Published online Aug 28, 2013. doi: 10.3748/wjg.v19.i32.5212
Revised: May 15, 2013
Accepted: May 18, 2013
Published online: August 28, 2013
Processing time: 153 Days and 18.8 Hours
Primary liver cancer is a global disease that is on the increase. Hepatocellular carcinoma (HCC) accounts for most primary liver cancers and has a notably low survival rate, largely attributable to late diagnosis, resistance to treatment, tumour recurrence and metastasis. MicroRNAs (miRNAs/miRs) are regulatory RNAs that modulate protein synthesis. miRNAs are involved in several biological and pathological processes including the development and progression of HCC. Given the poor outcomes with current HCC treatments, miRNAs represent an important new target for therapeutic intervention. Several studies have demonstrated their role in HCC development and progression. While many risk factors underlie the development of HCC, one process commonly altered is iron homeostasis. Iron overload occurs in several liver diseases associated with the development of HCC including Hepatitis C infection and the importance of miRNAs in iron homeostasis and hepatic iron overload is well characterised. Aberrant miRNA expression in hepatic fibrosis and injury response have been reported, as have dysregulated miRNA expression patterns affecting cell cycle progression, evasion of apoptosis, invasion and metastasis. In 2009, miR-26a delivery was shown to prevent HCC progression, highlighting its therapeutic potential. Several studies have since investigated the clinical potential of other miRNAs with one drug, Miravirsen, currently in phase II clinical trials. miRNAs also have potential as biomarkers for the diagnosis of HCC and to evaluate treatment efficacy. Ongoing studies and clinical trials suggest miRNA-based treatments and diagnostic methods will have novel clinical applications for HCC in the coming years, yielding improved HCC survival rates and patient outcomes.
Core tip: Hepatocellular carcinoma (HCC) has a high incidence and low survival rate, largely attributable to late diagnosis, resistance to treatment, tumour recurrence and metastasis. MicroRNAs (miRNAs) are regulatory RNAs that modulate protein synthesis and are involved in several biological and pathological processes including the development and progression of HCC. miRNAs represent important new targets for therapeutic intervention for HCC and have potential as diagnostic and prognostic HCC biomarkers. Ongoing studies and clinical trials suggest miRNA-based treatments and diagnostic methods will have clinical applications for HCC in the coming years, yielding improved HCC survival rates and patient outcomes.
- Citation: Greene CM, Varley RB, Lawless MW. MicroRNAs and liver cancer associated with iron overload: Therapeutic targets unravelled. World J Gastroenterol 2013; 19(32): 5212-5226
- URL: https://www.wjgnet.com/1007-9327/full/v19/i32/5212.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i32.5212
Hepatocellular carcinoma (HCC) accounts for 85%-90% of primary liver cancers; it ranks as the fifth most common cancer worldwide and the third leading cause of death from malignancy[1]. The development and progression of HCC is a multistage process, with transformation typically beginning in hepatocytes of livers undergoing chronic hepatitis or cirrhosis[2]. The major risk factor for HCC is chronic hepatitis due to infection with the hepatitis B or hepatitis C virus (HBV/HCV) accounting for 80%-90% of all HCC cases worldwide[3]. The other most important risk factors for hepatocarcinogenesis are alcoholic and non-alcoholic steatohepatitis-associated cirrhosis; less common risk factors include genetic conditions such as hereditary haemochromatosis (HH), alpha-1 antitrypsin deficiency[4,5] and aflatoxin B1 intake. Regardless of the underlying risk factor, hepatocytes progress through several hyperplastic and dysplastic stages before eventually acquiring a malignant phenotype, with subsequent intrahepatic metastasis and distant spread of HCC cells[6]. The 5-year survival rate of patients with HCC remains quite low, between 6%-11%. This is attributable to late diagnosis, resistance to treatment, tumour recurrence and metastasis[2].
Previously, studies investigating HCC development and progression have focused on the therapeutic potential of targeting various genes and proteins[7]. However, a new group of regulatory RNA molecules has more recently been identified, called microRNAs (miRNAs). Involvement of miRNAs in HCC development and progression has been demonstrated; as such miRNAs have considerable diagnostic and therapeutic potential for HCC. Here, the role of miRNAs in the pathogenesis of HCC is reviewed with a focus on their regulation of iron homeostasis and in the setting of iron overload, a common pathological event observed in several liver diseases associated with HCC development. The relevance of miRNAs to HCC progression with regard to hepatic fibrosis and response to injury, as well as their contribution to cell cycle progression, evasion of apoptosis and metastasis is explored. Finally, the potential diagnostic and therapeutic value of miRNAs in HCC is discussed.
miRNAs are endogenous single stranded RNAs, approximately 22 nucleotides in length. They are non-coding but are important post-transcriptional regulators of gene expression. miRNAs were first discovered in 1993, and since then the considerable extent of the gene regulatory capacity of miRNAs has been investigated. These investigations have demonstrated that specific miRNAs have central roles in critical biological processes such as development, cell proliferation, apoptosis and oncogenesis. The mechanisms of action and biogenesis of miRNAs have been reviewed in detail[8,9].
Mature miRNAs enter the RNA-induced silencing complex (RISC) in the cytosol. In this complex miRNA can post-transcriptionally regulate gene expression. Their mechanism of action is determined by the level of complementarity between the miRNA and the 3’-untranslated region (UTR) target on the mRNA. In perfect complementarity, miRNA-mRNA binding induces mRNA cleavage and degradation by RISC. In imperfect complementarity, miRNA-mRNA binding represses target mRNA translation[10]. Occassionally, miRNAs can upregulate translation even in conditions of growth arrest[11]. However translation is more commonly inhibited and the target mRNAs are eventually degraded in cytoplasmic processing bodies[12].
