Published online Jan 14, 2015. doi: 10.3748/wjg.v21.i2.616
Peer-review started: March 12, 2014
First decision: April 2, 2014
Revised: May 10, 2014
Accepted: July 22, 2014
Article in press: July 22, 2014
Published online: January 14, 2015
Processing time: 312 Days and 23.2 Hours
AIM: To evaluate the association of known copy number variations (CNVs) in ulcerative colitis (UC) progressing to colorectal cancer.
METHODS: Microsatellite instability analysis using the National Cancer Institute’s panel of markers, and CNV association studies using Agilent 2 × 105 k arrays were done in tissue samples from four patient groups with UC: those at low risk (LR) or high risk of developing colorectal cancer, those with premalignant dysplastic lesions, and those with colitis-associated colorectal cancer (CAC). DNA from tissue samples of these groups were independently hybridized on arrays and analyzed. The data obtained were further subjected to downstream bioinformatics enrichment analysis to examine the correlation with CAC progression.
RESULTS: Microarray analysis highlighted a progressive increase in the total number of CNVs [LR (n = 178) vs CAC (n = 958), 5.3-fold], gains and losses [LR (n = 37 and 141) vs CAC (n = 495 and 463), 13.4- and 3.3-fold, respectively], size [LR (964.2 kb) vs CAC (10540 kb), 10.9-fold] and the number of genes in such regions [LR (n = 119) vs CAC (n = 455), 3.8-fold]. Chromosome-wise analysis of CNVs also showed an increase in the number of CNVs across each chromosome. There were 38 genes common to all four groups in the study; 13 of these were common to cancer genes from the Genetic Disease Association dataset. The gene set enrichment analysis and ontology analysis highlighted many cancer-associated genes. All the samples in the different groups were microsatellite stable.
CONCLUSION: Increasing numbers of CNVs are associated with the progression of UC to CAC, and warrant further detailed exploration.
Core tip: Ulcerative colitis (UC) confers an increased risk of colorectal cancer (CRC). The role of copy number variations (CNVs) in different cancers including sporadic CRC has been established but their association in the development of colitis-associated neoplasia is not well described. Reports to date are limited to only a particular stage (e.g., dysplasia or cancer) in the development of colitis-associated cancer. In this first study of its kind, we report the association of increased numbers of known CNVs with the progression of UC to colitis-associated cancer.
- Citation: Shivakumar BM, Rotti H, Vasudevan TG, Balakrishnan A, Chakrabarty S, Bhat G, Rao L, Pai CG, Satyamoorthy K. Copy number variations are progressively associated with the pathogenesis of colorectal cancer in ulcerative colitis. World J Gastroenterol 2015; 21(2): 616-622
- URL: https://www.wjgnet.com/1007-9327/full/v21/i2/616.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i2.616
Longstanding ulcerative colitis (UC) confers an increased risk of colorectal cancer (CRC)[1-6]. The frequency of colitis-associated colorectal cancer (CAC) in the Asia-Pacific region has been variously reported to be similar to or lower than that reported from the West, mostly based on retrospective studies[6-12]. Recent data from this low prevalence region suggest that premalignant lesions may not be uncommon in patients with longstanding UC, if dysplasia is methodically looked for[13].
The inflammation-dysplasia-carcinoma sequence defines carcinogenesis in UC and is accompanied by a series of molecular changes[14]. The major molecular pathways in the development of CRC involve chromosomal instability (CIN) and microsatellite instability (MSI), which are associated with an increase in the range of gene expression and phenotypic changes[15,16]. Studies have found that around 80% of the tumors with MSI have a near-diploid karyotype and a distinct genetic alteration distinguishable from those of microsatellite stable cancers[17].
Copy number variations (CNVs), a source of genetic diversity in humans under CIN and affecting gene dosage, are also believed to play a major role in human health and disease. CNVs can lead to altered expression of genes thereby contributing to cancer development. Profiling these can help in identifying tumor suppressor genes and oncogenes[18]. In the past decade, studies have established that even though common CNVs with low penetrance levels contribute only minimally or modestly to the causation of cancer, their collective impact on the predisposition to cancer must be considered while estimating the cancer risk[19]. CNVs constitute important genetic changes in various cancers including sporadic CRC, but their association with neoplasia in UC is not well described[20-23].
