Published online May 28, 2014. doi: 10.3748/wjg.v20.i20.6302
Revised: December 31, 2013
Accepted: January 20, 2014
Published online: May 28, 2014
Processing time: 206 Days and 20.2 Hours
AIM: To assess the distribution of human leukocyte antigen (HLA)-DQ2 and -DQ8 in Iranian celiac disease (CD) patients and compare them to healthy Iranian controls.
METHODS: To predict the HLA-DQA1 and -DQB1 genes, we used six previously reported HLA-tagging single nucleotide polymorphism to determine HLA genotypes in 59 Iranian patients with ‘biopsy-confirmed’ CD and in 151 healthy Iranian individuals. To test the transferability of the method, 50 cases and controls were also typed using a commercial kit that identifies individual carriers of DQ2, DQ7 and DQ8 alleles.
RESULTS: In this pilot study 97% of CD cases (n = 57) and 58% of controls (n = 87) were carriers of HLA-DQ2 and/or HLA-DQ8 heterodimers, either in the homozygous or heterozygous state. The HLA-DQ pattern of these 57 CD patients: heterozygous DQ2.2 (n = 14) and homozygous DQ2.2 (n = 1), heterozygous DQ2.5 (n = 33) and homozygous DQ2.5 (n = 8), heterozygous DQ8 (n = 13) and homozygous DQ8 (n = 2). Two CD patients were negative for both DQ2 and DQ8 (3%).
CONCLUSION: The prevalence of DQ8 in our CD population was higher than that reported in other populations (25.4%). As reported in other populations, our results underline the primary importance of HLA-DQ alleles in the Iranian population’s susceptibility to CD.
Core tip: We describe, for the first time, the distribution of human leukocyte antigen (HLA)-DQ2/DQ8 alleles in Iranian celiac disease patients compared to healthy Iranian controls. To assess this distribution we applied a single nucleotide polymorphism-based approach, which was developed in European (Caucasian) study samples. We also demonstrate the transferability of such an approach to the Iranian population. Our results underline the primary importance of HLA-DQ alleles in the Iranian population’s susceptibility to celiac disease.
- Citation: Rostami-Nejad M, Romanos J, Rostami K, Ganji A, Ehsani-Ardakani MJ, Bakhshipour AR, Zojaji H, Mohebbi SR, Zali MR, Wijmenga C. Allele and haplotype frequencies for HLA-DQ in Iranian celiac disease patients. World J Gastroenterol 2014; 20(20): 6302-6308
- URL: https://www.wjgnet.com/1007-9327/full/v20/i20/6302.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i20.6302
Celiac disease (CD) is characterized by malabsorption of nutrients in the small intestine after ingestion of wheat gluten or related proteins from rye and barley. The disease is characterized by villous atrophy of the small intestinal mucosa. With adherence to a strict gluten-free diet, most CD patients show prompt clinical and histological improvement, with relapse of the symptoms if gluten is re-introduced[1-3]. Similar to Western studies, a high prevalence of CD has been reported in Iranian general population (about 1%)[3].
Many studies reflect the importance of genetic factors in the pathogenesis of CD[3]. A recent study by Anderson et al[4] suggested that a combination of human leukocyte antigen (HLA) typing and confirmatory serology could decrease the number of unnecessary endoscopies.
The HLA-DQ2 heterodimer, which is coded by alleles DQA1*05 and DQB1*02, is present in the vast majority of CD patients including approximately 95% of CD patients of Northern-European descent[4]. The most important risk factor for CD is the DQ2.5 haplotype[4]. Alpha and beta chains of this heterodimer are encoded together in cis (DQA1*05 and DQB1*02) on a DRB1*03 haplotype. Other DQ heterodimers that are encoded in trans on DR7 haplotypes are related to the DQ2.2 haplotype (DQA1*0201/DQB1*02)[5-13]. Those CD patients who do not carry the DQ2 heterodimer, and are encoded by the DR4 haplotype, carry HLA-DQ8 (α1*0301, β1*0302)[4]. HLA-DQ2 is common in Europeans and is expressed in 25%-30% of the healthy European population. Most of the CD patients who do not carry either DQ2.5 or DQ8, carry half of the DQ2.5 or DQ2.2 molecule (that is either HLA-DQA1*05 or HLA-DQB1*02), suggesting that carrying even some of the risk molecules still has functional implications for the risk of CD[13]. The majority of studies of HLA in CD are focused on Caucasian populations and detailed information on patient cohorts from diverse ethnic groups is lacking.