Functional target sites on mRNAs usually consist of a 6-8-nt long sequence complementary to the miRNA sequence (followed by an adenosine), this is termed the miRNA “seed” sequence and is located at the 5’ end of the miRNA[13]. The complementary sequence commonly referred to as a miRNA recognition element (MRE) is usually located in the 3’-UTR of the target mRNA. Some recent studies have shown miRNAs can also bind to MREs located in the 5’-UTR or the open reading frame[14-17]. Unusually miRNAs can act as decoys and bind to ribonucleoproteins independent of a seed sequence and RISC, thus interfering with roles requiring mRNA binding[18].
Given the considerable potential for variety in miRNA-mRNA interaction, it is not surprising that a single miRNA can target several genes[19-22]. In addition, approximately 60% of mRNAs carry at least one evolutionarily conserved MRE. Bioinformatic analysis predicts that the 3’-UTR of a single transcript is often targeted by several miRNAs, a prediction that has been validated experimentally for many genes[22]. The complex, widespread and co-operative regulation of gene expression by miRNAs is an important consideration when studying normal and pathological processes in terms of understanding the processes themselves and identifying potential biomarkers. Recently investigators have begun to study the role of miRNAs in the pathogenesis of HCC. In particular, several studies have demonstrated a role for miRNAs in HCC development and progression, wherein the importance of miRNAs in iron homeostasis and hepatic iron overload were highlighted.
Many risk factors underlie the development of HCC and one process commonly altered is iron homeostasis. Iron overload in the liver occurs in several liver diseases associated with the development of HCC, including chronic hepatitis due to HCV infection and also due to genetic conditions such as HH. Hepatic iron overload is an independent risk factor for the development of HCC[23] and emerging evidence points towards miRNAs as central regulators of iron homeostasis
Hepatocytes act as the principal site of iron storage in the body, storing iron as ferric oxyhydroxyapatite in the core of ferritin. During iron overload, the ability of hepatocytes to safely sequester iron is exceeded, denaturation of ferritin subunits occurs leading to ionic iron release into the hepatocyte cytoplasm[24]. The effects of hepatic iron overload have been particularly well studied in patients with the inherited iron metabolism disorder, HH and in Africans with dietary iron overload.
Patients with HH, without timely appropriate treatment, almost always develop hepatic fibrosis and cirrhosis due to hepatic iron accumulation[25]. Similarly patients with African dietary iron overload can develop cirrhosis, albeit less often[26,27]. HCC is a potential complication in untreated HH patients associated with premature death[28,29]. Comparison studies have showed that cirrhosis plays a role in the development of HCC in HH[30,31] however, HCC can also develop in HH patients without cirrhosis, albeit rarely[32-37]. Together this suggests that hepatic iron storage could directly contribute to HCC development[38,39], in addition to its indirect effect as a cause of cirrhosis. This concept is in keeping with a study comparing cirrhosis incidence in HH and non-iron related liver diseases, where the risk of HCC was greater in HH[40]. Interestingly, despite HCC initially being thought not to occur in dietary iron overload, three case/control studies have demonstrated a causal association between African dietary iron overload and HCC, even after allowing for the confounding effects of cirrhosis, chronic HBV and HCV infection and prolonged aflatoxin B1 exposure[41-43]. Dietary iron overload resulting in HCC has also been reported in animal models[44,45] supporting the directly hepatocarcinogenic effects of hepatic iron accumulation.
HCC can also develop with other causes of hepatic iron accumulation namely, thalassaemia major, sideroblastic anaemia and hereditary spherocytosis[46-48]. Lesser degrees of hepatic iron accumulation are seen in other liver diseases, such as chronic HCV hepatitis and alcoholic liver disease. Nonetheless, it is thought to have an important role in these diseases[24]. One area of recent interest is hepatic iron accumulation with HCV infection. As the main risk factor for HCC development, HCV is particularly relevant to HCC. Iron promotes the initiation of HCV translation by increasing expression of eukaryotic initiation factor 3a and La protein, whereas inhibiting expression of these proteins suppresses HCV translation[49,50]. Interestingly the expression of the chief iron regulatory hormone, hepcidin, is suppressed in chronic HCV infected patients. Given that hepcidin expression has direct anti-viral activity against HCV in cell culture[51] this represents an exciting area of ongoing research.
Hepatic iron accumulation has also been implicated in non-alcoholic fatty liver disease (NAFLD). Hyperferritinemia is associated with higher hepatic iron and fat content in NAFLD[52], and is also an independent predictor of liver damage in NAFLD patients[53]. As altered iron trafficking is frequent in patients with NAFLD, one recent study investigated the role of the Ala736Val polymorphism of TMPESS6 (an inhibitor of hepcidin expression) in NALFD-associated hepatic iron accumulation[54]. Homozygosity for this polymorphism was associated with low hepatic iron stores and was negatively associated with hepatic iron accumulation independent of age, gender, human haemochromatosis (HFE) genotype and beta thalassaemia trait.
A recent animal study examined the long-term effects of iron overload in HCC[44]. A high-iron diet was given to Wistar albino rats over 16 mo to induce hepatic iron overload. Altered hepatic foci developed in many animals by 20 mo. By 28 mo, these foci were more numerous and had become identical to the iron-free preneoplastic nodules seen in HH patients who develop HCC[55]. HCC was evident at 32 mo in the absence of portal fibrosis or cirrhosis. The mechanisms by which free iron induces hepatocarcinogenesis are not yet fully characterised but are likely due to the generation of reactive oxygen intermediates (ROI) and oxidative stress which damages DNA, lipids, and proteins resulting in both necrosis and apoptosis within hepatocytes[56-60]. Oxidative DNA damage correlates with cell immortalisation in HCC through induction of telomerase activity. This process has been associated with miR-92 over expression, a miRNA affecting specific cell proliferation and apoptosis pathways[61]. Iron overload leading to lipid peroxidation is also thought to contribute to HCC development[62-66]. Moreover, excess hepatic iron may induce immunologic alterations, leading to impaired immune surveillance of malignant transformation. Nontransferrin-bound iron can markedly suppress lymphocyte proliferation[67]. The same study showed that ferritin can inhibit lymphocyte proliferation. Indeed, the presence of both iron and ferritin were found to significantly reduce the tumouricidal function of macrophages[68]. In addition to its solitary effects, iron overload can act in tandem with other HCC risk factors to produce hepatocarcinogenesis. For example, dietary iron overload and aflatoxin B1 exposure have superadditive effects on mutagenesis rates[69]. Furthermore ROI generation and mutagenesis are synergistically increased in animal models with both risk factors, leading to greater DNA damage[70-73].
