (0.074-13.566) | |
| III | 37 | 1.445 (0.329-130.744) |
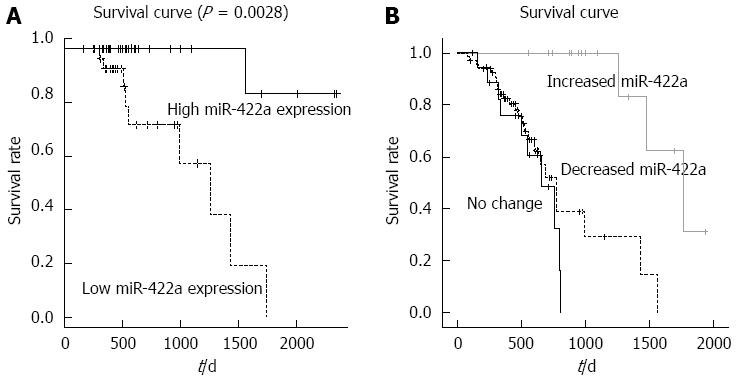
Our results demonstrated that 42 of 102 CRC patients died during the follow-up period. The 5-year overall survival rate was 58.8%. Patients were divided into high miR-422a expression and low miR-422a expression groups based on the median level. The prognosis was analyzed by Kaplan-Meier survival analysis, which revealed that patients with low miR-422a expression had a significantly poorer prognosis than those with high miR-422a (P = 0.0028; Figure 2A). Furthermore, patients were divided into three groups (decreased, no change, and increased) based on the change of miR-422a expression compared to adjacent normal mucosa. Kaplan-Meier survival analysis showed that patients with decreased miR-422a expression had a significantly poorer prognosis than those with increased miR-422a expression (P < 0.001; Figure 2B). There was no significant difference between the decreased miR-422a group and no change group. In addition, Cox regression multivariate analysis was performed to determine whether miR-422a was an independent factor of overall survival in CRC patients. The analysis revealed that miR-422a expression (HR = 0.568, 95%CI: 0.245-1.082; P = 0.015) and clinical TNM stage (HR = 2.942, 95%CI: 1.426-4.378; P = 0.003) were independent prognostic factors for overall survival in CRC patients (Table 2).
| Variable | Category | Univariate analysis | Multivariate analysis | ||||
| HR | 95%CI | P value | HR | 95%CI | P value | ||
| Gender | Male vs Female | 0.943 | 0.493-1.872 | 0.810 | |||
| Age | ≤ Median vs > Median | 1.436 | 0.528-3.157 | 0.148 | |||
| Tumor location | Colon vs Rectum | 0.862 | 0.484-1.706 | 0.603 | |||
| Tumor size | ≤ 5 cm vs > 5 cm | 1.093 | 0.568-2.194 | 0.873 | |||
| Differentiation | Well and moderate vs Poor | 1.018 | 0.462-2.180 | 0.160 | 0.986 | 0.486-2.006 | 0.074 |
| Local invasion | T1-T2 vs T3-T4 | 1.682 | 1.020-2.628 | 0.011 | 1.460 | 0.746-2.632 | 0.482 |
| Lymph node metastasis | Yes vs No | 1.182 | 0.634-2.530 | < 0.0011 | 1.262 | 0.584-2.680 | 0.306 |
| TNM stage | I and II vs III | 2.738 | 1.509-4.265 | < 0.0011 | 2.942 | 1.426-4.378 | 0.0031 |
| MiR-422a level | Low vs High | 0.306 | 0.108-0.763 | 0.0011 | 0.568 | 0.245-1.082 | 0.0151 |
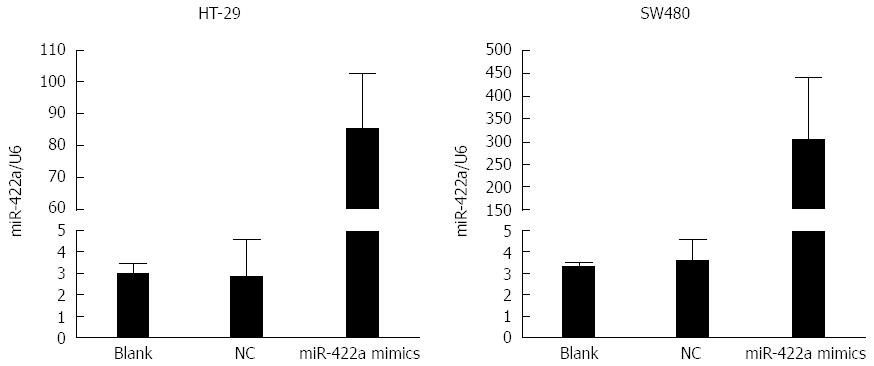