Copy number alterations detected by array-based Comparative Genomic Hybridization (aCGH) can be directly related to discovery of the underlying genes and/or molecular mechanisms involved with tumorigenesis, especially so with high or moderate penetrant CNVs[24,25]. Such discovery of altered regions associated with cancer may help in classifying the cancer patient at the molecular level along with the clinico-pathological features. With this background, the present study was aimed at elucidating the CNVs associated with the pathogenesis of CAC, a complex disease.
This study was approved by the Ethics Committee of Kasturba Hospital, Manipal. All the patients provided informed consent before participation.
Patients with UC were recruited into 4 groups: UC-low risk (LR): UC patients with disease duration less than 7 years; UC-high risk (HR): UC patients with disease duration more than 7 years in case of extensive colitis or more than 10 years for left sided colitis; UC-premalignant (PM): UC patients who had any type of dysplasia (low grade or high grade); and UC-CAC: UC patients who were found to have cancer. Fresh biopsy specimens were immediately digested with proteinase K (0.1 mg/mL) in the presence of 1% sodium dodecyl sulphate (Sigma-Aldrich, United States). DNA was extracted using phenol-chloroform, followed by ethanol precipitation. DNA was checked for purity and stored at -20 °C until further analysis.
MSI status was examined using 5 microsatellite markers [National Cancer Institute (NCI), Bethesda Panel]. The assay was carried out using appropriate primer sequences and the corresponding fluorescent dyes and polymerase chain reaction as described elsewhere[26].
DNA from appropriate colonic tissue samples in these groups were independently hybridized on 2 × 105 k CNV association microarray slides (Agilent Technologies, CA, United States) and analyzed according to the manufacturer’s protocol. Briefly, genomic DNA samples were sheared using a cycle of 15 s “on” and 15 s “off” for 15 min in an ultrasonic processor (Thomas Scientific, NJ, United States) with a 2 mm probe with amplitude set at 40. The purified sheared DNA samples were differentially labeled; test DNA (test genome) with fluorescent Cy5 and the pooled normal reference DNA (reference genome) with Cy3 dyes. Hybridization, washing and scanning of the arrays were performed according to the manufacturer’s protocol. Feature extracted data were analyzed with Genomic Workbench v5.0 software (Agilent Technologies, CA, United States) using the ADM-2 aberration detection algorithm (threshold 5.0) and the log2 ratios (± 0.25) as cut-off values with genomic boundaries switched on as track file of 022837. All genomic data reported in the present study were based on NCBI build 36 (hg18) of the human genome[27].
Bioinformatics scanning approaches such as DAVID, Gene Set Enrichment Analysis (GSEA), Genetic Disease Association dataset (GAD), etc., were used to explore the significance of a large variety of biological mechanisms and functional importance including associations with various cancer datasets in order to find the important set of enriched genes with significant functions in developing CRC.
Samples were included from the following patient groups: UC-LR: n = 20; 10 male, 10 female, median age: 42 years; UC-HR: n = 20; 10 male, 10 female, median age: 45 years; UC-PM: n = 6; 4 male, 2 female, median age: 41 years; UC-CAC: n = 2; 1 male, 1 female, median age: 38 years. Subjects undergoing colonoscopy and found to have a normal examination and normal histology (n = 20; 10 male, 10 female, median age: 49 years) were included as controls. There was no statistically significant difference between the different groups in terms of age and sex in the study.
Samples in all the groups did not show any instability in the microsatellites analyzed and all were microsatellite stable.
The number of CNV regions progressively increased by up to 5-fold with advancing stages of disease (LR to CAC): 178 in LR, 271 in HR, 616 in PM and 958 in CAC. CNV coverage (size) was found to increase 10-fold with progressive stages from LR (total of 964 kb) to CAC (10540 kb). While the number of CNV regions showing gains increased with the advancing stages of disease, regions showing loss did not follow any particular pattern (Table 1). The number of genes encompassed within the CNV regions in each group increased substantially from 119 (LR) to 455 (CAC).