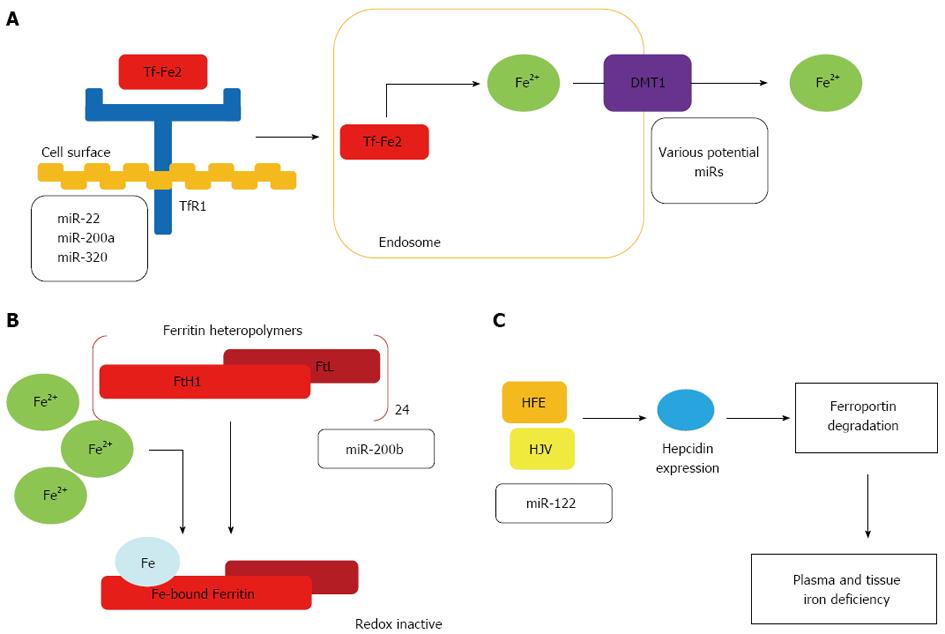
Control of cellular iron uptake by miRNAs: Most cells obtain iron from plasma via iron-bound transferrin (Tf-Fe2) uptake. Tf-Fe2 binds to TfR1 on the cell surface and the complex is internalised by clathrin-dependent endocytosis. Acidification of early endosomes aids iron release from transferrin[74], so that it can be reduced to Fe2+ by metalloreductases[75]. Transport into the cytoplasm occurs via endosomally-expressed Divalent metal transporter 1 (DMT1). Cell surface TfR1 levels reflect cellular iron requirements, with regulation of TfR1 expression mainly achieved by the IRE/IRP regulatory system[76]. However, recent studies have shown that the transferrin cycle is also controlled by miRNAs, at two separate steps (Figure 1A).
Cancerous cells have elevated TfR1 expression to meet the increased iron requirements of rapid cellular proliferation[77,78]. Conversely, differentiation of a human leukaemia cell line decreases TfR1 expression[79]; this is accompanied by reciprocal increases in miRNAs predicted to bind to the TfR1 3’-UTR (miR-22, miR-200a and miR-320). Of these, miR-320 was demonstrated to suppress the activity of a luciferase reporter vector under the control of the TfR1 3’-UTR[80]. Similarly, enforced miR-320 expression in a lung carcinoma cell line can reduce TfR1 expression and slow cell cycle progression and cell growth. This growth inhibitory effect can be reversed by treatment with a soluble iron solution suggesting that reduced TfR1 expression in miR-320-overexpressing cells lowers iron availability and reduces cell proliferation[81]. Currently, it is unknown whether miR-320-mediated TfR1 regulation is limited to cancer cells or whether it has a role under normal physiological conditions.
In addition to miRNA-dependent TfR1 regulation, miRNAs control the transferrin cycle at the release of iron from the endosome via DMT1. The gene coding for DMT1 (SLC11A2) produces four variant mRNA transcripts. These differ either at their 5’ end due to alternative promoter usage (DMT1A and 1B isoforms), or at the 3’ end, due to alternative splicing determining the presence or absence of an IRE sequence motif[82]; only the IRE-containing isoforms are controlled in response to cellular iron levels by IRP binding[83]. All DMT1 isoforms can transport iron and, with the exception of the duodenal 1A isoform, are ubiquitously expressed[84]. Of note, miRNA-controlled DMT1 expression by let-7d can contribute to the uptake of non-transferrin bound iron[85]. Further studies are needed to determine how miRNA-dependent control of DMT-1 expression is integrated with additional DMT-1 control mechanisms.
Importantly, as miRNA maturation requires iron in the form of heme[86], the finding that miRNAs control cellular iron uptake suggests a possible regulatory loop in which iron is needed for the efficient synthesis of mature miRNAs, while certain mature miRNAs control cellular iron uptake.