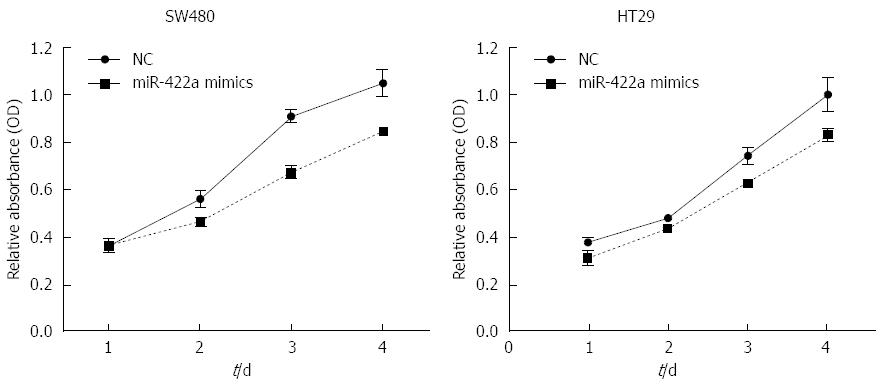
In our preliminary experiments, we detected the expression levels of miR-422a endogenously in several CRC cell lines, including HT-29, SW480, SW620 and HCT-116. The results showed that the levels of miR-422a were quite low in these CRC cells, and there were no significant differences in the expression level of miR-422a among these cell lines (data not shown). To measure the biological properties of miR-422a in CRC cells, miR-422a mimics were transiently transfected into HT-29 and SW480 cells, respectively. Subsequently, real-time RT-qPCR was performed, which showed high transfection efficiencies in both cell lines (Figure 3). The results of MTT assay showed that transfection of miR-422a mimics could significantly decrease cell number in SW480 and HT29 CRC cells (P < 0.05; Figure 4). It means that overexpression of miR-422a can inhibit the proliferation potential of SW480 and HT29 CRC cells.
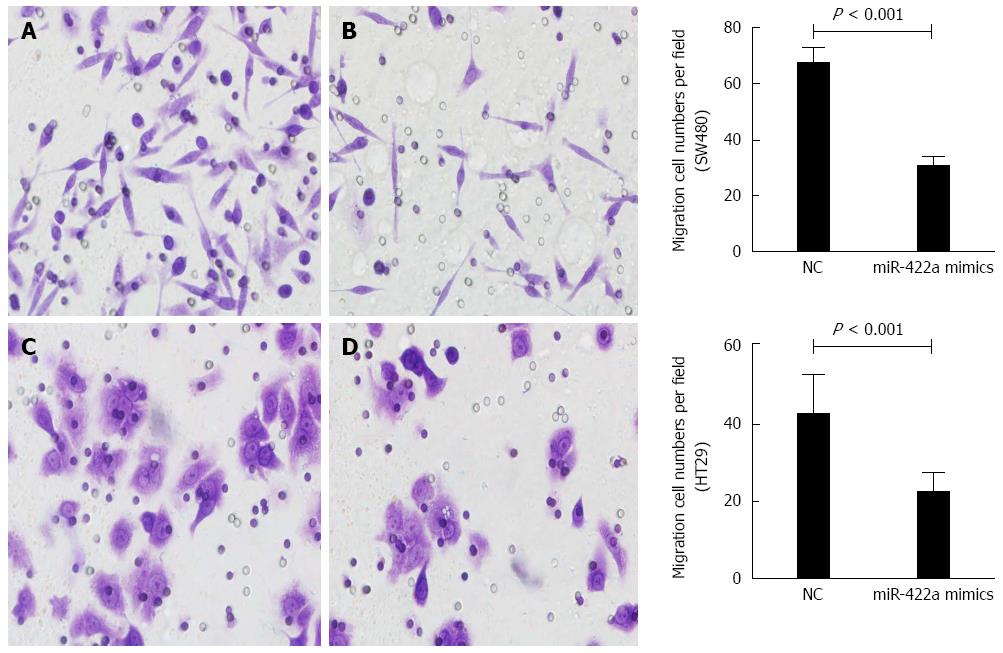
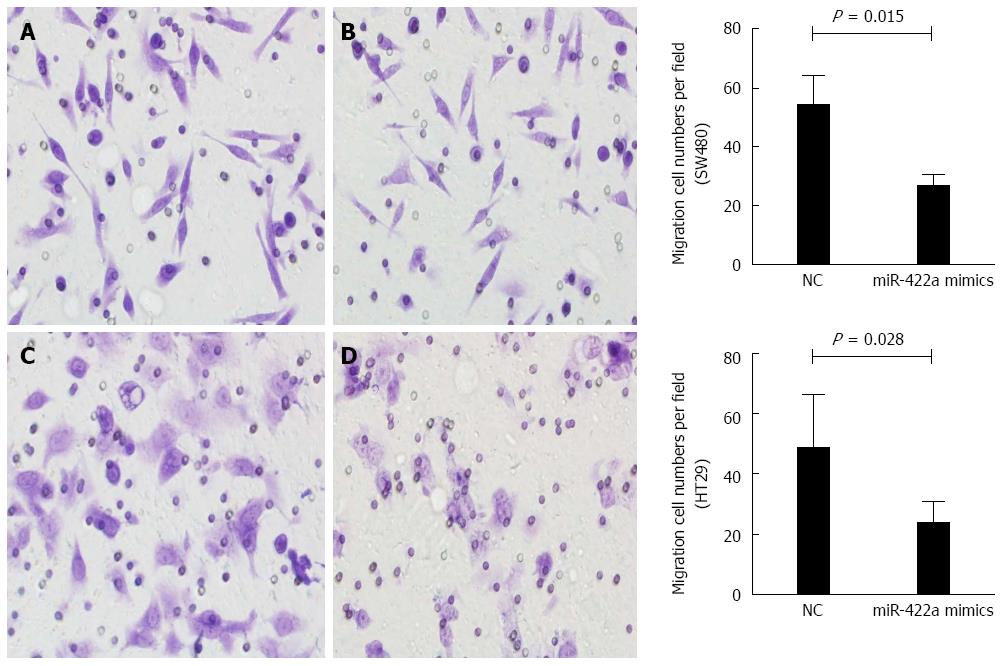
The transwell assays with and without Matrigel were performed to determine the effects of miR-422a on the migration and invasiveness of CRC cells. Overexpression of miR-422a significantly inhibited the migration of SW480 (P = 0.017; Figure 5A and B) and HT-29 cells (P = 0.007; Figure 5C and D). Furthermore, transwell assay with Matrigel showed that the invasiveness of these cells was significantly suppressed in cells transfected with miR-422a compared with miR control (Figure 6A-D).
Tumor progression in CRC is a multi-step process involving a large number of genetic and epigenetic alterations[17-19]. Numerous studies have showed that miRNA target genes are involved in CRC tumorigenesis[6-8,20]. Currently, there is an ongoing effort to elucidate new deregulated miRNAs and their roles in CRC progression.
miR-422a is encoded by gene MIR422A on 15q22.31 (64, 163, 129-64, 163, 218bp). Recently, several studies have studied the functional role of miR-422a in osteosarcoma, osteoporosis, HIV infection, and other tumor types[21-25]. Our previous study demonstrated that serum miR-422a was significantly downregulated in CRC patients and is a potential biomarker with a high diagnostic accuracy[15]. However, the expression and role of miR-422a in CRC tissues, as well as its clinicopathological