| UC-LR | UC-HR | UC-PM | CAC | |
| Total number of CNVs | 178 | 271 | 616 | 958 |
| Number of CNVs with gain | 37 | 190 | 465 | 495 |
| Number of CNVs with loss | 141 | 81 | 151 | 463 |
| Overall CNV coverage (in kb) | 964.2 | 2368.5 | 4875 | 10540.1 |
| Number of genes within CNVs | 119 | 141 | 318 | 455 |
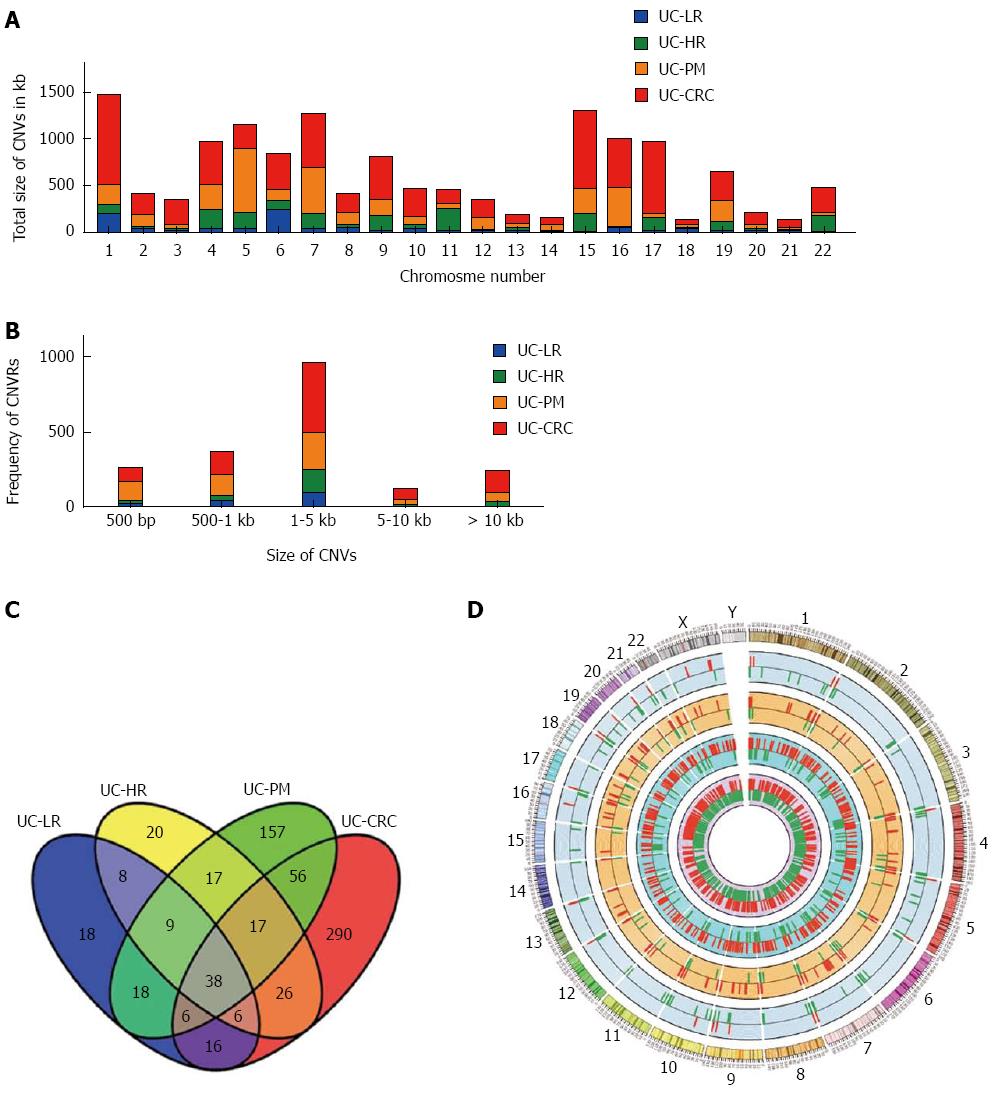
The chromosome-wise distribution of gains and losses of CNVs also showed an increase in number and size with disease progression (Figure 1A and D). The average number of CNVs per chromosome was < 5 in LR, increasing to > 30 in CAC. In addition, only two chromosomes (1 and 6) had altered CNVs > 100 kb in length in LR, while in CAC all but chromosomes 14, 18 and 21 harbored CNVs > 100 kb in length. LR showed individual CNVs up to 5 kb in size, but in the premalignant and malignant samples most CNVs were above the 5 kb range (Figure 1B).