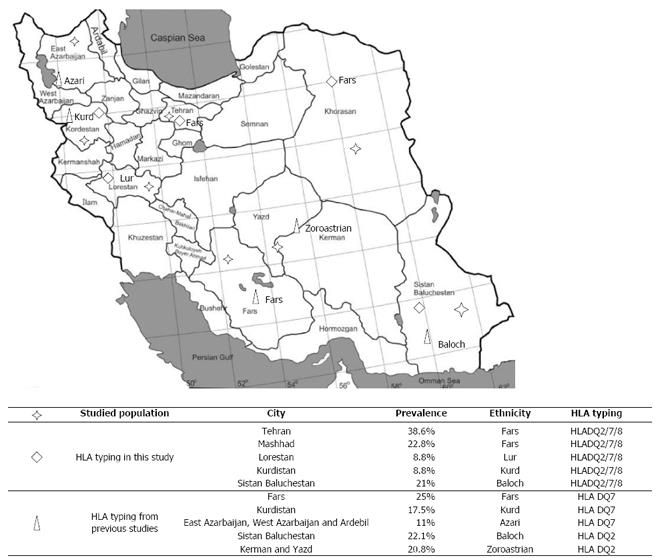
There are a number of ethnic groups in Iran, including Persians (Fars), Kurds, Lours, Arabs, Turkmen, Balochs, and Turks. Most Iranians are Muslims but Zoroastrians, Jews, Armenians, and Nestorians also live in this large country. Based on previous studies in Iran, the most common HLA haplotype in the different ethnic groups is DQ2 and DQ7, with reported prevalences of 22.1% and 25%, respectively[14-17] (Figure 1). These studies were performed in healthy populations in different parts of the country and found considerable similarities in the distribution of HLA class II haplotypes with European countries. However, Iranian CD patients have never been studied for HLA.
Recently, Monsuur et al[13] described an HLA-tagging single nucleotide polymorphism (SNP) method for detecting the HLA risk alleles for CD. Using six SNPs, HLA genotyping for specific CD genotypes could be performed in a high-throughput mode. The results of Monsuur et al[13] and Koskinen et al[12] showed that the sensitivity and specificity of this test in recognizing DQ2.2, DQ2.5, DQ7, and DQ8 haplotypes was > 99% in the Dutch population, and > 98% in the United Kingdom, Spanish, Finnish, Hungarian, and Italian populations.
The main aim of this study was to provide new information on HLA-DQ associations with CD in the Iranian patients, which has never been investigated before, by using the tagging SNP method.
A total of 59 CD patients (21 males and 38 females, median age 30.5 years, range 7-67 years) were included: 12 patients from East Iran (Zahedan Province, Baloch ethnicity), 13 from North Iran (Mashhad Province, Fars), 10 from West Iran (Lorestan and Kurdistan Provinces, Lour and Kurd ethnic groups), and 24 patients from Tehran, in the center of Iran (Fars). All patients had positive tTGA and/or EMA antibodies and histology according to the Rostami-Marsh classification (Marsh I-IIIc)[18].
A total of 151 healthy controls (78 females and 73 males, median age 32.7 years, range 5-83 years) were selected from those who were negative for CD serological screening in the study areas (25 from Zahedan Province, 30 from Mashhad, 26 from Lorestan and Kurdistan provinces, and 70 from Tehran) and matched by age and sex. None of them had a personal or family history of cancer or autoimmune diseases. The study was approved by the ethical committees of the Gastroenterology and Liver Diseases Research Centers, Shahid Beheshti University of Medical Sciences, Tehran, and all the participants (or their parents/guardians) were informed about the study according to the study protocol and gave their written informed consent.
DNA was extracted using the phenol chloroform method[19]. The six HLA-tagging SNPs reported by Monsuur et al[13] were genotyped using 5 ng of DNA from the Iranian participants, and using TaqMan chemistry and custom assays from Applied Biosystems (ABI, Foster City, CA, United States, http://www.appliedbiosystems.com), including DQ2.2 (rs2395182 and rs7775228 SNPs for DQ2.2, and rs4713586 SNP to exclude DQ4 from the DQ2.2 group), and SNPs rs2187668 (DQ2.5), rs4639334 (DQ7), and rs7454108 (DQ8). DNA was genotyped using a standard protocol provided by Applied Biosystems. The PCR assays and allelic discrimination were run using an ABI PRISM 7900HT Sequence Detection System (SDS) machine. All individuals were run on the same 384-well plate to avoid biased results due to technical issues. The data was analyzed using SDS program 2.3 (Applied Biosystems).