Control of cellular iron storage by miR-200b: Ferritin heteropolymers consist of 24 subunits of heavy (FtH1) and light (FtL) chains that bind iron from the cytoplasmic ‘‘labile iron pool’’[87]. The FtH1 subunit has ferroxidase activity necessary for iron deposition in ferritin. Ferritin detoxifies excess iron into a redox-inactive form, preventing chronic oxidative stress and subsequent cell and tissue damage. Ferritin also acts as an intracellular iron store mobilised via proteasomal and lysosomal degradation. One recent study showed that human breast cancer cells with an aggressive mesenchymal phenotype express significantly higher FtH1 and FtL mRNA and protein levels and have a smaller labile iron pool compared to breast cancer cells with a less aggressive epithelial phenotype[88]. High FtH1 concentrations correlated with low miR-200b expression, a miRNA that binds both FtH1 and FtL 3’UTRs (Figure 1B). Of clinical relevance, miR-200b transfection improved sensitivity of breast cancer cells to doxorubicin. Additionally, patients with higher plasma ferritin levels showed worse treatment outcomes, emphasising the clinical significance of this facet of iron regulation. These findings suggest that down regulation of miR-200b in human breast cancer contributes to increased cancer aggressiveness. Whether FtH1 and FtL are regulated by miR-200b in hepatocytes and if this has implications for HCC remains to be determined[89,90].
Control of systemic iron regulation by miR-122: The liver regulates systemic iron homeostasis via hepcidin and monitors systemic iron availability through genes involved in HH (e.g., HFE, hemojuvelin and TfR2), the bone morphogenetic protein (Bmp) 6 and the Smad4 protein. These all function in the regulation of hepcidin transcription. Low hepcidin activity due to mutations in HFE, hemojuvelin, TfR2 or hepcidin itself lead to the development of HH which is associated with increased iron uptake from the diet and increased iron release from macrophages.
miR-122 is selectively expressed in the liver. One recent study demonstrated that miR-122 expression is reduced in a mouse model of HFE-mutated HH[91]. Depletion of miR-122 in wild type mice led to low systemic iron levels, decreased plasma iron levels and lower transferrin iron binding capacity. These events in turn resulted in an insufficient iron supply to erythroid cells and a mild impairment of haematopoiesis[91]. Furthermore, the iron contents of the liver and spleen were also reduced. Interestingly, miR-122 depletion altered systemic iron homeostasis through changes in the level of expression of genes involved in the sensing of systemic iron levels (i.e., HFE, Hemojuvelin, and Bmpr1a), as well as genes that transmit signals via the Bmp/Smad signalling pathway, to regulate hepcidin transcription[91]. This study also validated HFE and hemojuvelin as direct targets of miR-122 (Figure 1C).
This suggests a miR-122-dependent regulatory loop that controls systemic iron homeostasis whereby depletion of miR-122 derepressed HFE and hemojuvelin expression, in turn increasing hepcidin transcription. As a result, high circulating hepcidin levels can enhance the degradation of ferroportin on target cells, leading to lower iron absorption from the diet and iron release from macrophages. This likely leads to plasma and tissue iron deficiency, with mild impairment of erythropoiesis. miR-122 levels are not regulated as a result of iron accumulation in the liver of HH patients, but more likely as a consequence of the signalling activities reduced by a lack of HFE which is known to attenuate BMP/Smad signalling in HH patients and its respective murine disease model[92].
The finding that miR-122 regulates systemic iron homeostasis is one of a growing number of functions known for this liver-specific miRNA. For example, miR-122 is necessary for HCV infection and replication, as well as for responsiveness to interferon therapy[93-95], all processes involving alterations in iron homeostasis[96]. miR-122 levels are reduced in cirrhosis[97] and HCC[98,99], two pathologies known to be exacerbated by increased liver iron levels[24]. Evidently, miRNAs have an important role in the maintenance of iron homeostasis, given their roles in controlling the level of cellular uptake of iron-bound transferrin, iron storage by ferritin, and hepatic control of systemic iron levels via hepcidin (Figure 1). Furthermore, tissue iron overload causes oxidative stress that itself has been shown to alter miRNA expression[100,101].
Overall, these findings suggest that miRNAs control large regulatory networks that link microenvironmental stress, such as oxidative stress and hypoxia to the regulation of iron metabolism. As the maintenance of iron homeostasis is critical for many essential cellular functions, it is expected that several more miRNAs that directly or indirectly control iron-related genes will be discovered. Given the role of miRNAs in regulating iron homeostasis and the significance of iron overload to the development of HCC, miRNAs likely play an important role in the pathogenesis of HCC (Table 1). However, further studies elucidating the full extent of miRNAs’ functions in iron homeostasis under normal conditions are needed to improve our understanding of the role of miRNAs in pathologies such as HCC.
| miRNAs | Function | Outcomes |
| miR-22 | Predicted to bind iron-bound transferrin receptor (TfR1) | Targets TfR1, DMT1 expression thereby inhibiting cell cycle progression and growth |
| miR-200a | ||
| miR-320 | ||
| miR-200b | Targets ferritin heteropolymers (FtH1, FtL) | Decreased miR-200b linked with enhanced cancer aggressiveness via increased iron indices |
| miR-122 | Targets HFE, hemojuvelin, BMPr1a, BMP/SMAD signalling, hepcidin | Control of systemic iron homeostasis. Decreased miR-122 corresponds to decreased HFE and hemojuvelin expression. This correlates with increased hepcidin expression |
In HCC miRNAs can act as oncogenes, promoting hepatocyte progression to HCC, or as tumour suppressors, preventing this process[2]. Increased oncogenic miRNA levels result in reduced translation of their gene targets, contributing to HCC development and progression. By contrast, miRNAs acting as tumour suppressors prevent the expression of their oncogenic targets and hence the downregulation of such miRNAs permits greater expression of these oncogenic genes, again contributing to HCC development and progression. Progression from normal hepatocytes to HCC is a multistage process. Several changes in the liver structure and in normal cell processes must occur for this progression to continue, mediated in part by altered miRNA expression profiles. These include liver fibrosis and hepatic stellate cell-mediated liver regeneration, while at the molecular level changes in cell cycle progression, susceptibility to apoptosis and capacity for invasion and metastasis are needed.