and prognostic value in CRC tumorigenesis remain undefined. In this study, our results demonstrated that miR-422a was down-regulated in CRC tissues compared with normal mucosa, which is consistent with a previous study[14]. We also found that the expression of miR-422a was significantly associated with lymph node metastasis and clinical stage in CRC patients, suggesting that down-regulation of miR-422a might participate in CRC progression. To further explore the value of miR-422a as a prognostic factor, we investigated the correlation between miR-422a and overall survival in CRC patients. Our results showed that patients with low expression of miR-422a and decreased miR-422a expression compared with normal mucosa had poorer survival, suggesting that miR-422a could serve as an independent prognostic factor in CRC patients. Overall, these results suggest that miR-422a plays a protective role against CRC and could be used to evaluate prognosis in CRC patients.
Until now, few studies have described the function of miR-422a[13,22,26]. A study has demonstrated that miR-422a and the mismatch repair protein Mutlα (MLH1) are involved in a feedback loop, leading to genome instability and tumorigenesis[13]. Another study showed that miR-422a significantly modulated the efficacy of IFN-α/RBV treatment in vivo. Exogenous IFN-α treatment led to decreased miR-422a and likely contributed to the IFN-mediated suppression of HIV-1, suggesting that restoring miR-422a treatment could be a potential therapeutic strategy for HIV-1 infection[26]. Moreover, overexpression of human telomerase reverse transcriptase (hTERT) has been associated with the invasion and metastasis of CRC cells. miR-138-5p and miR-422a were found to be hTERT-targeting miRNAs, potentially inhibiting hTERT expression[26]. Invasion and migration are required for tumor cells to spread from the primary site to lymph or blood vessels. In the present study, we found that miR-422a expression was associated with lymph node metastasis and overexpression of miR-422a inhibited CRC cell proliferation, migration and invasion. These results suggest that multiple signaling pathways regulating different aspects of tumorigenesis may be regulated by miR-422a expression, suggesting that miR-422a may be a new therapeutic target to repress cancer progression.