The HLA alleles for 20 CD cases and 30 controls were also typed using a commercial kit (BAG, Germany) which is commonly used in general diagnostic laboratories. The BAG kit makes it possible to identify individuals who are carriers of the DQB1*02, DQB1*0301, DQA1*05 or DQA1*0201 alleles for HLA-DQ2; DQB1*0302 or DQA1*03 for HLA-DQ8; and DQB1*0301 for HLA-DQ7. We could thus distinguish DQ2.5-positive individuals from those who have DQ2.2.
For the prediction of HLA alleles, we inferred the DQ types from the tag SNPs according to the method described by Monsuur et al[13]. Only individuals with complete histology and serology data were included in this study (59 cases). The statistical analysis was performed using the F-test. The correlation between HLA risk alleles and Marsh classification was also investigated. A P-value less than 0.05 was considered statistically significant. Power calculations were performed using the Quanto program (http://hydra.usc.edu/gxe/) assuming allele frequencies of 0.15-0.30 and an odds ratio of 6.
We first evaluated the 59 CD patients and 151 controls for prediction of related HLA genotypes by TaqMan SNPs. We observed that 83.03% of cases and 35.09% of controls were carriers of an HLA-DQ2 heterodimer, either in homozygous or heterozygous state. The patterns of the specific HLA-DQ haplotypes in the CD patients were as follows: 25.4% of CD patients were heterozygous DQ2.2 including DQ2.2/DQ2.5 compound heterozygosity, 1.7% of CD patients were homozygous DQ2.2/DQ2.2, 56% of CD patients were heterozygous DQ2.5 including DQ2.5/DQ8 compound heterozygosity, and 13.5% of CD patients were homozygous DQ2.5/DQ2.5. DQ8 heterozygosity was predicted in 15 patients, of which seven were DQ8/DQ2.5 carriers and two were homozygous DQ8/DQ8. The HLA-DQ pattern in the controls was as follows: heterozygous DQ2.2 in 28.5%, non-homozygous DQ2.2/DQ2.2, and heterozygous DQ2.5 in 16.5%, and homozygous DQ2.5/DQ2.5 in one. DQ8 was positive in the heterozygous state in 30 (19.9%) controls and in the homozygous state in four controls.
Table 1 shows the frequency of HLA-related CD genotypes in the healthy controls and the prevalence of the different HLA-related CD genotypes in our Iranian CD patients. DQ2.2 and DQ7 can only confer risk of CD when both are present together or with DQ2.5[13]. In our cohort, DQ2.2 and DQ7 haplotypes were present together or with DQ2.5 in 23.7% of CD patients and 11.9% of controls. A DQ2-DQ8-negative genotype was seen in 66 controls and two CD cases.
| Genotypes | Cases | Controls | P-value |
| DQ2.2/DQ2.2 | 1 (1.7) | 0 (0) | 0.28 |
| DQ2.2/DQ7 | 4 (6.8) | 8 (5.3) | 0.76 |
| DQ2.2/DQX | 3 (5.08) | 22 (14.5) | 0.06 |
| DQ2.5/DQ2.2 | 7 (11.9) | 5 (3.3) | 0.04 |
| DQ2.5/DQ2.5 | 8 (13.6) | 1 (0.6) | < 0.001 |
| DQ2.5/DQ7 | 3 (5.08) | 5 (3.3) | 0.69 |
| DQ2.5/DQ8 | 7 (11.9) | 5 (3.3) | 0.04 |
| DQ2.5/DQX | 16 (27.1) | 10 (6.6) | < 0.001 |
| DQ7/DQ7 | 0 (0) | 17 (11.2) | 0.004 |
| DQ7/DQX | 0 (0) | 17 (11.2) | 0.004 |
| DQ8/DQ2.2 | 0 (0) | 8 (5.3) | 0.11 |
| DQ8/DQ7 | 1 (1.7) | 9 (6) | 0.29 |
| DQ8/DQ8 | 2 (3.3) | 4 (2.6) | 0.67 |
| DQ8/DQX | 5 (8.5) | 8 (5.3) | 0.54 |
| DQX/DQX | 2 (3.3) | 32 (21.2) | 0.001 |
| Total | 59 (100) | 151 (100) |
DQ2.5/DQ2.2, DQ2.5/DQ2.5, DQ2.5/DQ8 and DQ2.5/DQX were detected as significant in the CD cases (P < 0.05) and DQ7/DQ7 and DQ7/DQX were detected as significant in the controls (P < 0.05). Accordingly, we can conclude that CD patients carry more HLA risk alleles than controls (Table 1).