miRNA expression profiles show considerable overlap in fibrotic disorders. The most significant mediators are the miR-29 family, important in regulating translation of extracellular matrix components and effectors of cellular differentiation[102]. Also important are miRs affecting translation of proteins involved in the pro-fibrotic transforming growth factor (TGF)-β/SMAD signalling pathway. Microarray analyses in a CCl4 rodent model of hepatic fibrosis have shown 31 differentially expressed miRs, 10 of which are over expressed in fibrotic tissue including miR-125-p, -199b, -221 and -302c[103]. This same study revealed a significant down regulation in 21 miRs, most notably the miR-29 family. Down regulation of miR-29b and miR-29c was independently confirmed in a bile duct ligation model and similar observations for miRs-29a/b/c have been reported in humans liver tissue samples of patients with a Desmet fibrosis score of 2-4[104].
Hepatic fibrosis is also affected by miR-132 levels. In two different models of hepatic fibrosis (BDL and CCl4), where a significant reduction in miR-132 levels was observed, this down regulation was found to alter the activity of hepatic stellate cells (HSCs). HSCs are the main effector cells of hepatic fibrosis, acting as the primary source for type I collagen deposition following liver injury. HSC activation occurs in response to hepatic insults including viral infection, alcohol consumption and obesity. During their activation, quiescent lipid-rich cells are transdifferentiated into fully activated myofibroblasts. The activated cells can secrete pro-fibrogenic mediators such as TGF-β, and produce extracellular matrix components[105]. Involvement of miRNAs in the process of HSC activation has been demonstrated. For example, let-7 family members are significantly up regulated in HSCs of BDL animals whereas miR-150, -187, -194 and -207 are down regulated[106]: over expression of miR-150 and miR-194 in human HSCs can inhibit HSC proliferation and prevent HSC transdifferentiation[106]. miR-150 together with another miR, miR-94, inhibits c-Myb and Rac-1, two proteins involved in pathways contributing to hepatic fibrosis development and progression. Further studies investigating differential miRNA expression in quiescent and activated rat HSCs showed that miR-15b and miR-16 are also implicated in HSC activation[107,108]. This process is also regulated by miR-27a and b which are up regulated and in turn repress RXRα[109]. Of interest miR-132 activates the methylCpG binding protein MeCP2 and components of the polycomb repressive process. Down regulation of miR-132, as seen in hepatic fibrosis, permits MeCP2 translation. This protein is subsequently recruited to the 5’UTR of PPARγ mRNA and through alteration of methylation patterns suppresses the quiescent profile of HSCs[110] - this is an example of a miRNA acting as an activator rather than an inhibitor of gene expression. Thus as our understanding of the role of miRNAs in the regulation of HSC differentiation improves so will the understanding of liver pathology and hepatic responsiveness to injury.
Aberrant cell cycle control is necessary for the development and progression of all human cancers, including HCC. Cell cycle regulation by oncoproteins and tumour suppressors is often defective resulting in increased cell proliferation. miRNAs targeting the main proliferation pathways have been identified in HCC. These miRNAs exert their effects through an interaction with essential regulators of the cell cycle, including cyclin-dependent kinase enzyme (CDK) complexes, Cip/Kip family proteins which act as cell cycle inhibitors, and the phosphoinositide 3-kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway, among others.
Cyclins are positive cell cycle regulators, controlling cell cycle stage advancement via activation of CDKs. Cyclin D2 and E2, mediators of cell cycle arrest, are directly targeted by miR-26a; low miR-26a levels are frequently found in HCC[111]. Modulation of cyclin G1 affects transcriptional activity and p53 protein stability, resulting in reduced G2-M phase and lower invasive capacity of HCC cells[112]. miR-122 inhibits hepatocyte growth by targeting cyclin G1 expression, however it is barely detectable in primary human HCC[113]. Levels of miR-122 are determined by several key regulatory molecules, including the transcription factors HNF1A, HNF3A and HNF3B[114]. Low miR-122 correlates with high serum response factor, a validated miR-122 target and important promoter of tumour development[115]. Expression of miR-195 is also reduced in HCC. Normally it regulates expression of cyclin D1, CDK6 and EnF3 however in its absence there is a failure to induce cell cycle arrest at the G1-S checkpoint[116]. CDK6 is also targeted by miR-124, a miRNA which blocks G1-S transition. miR-124 is silenced in HCC by CpG methylation, as is miR-203[117].
Another method by which oncogenic miRNAs contribute to cell cycle progression is via inhibition of cyclin-dependent kinase inhibitors (CDKIs), most notably the members of the Cip/Kip family. Both miR-106b and miR-93 are overexpressed in HCC and directly target p21 and promote cell cycle progression[118]. miR-221 and miR-222 both inhibit expression of p27 mRNA, another member of the Cip/Kip family[119] whilst miR-221 also regulates the CDKI p57[120]. Direct targeting of these two CDKIs leads to greater numbers of HCC cells in the S-phase thus promoting cell growth.
PI3K has an important role in balancing cell survival and apoptosis. Its activation leads to increased cell growth via phosphorylation of mTOR by AKT kinase, an effect that is inhibited by PTEN. mTOR is a target of miR-199a-3p; restoring normal levels of miR-199a-3p can cause cell cycle arrest in HCC by blocking the G1-S transition, sensitising cells to doxorubicin[121]. miR-221 and miR-222, in addition to their effect on p27 also target DNA damage-inducible transcription factor 4 (DDIT4), a modulator of mTOR signalling[122]. PTEN is directly targeted by miR-21, -221 and -222; all three are often found to be overexpressed in HCC[123,124]. As such, suppression of PTEN resulting in increased PI3K/AKT pathway activation is an important mediator of HCC cell survival.