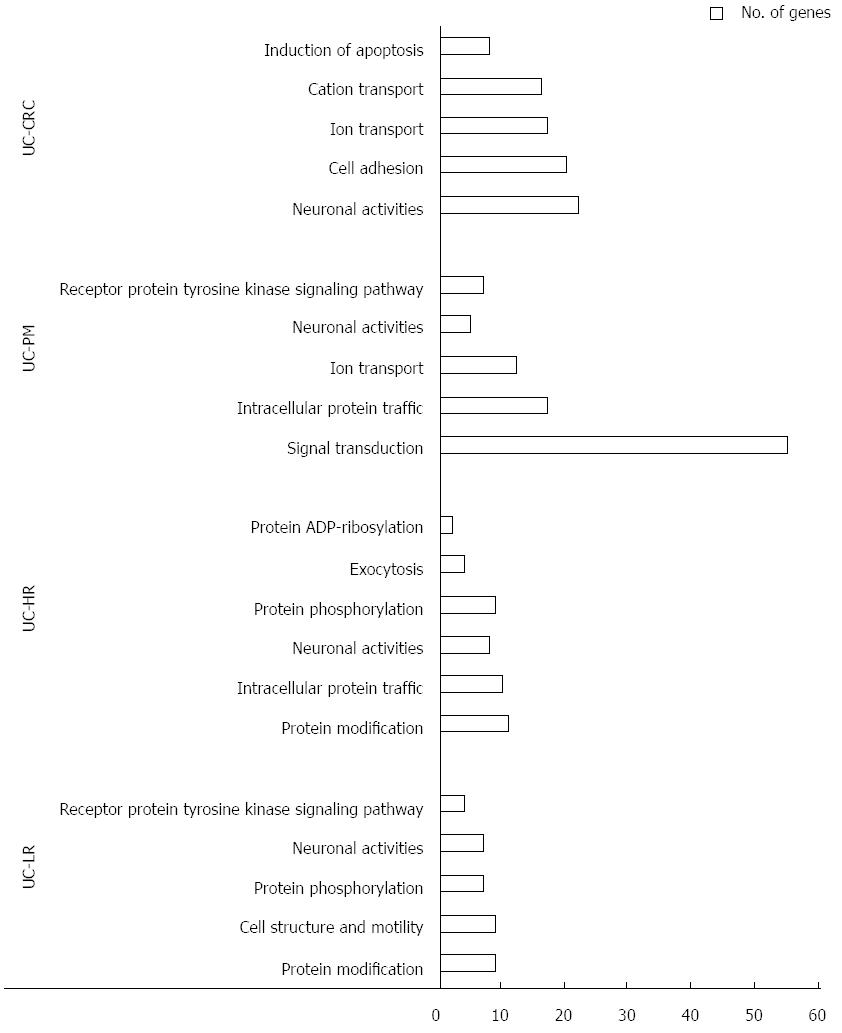
To gain further insight of these CNV regions and their functional significance, we analyzed chromosomal gains and losses across all 4 sample groups using various computational tools and databases. By using Venny tool analysis, 38 genes were found common to LR, HR, PM and CAC (Figure 1C). Common genes analysis using Venny for our 4 groups of genes with the reported human GAD genes specific to cancer yielded 13 genes. GSEA analysis for genes from the CNV regions showed an increase in the functionally significant families of genes, such as transcription factors, oncogenes and other cancer-related genes (Table 2). Gene ontology analysis also showed an increased enrichment of the genes involved in extracellular biological processes among the 4 groups of samples (Figure 2).
| UC-LR | UC-HR | UC-PM | CAC | |
| Cytokines and growth factors | 0 | 0 | 2 | 2 |
| Transcription factors | 7 | 8 | 18 | 25 |
| Cell differentiation markers | 2 | 2 | 6 | 8 |
| Protein kinases | 7 | 8 | 9 | 8 |
| Translocated cancer genes | 1 | 4 | 5 | 11 |
| Oncogenes | 2 | 5 | 7 | 11 |
| Tumor suppressor | 0 | 0 | 0 | 1 |
The major aim of this study was to analyze the comprehensive association of known CNVs during various stages of UC progressing to CRC and thereby to understand the role of CNVs in inflammation-associated cancer development. The study, one of the first of its kind using 2 × 105 k CNV association arrays on DNA extracted from tissue samples has shown a progressive involvement of CNVs at different levels as the disease progresses from UC to CAC. Increasing numbers of CNVs were found to be associated with the progression of the disease from earlier stages to cancer. Other factors such as the size of CNVs and number of genes from these CNV regions were similarly found to be correlated with neoplastic progression. Bioinformatics enrichment analysis of CNV genes also enumerated putative functionally important cancer-associated genes. Hence, the study highlights the importance of classifying UC patients into subgroups at various stages of progression using clinical details in the evaluation of molecular patho-mechanisms involved in CAC.