We typed 20 random CD cases and 30 controls using both the BAG kit and the tag SNPs to test the validity of the tag SNP approach in the Iranian population. Nine cases and three controls showed discordant results between the two tests. Eight CD samples, which the tag SNP approach classified as non-DQ2/DQ8, were typed as DQ8 with the BAG kit. The results of the validation showed that the sensitivity to detect DQ2 and DQ7 was 100% and 84.6% for DQ8, while the specificity was 97% for DQ2 and DQ7, and 86.4% for DQ8 (Table 2). These results suggest that DQ2 and DQ7 can be inferred using the tag-SNP method, but that the detection of DQ8 is less sensitive and less specific. Heterozygosity and homozygosity of the haplotypes could not be determined by the BAG kit for all samples, especially when the parallel haplotypes are unknown (DQX).
| Tag SNP | |||
| Celiac patients | Controls | ||
| Number of patients | 59 | 151 | |
| DQ 2.2 | Sensitivity | 100% | 100% |
| Specificity | 100% | 100% | |
| DQ 2.5 | Sensitivity | 100% | 100% |
| Specificity | 97.8% | 96.7% | |
| DQ 8 | Sensitivity | 86.4% | 100% |
| Specificity | 86.4% | 97.1% | |
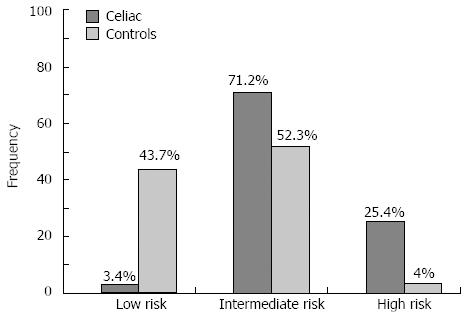
Based on the outcome of the HLA-DQ2/DQ8 prediction using tagging SNPs, we divided the Iranian population into three risk groups: low risk, which were DQ2/DQ8-negative (3.4% of cases, 43.7% of controls); intermediate risk, which were homozygous for DQ2.2 and DQ8, or heterozygous for DQ8, DQ2.5 or DQ2.2 (71.2% of cases, 52.3% of controls); and high risk, which were homozygous DQ2.5 or DQ2.5/DQ2.2) (25.4% of cases, 4% of controls) (Figure 2). The differences were statistically significant for all the risk groups when comparing cases to controls (P < 0.05). We also confirmed that CD patients carry more HLA risk alleles than healthy controls and this difference was statistically significant for the DQ2.5 haplotype distribution between cases and controls (P = 0.0001).
We included individuals from four areas with different ethnicities in Iran but did not find any statistical difference in the distribution of HLA-DQ2 and HLA-DQ7 genotypes in either the case or control groups (P > 0.05) (data not shown).
Fourteen patients with Marsh I histology and 12 with Marsh II were compared to 33 patients with Marsh III (including 14 Marsh IIIa, 7 Marsh IIIb and 13 Marsh IIIc) in relation to their HLA-DQ haplotypes. Table 3 shows the distribution of HLA haplotypes according to the Rostami-Marsh classification[18]. Of the 25 patients demonstrating only minor small bowel mucosal changes (i.e., Marsh I and II histology), two (3.4%) were negative for DQ2 and DQ8. Our result thus confirmed that patients with Marsh I or II histology carry low HLA risk alleles, such as HLA-DQ8. No statistically significant correlations were detected between the corresponded haplotypes and the different Marsh classifications.