Other important cell cycle regulators are known targets of aberrantly-expressed miRNAs in HCC. Let-7g down regulates c-Myc, an oncogenic transcription factor. This suppresses HCC cell proliferation through reduced c-Myc-induced miR-17-92 transcription, a tumour-promoting miR[125,126]. Others such as miR-1[127] and miR-375[128] suppress HCC cell proliferation whereas miR-18a stimulates proliferation via targeting the ESR1 gene thereby preventing oestrogen’s protective effects against HCC in females[129].
These studies emphasise the important role that miRNAs have in the progression of HCC by regulating oncogenes and tumour suppressors, and a number of miRNAs have now been identified in this context.
Evasion of apoptosis is another key step in malignant transformation and tumour progression. This allows cells to escape normal surveillance mechanisms, enabling continued survival in the tumour microenvironment. The tumour suppressor gene p53 increases miR-34 expression leading to cell cycle arrest and apoptosis, whereas low miR-34 levels, as are frequently seen in HCC, are believed to contribute to apoptosis evasion[130-133]. miRNAs directly target the Bcl-2 family of genes, their proteins being either pro-apoptotic (Bim, Bmf, Bax, Bak, Bid) or anti-apoptotic (Bcl-2, Bcl-W, Bcl-XL, Mcl-1)[134]. miR-122 and let-7b regulate Bcl-w and Bcl-XL, respectively, whilst Mcl-1 is regulated by miR-101 and miR-29[104,135-137]. Reduced levels of all of these miRNAs are often seen in HCC thus increasing resistance to apoptosis. Bcl-2 is also targeted by miR-29[104]; increasing miR-29 levels can sensitise HCC cells to pro-apoptotic signals, a finding of great therapeutic application potential. With respect to miRNA regulation of pro-apoptotic Bcl-2 family members, miR-221 and miR-25 are commonly over expressed in HCC and target Bmf and Bim, respectively[138,139]. miRNAs can also target other apoptosis-related genes. miR-602 is increased in HBV-related HCC, it targets RASSF1A to exert an anti-apoptotic effect[140].
Invasion and metastasis are two hallmarks of cancers and the leading causes of cancer-related mortality. Survival rates after curative resection of HCC are still poor due to high recurrence secondary to intrahepatic metastasis. Given this, a better understanding of the mechanisms underlying invasion and metastasis is critical to improvements in patient survival. Several metastasis-related genes important in HCC have been identified, and with them, several miRNAs promoting and preventing metastasis in HCC.
miRNAs promoting metastasis: As mentioned, levels of miR-21, -221 and -222 are increased in HCC[124]. These miRNAs directly target PTEN, contributing to cell growth but also mediating cell invasion. miR-221 and miR-222 also modulate the expression of TIMP3 and phosphatase 2A subunit B (PPP2R2A), thereby preventing inactivation of metalloproteases, important enzymes involved in cell migration and invasion, and activating the PI3K pathway[124,141]. miR-181b is induced by TGF-β and also targets TIMP3 on a functional level, increasing MMP2 and MMP9 activity[142]. The TGF-β-mediated metastasis pathway is well characterised, and this TGF-β/miR-181/TIMP3 axis may be an important component. One study has also shown a novel miRNA, miR-143 is induced by NFκB, promoting metastasis of HBV-related HCC by inhibiting expression of fibronectin[143]. High miR-17-5p levels are often found in HCC. This miRNA activates p38 mitogen-activated protein kinase and leads to greater heat shock protein 27 phosphorylation thereby promoting HCC invasion[144].
The chromosomal region 8q24 is implicated in metastasis in HCC. Two frequently amplified miRNAs contained within, miRNA-30d and miRNA-151, are involved in HCC invasion and metastasis[145,146]. An increased miR-30d expression is frequently seen in HCC enhancing metastasis through repression of G-αi2. This can contribute to metastasis both within the liver and to the lung. RhoGDIA, thought to be a suppressor of HCC metastasis is targeted by miR-151; with subsequent activation of Rac1, Cdc42 and Rho GTPases enhancing cell migration and invasion[134]. Moreover, this miRNA is often co-expressed with host gene focal adhesion kinase (FAK); it can function synergistically with FAK to increase HCC cell motility and spread[134].
miRNAs preventing metastasis: ADAM10 (a distintegrin and metalloprotease family 10), serum response factor (SRF), and insulin-like growth factor 1 receptor (Igf1R) promote tumorigenesis. These are validated targets of miR-122 and their expression is up regulated in primary human HCC due to decreased miR-122 levels[115,147]. Metastatic HCCs also show significantly lower let-7g levels, a miRNA that targets type I collagen a2 and when present at normal levels should prevent HCC spread[148].
The hepatocyte growth factor (HGF)/c-Met signalling cascade is considered a key pathway in HCC metastasis[134]. HGF interacts with the c-Met receptor tyrosine kinase to increase cell motility and invasion, while also conferring apoptotic protection. c-Met is associated with aggressive HCC and poor outcomes, and is regulated by miR-1, -34a, -23b and -199-3p levels of which are low in HCC[134]. Silencing of miR-1 inhibits HCC cell growth, and increases cell invasion, through c-Met down regulation[127]. Ectopic expression of miR-34a prevents HCC invasion and migration by reducing c-Met-induced phosphorylation of extracellular signal-related kinases 1 and 2 in HepG2 cells[149]. Likewise, over expression of miR-23b reduces levels of c-Met and urokinase-type plasminogen activator, a downstream target of HGF/c-Met signalling; this inhibits HCC proliferation and migration[150]. Regulation of cell cycle progression by restoring miR-199-3p levels to normal leads to induction of G1-phase cell cycle arrest (miR-199-3p targets c-Met and mTOR) thereby decreasing HCC cells’ invasive ability[121]. Finally, miR-101 is also downregulated in HCC and reduces HGF-induced cell invasion and migration via inhibition of FOS oncogene expression[151]. Taken together these studies highlight how miRNAs control the central processes of invasion, metastasis and apoptosis that contribute to malignant transformation and tumour progression.