The 2 × 105 k array has been successfully used in large sample-based studies for CNV association in common diseases[28]. A population-specific array (2 × 105 k CNV association array) with a target of analyzing the association of CNVs has shown promising results albeit with a limited number of inflammation-related and cancer-coordinating genes in the study. Being a tissue based study, the cells, and the CNVs therein being heterogeneous because of the pooling of the samples, could probably have affected the assessment of CNVs[29]. In this aspect, our results from tissue DNA samples have identified the CNV regions and the important genes situated in them that are associated with various stages of progression of UC to CAC. Unlike the present report, earlier studies on colitis-associated neoplasia used conventional methods (chromosomal genomic hybridization or bacterial artificial chromosome arrays) and were limited to only one stage of progression[21-23].
In another important observation, the genes encompassing these CNV regions matched with the cancer gene sets from various databases such as GAD or GSEA, and highlight the importance of these CNVs in carcinogenesis. Genes from amplified or gain CNV regions may act as oncogenes while the loss regions are likely to be embedded with tumor suppressor regions[18]. Gene ontology analysis further highlighted the significant number of genes involved in various molecular and biological functions from these CNV regions, increasing as the disease progressed to CAC. Thus, our data can also be used in future research to determine their definitive contribution to colorectal carcinogenesis, upon functional validation of genes from these CNV regions.
MSI is believed to play a role in the pathways of UC-associated and sporadic CRCs, contributing at a frequency of approximately 15%-20% compared to 80% CIN in the case of CRC[14]. The present study using the NCI panel of Bethesda markers found no instability in any of the samples. One reason for this difference could be the use of a cancer-specific panel and recruiting only 2 patients with CAC in the study[26]. Reports suggest that CIN is greater in microsatellite stable samples[17,30].
The study of CNVs and cancer is in its infancy, but recent advancements in and the availability of technology is ensuring that more studies are being reported in this area. There is tremendous scope for further studies considering the effect of this form of genetic variation on cancer predisposition and the association with cancer genes. To our knowledge, this is the only study available till now on the association of CNVs with UC stratified into different stages of evolution to CAC. In a first study of its kind, using the association arrays of higher resolution on tissue samples we have demonstrated the progressive changes in CNVs as UC advances to cancer, establishing the importance of such genomic alterations in the pathogenesis of CAC. These results clearly indicate a major role for CNVs in the pathogenesis of CAC, warranting further focused studies on the regions and genes identified.
The authors thank Agilent Corp (India) for their generosity in providing 2 × 105 k CNV association arrays.
Ulcerative colitis (UC) patients have a higher risk of developing cancer with increased duration of disease. The epidemiological data of colitis-associated colorectal cancer (CAC) in the Asia-Pacific region has been reported as being similar to or lower than that from the West. The accumulation of a series of molecular changes that accompany the progressive pathological changes in the inflammation-dysplasia-carcinoma sequence in UC has been well established. The role of copy number variations (CNVs) in different cancers including sporadic colorectal cancer (CRC) has been evaluated but their association with the stages of development CAC is not well described.
CNVs in different cancers including sporadic CRC has been established but their association with the development of UC to CAC is not well described and previous studies are generally limited to a particular stage of disease. In this study, the authors have attempted to elucidate the association of CNVs with the pathogenesis of CAC carcinogenesis in a stage-wise manner for the first time.
Recent studies have highlighted the importance of structural variations such as CNVs in different cancers, including sporadic CRC, but there is a lack of similar types of studies in CAC. This is the only study to date that has stratified UC patients as low risk, high risk, premalignant and malignant for evaluating their CNV association. Progressive changes in CNVs are shown as UC advances to cancer establishing the importance of such genomic alterations in the pathogenesis of CAC.
By exploring and understanding these CNVs in the progression of CRC in UC through various stages, this study paves the way for future studies to evaluate the contribution of specific genes in colorectal carcinogenesis, with potential future possibilities for them as diagnostic markers and possible therapeutic targets.