A growing number of studies have shown that deregulated miRNAs play critical roles in tumorigenesis and progression of colorectal cancer (CRC). Nevertheless, an ongoing effort to elucidate new deregulated miRNAs and their roles in CRC is still urgently needed. Several studies have demonstrated the importance of miR-422a in different types of human diseases. In our previous study, miR-422a was found to be down-regulated in serum and could be used as a potential biomarker for CRC. Therefore, it is necessary to further determine the expression of miR-422a in CRC tissues and to explore its clinicopathological, prognostic value and role in CRC carcinogenesis.
Several studies have determined the importance of miR-422a in human diseases such as cancer, multiple sclerosis, and postmenopausal osteoporosis. miR-422a could inhibit signaling pathways regulating tumor cell proliferation in osteosarcoma. It was also shown that miR-422a targeted key mismatch repair protein (MutLα) by suppressing MLH1 expression, resulting in genome instability and tumorigenesis. Previously, we identified a serum 4-miRNA panel that included miR-19a-3p, miR-92a-3p, miR-223-3p, and miR-422a. This miRNA panel served as biomarkers for early diagnosis of colorectal adenocarcinoma.
The present study indicated that down-regulation of miR-422a might serve as an independent prognosis factor in CRC. miR-422a functions as a tumor suppressor and regulates progression of CRC.
By understanding the differential expression of miR-422a in CRC patients and its relationship with clinicopathological characteristics, prognosis and in vitro function, the present study may provide a new prognostic factor of CRC and further reveal the mechanism of miR-422a participating in CRC carcinogenesis.
MicroRNAs are a class of short non-coding RNAs that regulate up to 30% of human genes and play a pivotal role in various cellular processes. A growing number of miRNAs have been implicated in the initiation and progression of tumors including miR-422a. miR-422a is encoded by gene MIR422A on 15q22.31 (64,163,129-64,163,218bp), which has been found to be important in human diseases such as cancer, multiple sclerosis, and postmenopausal osteoporosis.
The authors presented a study analyzing the prognostic value and function of miR-422a in CRC carcinogenesis. The results demonstrated that miR-422a was dramatically reduced in CRC tissues and significantly correlated with local invasion and lymph node metastasis. miR-422a expression was an independent prognostic factor for overall survival. In vitro experiments showed that overexpression of miR-422a inhibited the proliferation, migration, and invasion of SW480 and HT-29 cells. These data indicated that miR-422a might serve as an independent prognosis factor and regulate progression of CRC by functioning as a tumor suppressor.
| 1. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21466] [Article Influence: 1951.5] [Reference Citation Analysis (6)] |
| 2. | Haggar FA, Boushey RP. Colorectal cancer epidemiology: incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22:191-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1215] [Cited by in RCA: 1424] [Article Influence: 94.9] [Reference Citation Analysis (0)] |
| 3. | Lieberman DA. Clinical practice. Screening for colorectal cancer. N Engl J Med. 2009;361:1179-1187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 192] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 4. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16308] [Article Influence: 959.3] [Reference Citation Analysis (2)] |
| 5. | Mirghasemi A, Taheriazam A, Karbasy SH, Torkaman A, Shakeri M, Yahaghi E, Mokarizadeh A. Down-regulation of miR-133a and miR-539 are associated with unfavorable prognosis in patients suffering from osteosarcoma. Cancer Cell Int. 2015;15:86. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 6. | Xu W, Liu M, Peng X, Zhou P, Zhou J, Xu K, Xu H, Jiang S. miR-24-3p and miR-27a-3p promote cell proliferation in glioma cells via cooperative regulation of MXI1. Int J Oncol. 2013;42:757-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 101] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 7. | Lin C, Huang F, Li QZ, Zhang YJ. miR-101 suppresses tumor proliferation and migration, and induces apoptosis by targeting EZH2 in esophageal cancer cells. Int J Clin Exp Pathol. 2014;7:6543-6550. [PubMed] |
| 8. | Yunqiao L, Vanke H, Jun X, Tangmeng G. MicroRNA-206, down-regulated in hepatocellular carcinoma, suppresses cell proliferation and promotes apoptosis. Hepatogastroenterology. 2014;61:1302-1307. [PubMed] |