miRNAs are predominantly down regulated in tumour tissues[152], a pattern also seen in HCC. Several issues affect the identification and quantification of aberrantly expressed miRNAs in clinical samples confounding their potential as biomarkers. Despite these issues, several consistently dysregulated miRNAs have been identified in HCC (Table 2). Numerous studies have shown that circulating miRNA levels are altered in HCC progression. For example, serum miR-221 concentrations are 4.8-fold higher in HCC patients; high miR-221 levels correlate positively with cirrhosis, tumour size and tumour stage, and negatively correlate with overall survival[153]. Currently, there are few clinically useful serum HCC markers; α-fetoprotein (AFP), Lens culinaris agglutinin-reactive AFP (AFP-L3) and des-γ-carboxyprothrombin (DCP) are of limited use[154]. The American Association for the Study of Liver Diseases discarded AFP as a marker for HCC surveillance and diagnosis in its July 2010 Practice Guidelines, highlighting the need for new biomarkers. miRNAs may have this potential. However, their use is complicated by the need for appropriate controls, as HCC usually develops from an underlying liver condition. For example, one study compared the miRNA expression profiles of three patient groups: one with HCC, one with chronic liver disease and one consisting of normal controls[155]. This study also showed that serum miR-16 and miR-199a concentrations were reduced and significantly associated with HCC[155]; of potential clinical relevance, miR-16 was more sensitive for detection of HCC than the three currently used biomarkers. Overall, these findings show the feasibility of miRNAs as serum markers for diagnosis of HCC. Should they continue to outperform current HCC markers in further studies, circulating miRNAs could be used in first-line testing of HCC patients. However, the study of circulating miRNAs as HCC biomarkers is a relatively recent concept, with further studies and validation of results in larger patient cohorts needed before miRNAs are used in the clinical setting. In particular, the discovery of a miRNA which sensitively and reliably diagnose early stage HCC would greatly enhance their potential for clinical use.
| miRNAs | Detail | Relevance |
| miR-221 | 4.8 fold higher in HCC patients, positively correlates with cirrhosis, tumour size and stage. Negatively correlates with overall survival | Potential circulating biomarker |
| miR-199a | Reduced and significantly associated with HCC | Potential circulating biomarker |
| miR-16 | Reduced and significantly associated with HCC | Potential circulating biomarker |
| miR-26 | Low levels associated with high IL-6 and shorter survival | Potential biomarker to assess prognosis of HCC |
| miR-375 | Lower than normal levels associated with β-catenin mutation | Potential for HCC classification system, determine treatment allocation |
| miR-107 | Reduced levels associated with HFN 1α | Potential for HCC classification system, use to determine treatment allocation |
| miR-122 | Expression inhibited using Miravirsen LNA-modified oligonucleotides | Direct effect in chimpanzee model in reducing HCV replication and viraemia |
| miR-196 | Selective target for intervention | Implications for treatment |
| miR-26a | Deliverable to HCC sites using adeno-associated virus serotype 8 | Decreased proliferation and induced tumour-specific apoptosis |
| miR-124 | Induces tumour-specific apoptosis | Prevents and suppresses HCV development in murine model |
miRNA expression profiles can also be used to assess prognosis. For example, low miR-26 expression is associated with high interleukin-6 expression and shorter survival[156]; better response to interferon treatment also occurs in patients with low miR-26 levels. Furthermore, a 20-miRNA signature which accurately predicts survival and recurrence of HCC has been developed[157]. These studies suggest that miRNA profiling may play an important role in HCC management in the clinic, both for classification of HCC into subtypes determining treatment and in assessment of prognosis. Patterns of dysregulated miRNAs distinguish tumours based on molecular characteristics. For example, β-catenin mutation is associated with reduced miR-375 levels, and reduced miR-107 levels with HNF1α[158]. Such findings led to the proposal of a miRNA-based HCC classification system[159]; this could be used to determine treatment allocation, based on molecular pathology.
Efficacy of miRNA-based gene therapy in HCC treatment has been demonstrated (Table 2). In one study, miR-122 expression was inhibited in chimpanzees using SPC3649 LNA-modified oligonucleotides. As miR-122 up regulates HCV replication in infected hepatocytes, its inhibition reduced HCV RNA production and decreased viraemia[160]. A phase I trial for SPC3649 (Miravirsen) resulted, becoming the first miRNA-targeted drug to enter human clinical trials. Miravirsen was well-tolerated and is currently undergoing phase II trials in HCV null responders to pegylated interferon-α and ribavirin. However, issues regarding possible viral escape are arising, with one study showing that mutations in the miR-122 binding site in HCV 5’-UTR decreases Miravirsen efficacy[161]. Similarly, therapeutic miR-196 targeting has been investigated, with the results of these and similar studies likely to have significant implications for future treatment of HCV infection and HCC[162]. Recently it was demonstrated that HNF4α, a key regulator of hepatocellular carcinogenesis, becomes stably inhibited during hepatocellular transformation. Perturbation of this event through miR-124 systemic administration can prevent and suppress HCC development in a murine liver cancer model by inducing tumour-specific apoptosis without toxic side effects[163]. Thus miR-124 has therapeutic potential for treating liver cancer.