CNVs: Structural variations in the genome of approximately 1 kb or larger in size, including genomic imbalances such as amplifications and deletions, balanced translocations or inversions, are altogether universally referred to as CNVs. MSI: Microsatellites are repeated sequences of DNA and instability is the result of defective mismatch repair in the cells which is more commonly found in cancerous cells.
In this study, the authors reported CNVs in the tissue samples from various stages of progression of UC through to CAC, and known CNVs were found to be increasingly associated with the progression of UC to CAC. Overall, these findings are well written with interests.
| 1. | Eaden JA, Abrams KR, Mayberry JF. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. 2001;48:526-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1985] [Cited by in RCA: 2128] [Article Influence: 85.1] [Reference Citation Analysis (2)] |
| 2. | Xie J, Itzkowitz SH. Cancer in inflammatory bowel disease. World J Gastroenterol. 2008;14:378-389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 215] [Cited by in RCA: 227] [Article Influence: 12.6] [Reference Citation Analysis (1)] |
| 3. | Jess T, Rungoe C, Peyrin-Biroulet L. Risk of colorectal cancer in patients with ulcerative colitis: a meta-analysis of population-based cohort studies. Clin Gastroenterol Hepatol. 2012;10:639-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 581] [Cited by in RCA: 691] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 4. | Farraye FA, Odze RD, Eaden J, Itzkowitz SH. AGA technical review on the diagnosis and management of colorectal neoplasia in inflammatory bowel disease. Gastroenterology. 2010;138:746-774, 774.e1-e4; quiz e12-e13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 345] [Article Influence: 21.6] [Reference Citation Analysis (1)] |
| 5. | Itzkowitz SH, Present DH. Consensus conference: Colorectal cancer screening and surveillance in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:314-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 431] [Cited by in RCA: 397] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 6. | Ooi CJ, Fock KM, Makharia GK, Goh KL, Ling KL, Hilmi I, Lim WC, Kelvin T, Gibson PR, Gearry RB. The Asia-Pacific consensus on ulcerative colitis. J Gastroenterol Hepatol. 2010;25:453-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 130] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 7. | Kim BJ, Yang SK, Kim JS, Jeen YT, Choi H, Han DS, Kim HJ, Kim WH, Kim JY, Chang DK. Trends of ulcerative colitis-associated colorectal cancer in Korea: A KASID study. J Gastroenterol Hepatol. 2009;24:667-671. [PubMed] |
| 8. | Kochhar R, Goenka MK, Kaushik SP, Gupta NM, Nagi B, Mehta SK. Colorectal carcinoma in Indian patients with idiopathic ulcerative colitis. Eur J Cancer Prev. 1992;1:293-296. [PubMed] |
| 9. | Ray G. Inflammatory bowel disease in India--changing paradigms. Int J Colorectal Dis. 2011;26:635-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 15] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 10. | Venkataraman S, Mohan V, Ramakrishna BS, Peter S, Chacko A, Chandy G, Kurian G, Kurian S, Mathan M, Mathan VI. Risk of colorectal cancer in ulcerative colitis in India. J Gastroenterol Hepatol. 2005;20:705-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 11. | Gong W, Lv N, Wang B, Chen Y, Huang Y, Pan W, Jiang B. Risk of ulcerative colitis-associated colorectal cancer in China: a multi-center retrospective study. Dig Dis Sci. 2012;57:503-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (1)] |
| 12. | Ramakrishna BS, Makharia GK, Abraham P, Ghoshal UC, Jayanthi V, Agarwal BK, Ahuja V, Bhasin DK, Bhatia SJ, Choudhuri G. Indian Society of Gastroenterology consensus on ulcerative colitis. Indian J Gastroenterol. 2012;31:307-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 13. | Shivakumar BM, Lakshmankumar B, Rao L, Bhat G, Suvarna D, Pai CG. Colorectal neoplasia in long-standing ulcerative colitis - a prospective study from a low-prevalence area. Colorectal Dis. 2013;15:e462-e468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 14. | Itzkowitz SH. Molecular biology of dysplasia and cancer in inflammatory bowel disease. Gastroenterol Clin North Am. 2006;35:553-571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 127] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 15. | Pancione M, Remo A, Colantuoni V. Genetic and epigenetic events generate multiple pathways in colorectal cancer progression. Patholog Res Int. 2012;2012:509348. [PubMed] |