| Haplotype | Rostami-Marsh classification | Total | P-value | |||
| Marsh I | Marsh II | Marsh III | ||||
| HLA- | DQ2.2/DQ2.2 | 0 | 0 | 1 | 1 | 0.73 |
| DQ2.2/DQ7 | 0 | 2 | 2 | 4 | 0.46 | |
| DQ2.2/DQX | 1 | 0 | 2 | 3 | 0.2 | |
| DQ2.5/DQ2.2 | 1 | 3 | 3 | 7 | 0.06 | |
| DQ2.5/DQ2.5 | 1 | 2 | 5 | 8 | 0.46 | |
| DQ2.5/DQ7 | 2 | 0 | 1 | 3 | 0.2 | |
| DQ2.5/DQ8 | 1 | 1 | 5 | 7 | 0.78 | |
| DQ2.5/DQX | 5 | 2 | 9 | 16 | 0.96 | |
| DQ8/DQ7 | 0 | 1 | 0 | 1 | 0.04 | |
| DQ8/DQ8 | 0 | 0 | 2 | 2 | 0.53 | |
| DQ8/DQX | 0 | 2 | 3 | 5 | 0.13 | |
| DQX/DQX | 1 | 0 | 1 | 2 | 0.64 | |
| Total | 12 | 13 | 34 | 59 | ||
Several epidemiological studies indicated that CD is a common disorder in the Iranian population with a prevalence of 1%[20-22], but there are no data on the frequency of HLA-related CD-predisposing alleles. We examined the frequency of the HLA-DQA1 and HLA-DQB1 haplotypes in Iranian CD patients and compared them to a matched, healthy control group.
A simple PCR-based SNP approach has been developed for large-scale population screening[13] and this method can be applied to different Caucasian populations[12]. A major advantage of this PCR-based SNP method is its ability to distinguish HLA-DQ2 and HLA-DQ8 homozygotes and heterozygotes which, in turn, translates into differences in the risk carried for CD. We tested the transferability of this method to the Iranian population and found that the sensitivity and specificity to detect the CD risk alleles was 100% and 97% for HLA-DQ2 and HLA-DQ7, respectively, but slightly lower for HLA-DQ8 (86.4%).
The lower sensitivity and specificity for HLA-DQ8 in the Iranian population might be attributable to differences in the patterns of linkage disequilibrium blocks in Iranians and Europeans. The frequency of the HLA haplotypes in Iran is not necessarily different from Europeans, but the tagging SNP method might not be the ideal test to establish the frequency of CD-associated HLA-DQ8 haplotypes in this population.
The false positives or false negatives observed with the tagging SNP method may be due to the presences of rare alleles (DQX) either in heterozygous or homozygous state that cannot be detected by the BAG kit.
As reported for other populations[23-25], the most frequent haplotype in Iranian CD patients was HLA-DQ2.5 (69.5%) and its frequency is very similar to that seen in European CD populations.
Surprisingly, in contrast to the low prevalence of HLA-DQ8 in CD population reported in the literature (0.6% in Cameroon, 2% in Italy, 2.3% in Hungary, 4.2% in United States, 6.4% in Finland and 7.6% in Japan)[12,26], we found a higher prevalence (25.4%) of this genotype in the Iranian CD population. Our result is comparable with those presented by Catassi et al[27] in 2009 for Turks (22%), North American Indians (25.3%), Mexicans (28.3%) and Bushmen (ethnic groups in Southern Africa) (30%).
Karell et al[28] and Polvi et al[29] suggested that only a small number of CD patients (6%) carry neither DQ2 nor DQ8. Our study supports these figures, as we only found two CD patients (3.4%) who were DQ2-DQ8-negative.
Several studies have shown that individuals homozygous for HLA-DQ2.5 or heterozygous for HLA-DQ2.5/DQ2.2 genotypes have an increased risk for CD compared to those homozygous for HLA-DQ2.2 or heterozygous for HLA-DQ2.5 or HLA-DQ2.2[30-32]. Similarly, our results show that 25.4% of cases lie in the high risk group for CD compared to 3.4% in the low risk group. The frequencies of intermediate risk and high risk groups were, as expected, higher in cases than controls (Figure 2).
The frequency of DQ8/DQ2.2 in the cases was zero but this number may be less reliable. This may be due to differences in the patterns of linkage disequilibrium blocks in Iranians or may result from low concentrations of DNA samples (the concentration of the samples was reduced when they were sent to the UMCG for genetic analysis). We concluded that DQ2 worked well, but DQ8 needs to be interpreted more carefully when identified using tagging SNPs.