| 9. | Wang J, Zhang X, Wang L, Yang Y, Dong Z, Wang H, Du L, Wang C. MicroRNA-214 suppresses oncogenesis and exerts impact on prognosis by targeting PDRG1 in bladder cancer. PLoS One. 2015;10:e0118086. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 10. | Kavitha N, Vijayarathna S, Jothy SL, Oon CE, Chen Y, Kanwar JR, Sasidharan S. MicroRNAs: biogenesis, roles for carcinogenesis and as potential biomarkers for cancer diagnosis and prognosis. Asian Pac J Cancer Prev. 2014;15:7489-7497. [PubMed] |
| 11. | Lyra-González I, Flores-Fong LE, González-García I, Medina-Preciado D, Armendáriz-Borunda J. MicroRNAs dysregulation in hepatocellular carcinoma: Insights in genomic medicine. World J Hepatol. 2015;7:1530-1540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 12. | Gougelet A, Pissaloux D, Besse A, Perez J, Duc A, Dutour A, Blay JY, Alberti L. Micro-RNA profiles in osteosarcoma as a predictive tool for ifosfamide response. Int J Cancer. 2011;129:680-690. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 109] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 13. | Mao G, Lee S, Ortega J, Gu L, Li GM. Modulation of microRNA processing by mismatch repair protein MutLα. Cell Res. 2012;22:973-985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 14. | Faltejskova P, Svoboda M, Srutova K, Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K, Kiss I. Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med. 2012;16:2655-2666. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 15. | Zheng G, Du L, Yang X, Zhang X, Wang L, Yang Y, Li J, Wang C. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer. 2014;111:1985-1992. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 125] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 16. | Zheng G, Wang H, Zhang X, Yang Y, Wang L, Du L, Li W, Li J, Qu A, Liu Y. Identification and validation of reference genes for qPCR detection of serum microRNAs in colorectal adenocarcinoma patients. PLoS One. 2013;8:e83025. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 99] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 17. | Guda K, Veigl ML, Varadan V, Nosrati A, Ravi L, Lutterbaugh J, Beard L, Willson JK, Sedwick WD, Wang ZJ. Novel recurrently mutated genes in African American colon cancers. Proc Natl Acad Sci USA. 2015;112:1149-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 101] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 18. | Vaiopoulos AG, Athanasoula KCh, Papavassiliou AG. Epigenetic modifications in colorectal cancer: molecular insights and therapeutic challenges. Biochim Biophys Acta. 2014;1842:971-980. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 19. | Bardhan K, Liu K. Epigenetics and colorectal cancer pathogenesis. Cancers (Basel). 2013;5:676-713. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 193] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 20. | Li J, Du L, Yang Y, Wang C, Liu H, Wang L, Zhang X, Li W, Zheng G, Dong Z. MiR-429 is an independent prognostic factor in colorectal cancer and exerts its anti-apoptotic function by targeting SOX2. Cancer Lett. 2013;329:84-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 21. | Cao Z, Moore BT, Wang Y, Peng XH, Lappe JM, Recker RR, Xiao P. MiR-422a as a potential cellular microRNA biomarker for postmenopausal osteoporosis. PLoS One. 2014;9:e97098. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 22. | Abdel-Mohsen M, Deng X, Danesh A, Liegler T, Jacobs ES, Rauch A, Ledergerber B, Norris PJ, Günthard HF, Wong JK. Role of microRNA modulation in the interferon-α/ribavirin suppression of HIV-1 in vivo. PLoS One. 2014;9:e109220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 23. | Bidzhekov K, Gan L, Denecke B, Rostalsky A, Hristov M, Koeppel TA, Zernecke A, Weber C. microRNA expression signatures and parallels between monocyte subsets and atherosclerotic plaque in humans. Thromb Haemost. 2012;107:619-625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 99] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 24. | Siegel SR, Mackenzie J, Chaplin G, Jablonski NG, Griffiths L. Circulating microRNAs involved in multiple sclerosis. Mol Biol Rep. 2012;39:6219-6225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 142] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 25. | Song KH, Li T, Owsley E, Chiang JY. A putative role of micro RNA in regulation of cholesterol 7alpha-hydroxylase expression in human hepatocytes. J Lipid Res. 2010;51:2223-2233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 68] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Qin YZ, Xie XC, Liu HZ, Lai H, Qiu H, Ge LY. Screening and preliminary validation of miRNAs with the regulation of hTERT in colorectal cancer. Oncol Rep. 2015;33:2728-2736. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Voutsadakis IA S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Wang CH