| 16. | Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 2008;135:1079-1099. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 717] [Cited by in RCA: 730] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 17. | Lin CH, Lin JK, Chang SC, Chang YH, Chang HM, Liu JH, Li LH, Chen YT, Tsai SF, Chen WS. Molecular profile and copy number analysis of sporadic colorectal cancer in Taiwan. J Biomed Sci. 2011;18:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Krepischi AC, Pearson PL, Rosenberg C. Germline copy number variations and cancer predisposition. Future Oncol. 2012;8:441-450. [PubMed] |
| 19. | Kuiper RP, Ligtenberg MJ, Hoogerbrugge N, Geurts van Kessel A. Germline copy number variation and cancer risk. Curr Opin Genet Dev. 2010;20:282-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 85] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 20. | Trautmann K, Terdiman JP, French AJ, Roydasgupta R, Sein N, Kakar S, Fridlyand J, Snijders AM, Albertson DG, Thibodeau SN. Chromosomal instability in microsatellite-unstable and stable colon cancer. Clin Cancer Res. 2006;12:6379-6385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 76] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 21. | Aust DE, Willenbucher RF, Terdiman JP, Ferrell LD, Chang CG, Moore DH, Molinaro-Clark A, Baretton GB, Loehrs U, Waldman FM. Chromosomal alterations in ulcerative colitis-related and sporadic colorectal cancers by comparative genomic hybridization. Hum Pathol. 2000;31:109-114. [PubMed] |
| 22. | Willenbucher RF, Aust DE, Chang CG, Zelman SJ, Ferrell LD, Moore DH, Waldman FM. Genomic instability is an early event during the progression pathway of ulcerative-colitis-related neoplasia. Am J Pathol. 1999;154:1825-1830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 130] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 23. | van Dieren JM, Wink JC, Vissers KJ, van Marion R, Hoogmans MM, Dinjens WN, Schouten WR, Tanke HJ, Szuhai K, Kuipers EJ. Chromosomal and microsatellite instability of adenocarcinomas and dysplastic lesions (DALM) in ulcerative colitis. Diagn Mol Pathol. 2006;15:216-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 1.5] [Reference Citation Analysis (1)] |
| 24. | Shlien A, Malkin D. Copy number variations and cancer. Genome Med. 2009;1:62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 25. | Bernstein C, Bernstein H, Payne CM, Dvorak K, Garewal H. Field defects in progression to gastrointestinal tract cancers. Cancer Lett. 2008;260:1-10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 26. | Shivakumar BM, Kumar BL, Bhat G, Suvarna D, Rao L, Pai CG, Satyamoorthy K. Molecular alterations in colitis-associated colorectal neoplasia: study from a low prevalence area using magnifying chromo colonoscopy. J Crohns Colitis. 2012;6:647-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 12] [Article Influence: 0.9] [Reference Citation Analysis (1)] |
| 27. | Chakrabarty S, D’Souza RR, Bellampalli R, Rotti H, Saadi AV, Gopinath PM, Acharya RV, Govindaraj P, Thangaraj K, Satyamoorthy K. Comprehensive DNA copy number profile and BAC library construction of an Indian individual. Gene. 2012;500:186-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 28. | Craddock N, Hurles ME, Cardin N, Pearson RD, Plagnol V, Robson S, Vukcevic D, Barnes C, Conrad DF, Giannoulatou E. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 2010;464:713-720. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 664] [Cited by in RCA: 604] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 29. | Leedham SJ, Graham TA, Oukrif D, McDonald SA, Rodriguez-Justo M, Harrison RF, Shepherd NA, Novelli MR, Jankowski JA, Wright NA. Clonality, founder mutations, and field cancerization in human ulcerative colitis-associated neoplasia. Gastroenterology. 2009;136:542-550.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 136] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 30. | Boardman LA, Johnson RA, Petersen GM, Oberg AL, Kabat BF, Slusser JP, Wang L, Morlan BW, French AJ, Smyrk TC. Higher frequency of diploidy in young-onset microsatellite-stable colorectal cancer. Clin Cancer Res. 2007;13:2323-2328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Capasso R, Lee HW, Sier C, Wang K S- Editor: Ma N L- Editor: Cant MR E- Editor: Liu XM