In conclusion, this pilot study has provided information on the genetic background of CD in Iranians. This should prove useful to other researchers and to future work on this disease. Despite our small cohort, the data we present offer a first step towards defining the genetic structure of HLA in the Iranian population with CD and provide information that can be used in family screening. In addition, our data suggest that the Iranian population, as a non-European Caucasian population, does share certain HLA class II genetic components with the CD populations of European ancestry. We found that Iranians are most similar to non-European Caucasian CD patients, given their increased frequency of HLA-DQ8 carriers. Further works, with larger sample sizes, on HLA polymorphisms among people living in different parts of Iran will provide more detailed information about the genetic background of the Iranian population and its relationship to other ethnic populations in the world.
This work was performed within the framework of an agreement between the University Medical Center Groningen (UMCG, Groningen, the Netherlands) and the Gastroenterology and Liver Diseases Research Center, Shahid Beheshti University of Medical Sciences, Tehran (Iran). It was financed by the UMCG, by grants from the Dutch Celiac Disease Consortium (an innovative cluster approved by the Netherlands Genomics Initiative and partly funded by the Dutch Government (grant BSIK03009 to C.W.), the Netherlands Organization for Scientific Research (NWO-VICI grant 918.66.620 to C.W.), the Dutch Digestive Disease Foundation (MLDS WO11-30 to C.W.), and the Iran National Science Foundation (INSF). We sincerely thank R. Booij and S. Medema-Jankipersadsing (UMCG) for their help in genotyping, Dr. M.A. Pourhosseingholi for his assistance in statistical analysis and J.L. Senior and K. Mc Intyre (UMCG) for editing the manuscript. This paper resulted from part of the PhD research of Mohammad Rostami-Nejad.
Celiac disease (CD) is caused by an immune response to gluten in genetically predisposed individuals. This disease is strongly associated with human leukocyte antigen (HLA)-DQ2 or DQ8 genotypes. HLA molecules are postulated to present gluten antigens to T-cells, which in turn induce tissue damage. Approximately 95% of patients with CD have the HLA-DQ2 heterodimer encoded by the DQA1*05 and DQB1*02 alleles, while close to 5% have the HLA-DQ8 heterodimer encoded by the DQA1*03 and DQB1*0302 alleles. The HLA-DQ alleles are also found in 48%-65% of first-degree relatives of patients with CD and the general population.
HLA-DQ2 and -DQ8 determination is useful in the exclusion, probably lifelong, of celiac disease in individuals with an equivocal small bowel histological finding.
This describe, for the first time, the distribution of HLA-DQ2/DQ8 alleles in Iranian celiac disease patients compared to healthy Iranian controls. To assess this distribution their authorized a novel SNP method to assess HLA distribution. They also demonstrate the transferability of such an approach to the Iranian population. The results underline the primary importance of HLA-DQ alleles in the Iranian population’s susceptibility to celiac disease. This show that the frequency of HLA-DQ2 is higher in Iranian celiac disease patients than in controls and that the prevalence of HLA-DQ8 is higher than that reported in other international populations. The data presented in this study offer the first step towards defining the genetic structure of HLA in the Iranian population with celiac disease and provide information that can be used in family screening. In addition, the data suggest that the Iranian population, as a non-European Caucasian population, does share certain HLA class II genetic components with the populations of European countries. They therefore suggest that performing readily available serological tests in combination with HLA typing will significantly reduce the burden of celiac disease in the at-risk population by providing more accurate diagnoses and enabling timely treatment with a gluten-free diet.
The main importance of HLA mapping is to combine the less-invasive methods into a diagnostic algorithm that can be a substitute for the gold standard of biopsy sampling. Carrying HLA risk alleles can be a predictor of susceptibility to celiac disease.
HLA: the loci of genes that encode for major histocompatibility complex in humans; tissue transglutaminase IgA and/or IgG test: autoantibodies against the transglutaminase protein used as part of an evaluation for certain autoimmune conditions, most notably celiac disease.
The authors have aimed to identify the prevalence of HLA-DQ2 and -DQ8 alleles in celiac patients and a healthy population in Iran and have compared the results with those determined by previously described tagging single nucleotide polymorphism (SNP) methods and commercial kits. The reviewed article describes a nice study. Indeed, there were no data available on the HLA distribution in celiac patients in Iran. Furthermore, the approach based on HLA-tagging SNPs that was adopted in this study is the best described method for determining HLA alleles. The article is well written, and the tables and figures are of outstanding quality. It is thus a great summary of a pilot study conducted in Iranian celiac patients.
| 1. | Rostami-Nejad M, Villanacci V, Hogg-Kollars S, Volta U, Manenti S, Reza-Zali M, Caio G, Giovenali P, Barakauskiene A, Kazenaite E. Endoscopic and histological pitfalls in the diagnosis of celiac disease: A multicentre study assessing the current practice. Rev Esp Enferm Dig. 2013;105:326-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 2. | Rostami K, Villanacci V. Microscopic enteritis: novel prospect in coeliac disease clinical and immuno-histogenesis. Evolution in diagnostic and treatment strategies. Dig Liver Dis. 2009;41:245-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 3. | Schuppan D. Current concepts of celiac disease pathogenesis. Gastroenterology. 2000;119:234-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 326] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 4. | Anderson RP, Henry MJ, Taylor R, Duncan EL, Danoy P, Costa MJ, Addison K, Tye-Din JA, Kotowicz MA, Knight RE. A novel serogenetic approach determines the community prevalence of celiac disease and informs improved diagnostic pathways. BMC Med. 2013;11:188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 5. | Petronzelli F, Bonamico M, Ferrante P, Grillo R, Mora B, Mariani P, Apollonio I, Gemme G, Mazzilli MC. Genetic contribution of the HLA region to the familial clustering of coeliac disease. Ann Hum Genet. 1997;61:307-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 82] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 6. | Lundin KE, Scott H, Hansen T, Paulsen G, Halstensen TS, Fausa O, Thorsby E, Sollid LM. Gliadin-specific, HLA-DQ(alpha 1*0501,beta 1*0201) restricted T cells isolated from the small intestinal mucosa of celiac disease patients. J Exp Med. 1993;178:187-196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 434] [Cited by in RCA: 436] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 7. | Sollid LM, Thorsby E. HLA susceptibility genes in celiac disease: genetic mapping and role in pathogenesis. Gastroenterology. 1993;105:910-922. [PubMed] |
| 8. | Kaukinen K, Partanen J, Mäki M, Collin P. HLA-DQ typing in the diagnosis of celiac disease. Am J Gastroenterol. 2002;97:695-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 152] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 9. | Zubillaga P, Vidales MC, Zubillaga I, Ormaechea V, García-Urkía N, Vitoria JC. HLA-DQA1 and HLA-DQB1 genetic markers and clinical presentation in celiac disease. J Pediatr Gastroenterol Nutr. 2002;34:548-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 54] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 10. | Sumník Z, Kolousková S, Cinek O, Kotalová R, Vavrinec J, Snajderová M. HLA-DQA1*05-DQB1*0201 positivity predisposes to coeliac disease in Czech diabetic children. Acta Paediatr. 2000;89:1426-1430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 11. | Freeman HJ. Risk factors in familial forms of celiac disease. World J Gastroenterol. 2010;16:1828-1831. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Koskinen L, Romanos J, Kaukinen K, Mustalahti K, Korponay-Szabo I, Barisani D, Bardella MT, Ziberna F, Vatta S, Széles G. Cost-effective HLA typing with tagging SNPs predicts celiac disease risk haplotypes in the Finnish, Hungarian, and Italian populations. Immunogenetics. 2009;61:247-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 13. | Monsuur AJ, de Bakker PI, Zhernakova A, Pinto D, Verduijn W, Romanos J, Auricchio R, Lopez A, van Heel DA, Crusius JB. Effective detection of human leukocyte antigen risk alleles in celiac disease using tag single nucleotide polymorphisms. PLoS One. 2008;3:e2270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 125] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 14. | Amirzargar A, Mytilineos J, Farjadian S, Doroudchi M, Scherer S, Opelz G, Ghaderi A. Human leukocyte antigen class II allele frequencies and haplotype association in Iranian normal population. Hum Immunol. 2001;62:1234-1238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 51] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 15. | Farjadian S, Naruse T, Kawata H, Ghaderi A, Bahram S, Inoko H. Molecular analysis of HLA allele frequencies and haplotypes in Baloch of Iran compared with related populations of Pakistan. Tissue Antigens. 2004;64:581-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 48] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 16. | Farjadian S, Moqadam FA, Ghaderi A. HLA class II gene polymorphism in Parsees and Zoroastrians of Iran. Int J Immunogenet. 2006;33:185-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Farjadian S, Ghaderi A. HLA class II similarities in Iranian Kurds and Azeris. Int J Immunogenet. 2007;34:457-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 18. | Rostami K, Kerckhaert J, Tiemessen R, von Blomberg BM, Meijer JW, Mulder CJ. Sensitivity of antiendomysium and antigliadin antibodies in untreated celiac disease: disappointing in clinical practice. Am J Gastroenterol. 1999;94:888-894. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 327] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 19. | Sambrook J, Russell DW, editors . Molecular Cloning: a Laboratory Manual. New York: Cold Spring Harbor Laboratory Press 2001; . |
| 20. | Shahbazkhani B, Malekzadeh R, Sotoudeh M, Moghadam KF, Farhadi M, Ansari R, Elahyfar A, Rostami K. High prevalence of coeliac disease in apparently healthy Iranian blood donors. Eur J Gastroenterol Hepatol. 2003;15:475-478. [PubMed] |
| 21. | Rostami-Nejad M, Villanacci V, Mashayakhi R, Molaei M, Bassotti G, Zojaji H, Mirstatari D, Rostami K, Zali MR. Celiac disease and Hp infection association in Iran. Rev Esp Enferm Dig. 2009;101:850-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 22. | Rostami Nejad M, Rostami K, Pourhoseingholi MA, Nazemalhosseini Mojarad E, Habibi M, Dabiri H, Zali MR. Atypical presentation is dominant and typical for coeliac disease. J Gastrointestin Liver Dis. 2009;18:285-291. [PubMed] |
| 23. | Sollid LM. Coeliac disease: dissecting a complex inflammatory disorder. Nat Rev Immunol. 2002;2:647-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 686] [Cited by in RCA: 661] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 24. | Kaukinen K, Collin P, Maki M. Natural History of Celiac Disease. Frontiers in Celiac Disease. Basel, Karger: Pediatr Adolesc Med 2008; 12-17. [DOI] [Full Text] |
| 25. | Karinen H, Kärkkäinen P, Pihlajamäki J, Janatuinen E, Heikkinen M, Julkunen R, Kosma VM, Naukkarinen A, Laakso M. Gene dose effect of the DQB1*0201 allele contributes to severity of coeliac disease. Scand J Gastroenterol. 2006;41:191-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 26. | Alarida K, Harown J, Di Pierro MR, Drago S, Catassi C. HLA-DQ2 and -DQ8 genotypes in celiac and healthy Libyan children. Dig Liver Dis. 2010;42:425-427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (1)] |
| 27. | Catassi C, Yachha SK. The global village of celiac disease. Frontiers in Celiac Disease. Basel, Karger: Pediatr Adolesc Med 2008; 23-31. [DOI] [Full Text] |
| 28. | Karell K, Louka AS, Moodie SJ, Ascher H, Clot F, Greco L, Ciclitira PJ, Sollid LM, Partanen J. HLA types in celiac disease patients not carrying the DQA1*05-DQB1*02 (DQ2) heterodimer: results from the European Genetics Cluster on Celiac Disease. Hum Immunol. 2003;64:469-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 417] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 29. | Polvi A, Arranz E, Fernandez-Arquero M, Collin P, Mäki M, Sanz A, Calvo C, Maluenda C, Westman P, de la Concha EG. HLA-DQ2-negative celiac disease in Finland and Spain. Hum Immunol. 1998;59:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 30. | Vader W, Stepniak D, Kooy Y, Mearin L, Thompson A, van Rood JJ, Spaenij L, Koning F. The HLA-DQ2 gene dose effect in celiac disease is directly related to the magnitude and breadth of gluten-specific T cell responses. Proc Natl Acad Sci USA. 2003;100:12390-12395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 282] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 31. | Congia M, Cucca F, Frau F, Lampis R, Melis L, Clemente MG, Cao A, De Virgiliis S. A gene dosage effect of the DQA1*0501/DQB1*0201 allelic combination influences the clinical heterogeneity of celiac disease. Hum Immunol. 1994;40:138-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 71] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 32. | Romanos J, van Diemen CC, Nolte IM, Trynka G, Zhernakova A, Fu J, Bardella MT, Barisani D, McManus R, van Heel DA. Analysis of HLA and non-HLA alleles can identify individuals at high risk for celiac disease. Gastroenterology. 2009;137:834-40, 840.e1-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 102] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
P- Reviewers: Cseh A, Nejad MR, Saadah OI S- Editor: Qi Y L- Editor: A E- Editor: Zhang DN