Several virally-delivered ‘‘classical’’ gene therapy products developed for HCC are currently progressing through clinical trial phases; however, virus-delivered miRNA-based gene therapies have yet to be tested in clinical trials[2]. Accurate assessment of this method’s potential risks must be performed before further progress can be achieved. Nevertheless, early results from studies investigating the therapeutic delivery of miRNAs are showing promise. One such study in mice used self-complementary AAV serotype 8 (scAAV8) to deliver miR-26a to the HCC site; this delivery restored miR-26a expression in HCC cells, specifically decreasing cancer cell proliferation, inducing tumour-specific apoptosis, and protecting from HCC progression without toxicity[112]. 80% of treated mice had no or small tumours at 3 wk post-transduction, while most liver tissue in the untreated control group was replaced with HCC tumours. This study is of critical importance to the future of HCC treatment in that it was the first to demonstrate the therapeutic potential of restoration of expression of a dysregulated miRNA in the liver. Despite this, the relevance of therapeutic miRNA delivery to human HCC patients remains to be determined, emphasising the considerable amount of research needed in this field before clinical applications can be made. Nevertheless, the early successes of RNA-based therapies in clinical trials demonstrate that miRNAs and their inhibitors show great therapeutic promise for HCC. Future studies will no doubt shed light on how best miRNAs have the potential to alter survival rates of HCC patients.
Findings have also pointed towards long non-coding RNAs (lncRNA) as important tumorigenic candidates actively involved in gene regulation, with lncRNAs suggested as a link in carcinogenesis. Morover, lncRNAs can act as negative regulators of miRNAs and therefore may become important factors to consider when developing miRNA therapeutics. Several reports demonstrate an association of lncRNA with the development, progression, metastasis and poor prognosis in HCC patients[164-168].
In summary, studies have demonstrated unequivocally that miRNAs are important modulators of mRNA and protein expression. They are known to be involved in a variety of biological and pathological processes, such as the regulation of iron homeostasis and in HCC development and progression. As predicted by bioinformatic analysis and confirmed by numerous studies, some miRNAs target multiple genes involved in HCC progression. Similarly, several miRNAs often regulate a single aberrantly expressed gene. From these findings, we see that HCC progression is determined by a complex interaction of dysregulated miRNAs and their target mRNAs. This must be kept in mind when investigating the therapeutic potential of miRNAs, as changing the expression of a single miRNA may not be adequate to alter expression of the target gene.
Investigations into the potential clinical uses of miRNAs are ongoing, most notably in the early diagnosis and treatment of HCC. In addition, using miRNAs to subdivide HCC cases based on molecular pathology has been proposed; this system could also determine treatment allocation and aid in prognostic assessment. Overall, it seems likely that miRNAs will play an increasingly important role in the diagnosis and treatment of liver diseases associated with HCC over the coming years, leading to improved patient survival rates and better patient outcomes.
| 1. | Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674-687. [PubMed] |
| 2. | Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56:1371-1383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 196] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 3. | Bosch FX, Ribes J, Cléries R, Díaz M. Epidemiology of hepatocellular carcinoma. Clin Liver Dis. 2005;9:191-211, v. [PubMed] |
| 4. | Lawless MW, Mankan AK, Gray SG, Norris S. Endoplasmic reticulum stress--a double edged sword for Z alpha-1 antitrypsin deficiency hepatoxicity. Int J Biochem Cell Biol. 2008;40:1403-1414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 5. | Lawless MW, Greene CM, Mulgrew A, Taggart CC, O’Neill SJ, McElvaney NG. Activation of endoplasmic reticulum-specific stress responses associated with the conformational disease Z alpha 1-antitrypsin deficiency. J Immunol. 2004;172:5722-5726. [PubMed] |
| 6. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. [PubMed] |
| 7. | Aravalli RN, Steer CJ, Cressman EN. Molecular mechanisms of hepatocellular carcinoma. Hepatology. 2008;48:2047-2063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 513] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 8. | Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175-205. [PubMed] |
| 9. | Zhang B, Wang Q, Pan X. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210:279-289. [PubMed] |
| 10. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16302] [Article Influence: 958.9] [Reference Citation Analysis (2)] |
| 11. | Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931-1934. [PubMed] |
| 12. | Liu J, Rivas FV, Wohlschlegel J, Yates JR, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261-1266. [PubMed] |
| 13. | Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J. 2012;18:215-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 245] [Article Influence: 17.5] [Reference Citation Analysis (1)] |
| 14. | Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci USA. 2007;104:9667-9672. [PubMed] |
| 15. | Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5’ untranslated region or the open reading frame. RNA. 2010;16:2493-2502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 119] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 16. | Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 918] [Cited by in RCA: 1037] [Article Influence: 57.6] [Reference Citation Analysis (0)] |
| 17. | Qin W, Shi Y, Zhao B, Yao C, Jin L, Ma J, Jin Y. miR-24 regulates apoptosis by targeting the open reading frame (ORF) region of FAF1 in cancer cells. PLoS One. 2010;5:e9429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 173] [Cited by in RCA: 198] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 18. | Beitzinger M, Meister G. Preview. MicroRNAs: from decay to decoy. Cell. 2010;140:612-614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 19. | Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149-D153. [PubMed] |
| 20. | Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5833] [Cited by in RCA: 6651] [Article Influence: 369.5] [Reference Citation Analysis (0)] |
| 21. | Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495-500. [PubMed] |
| 22. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [PubMed] |
| 23. | Nahon P, Ganne-Carrié N, Trinchet JC, Beaugrand M. Hepatic iron overload and risk of hepatocellular carcinoma in cirrhosis. Gastroenterol Clin Biol. 2010;34:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 24. | Kew MC. Hepatic iron overload and hepatocellular carcinoma. Cancer Lett. 2009;286:38-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 67] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 25. | Strohmeyer G, Niederau C, Stremmel W. Survival and causes of death in hemochromatosis. Observations in 163 patients. Ann N Y Acad Sci. 1988;526:245-257. [PubMed] |
| 26. | Bothwell TH, Bradlow BA. Siderosis in the Bantu. A combined histopathological and chemical study. Arch Pathol. 1960;70:279-292. [PubMed] |
| 27. | Bothwell TH, Isaacson C. Siderosis in the bantu. A comparison of incidence in males and females. Br Med J. 1962;1:522-524. [PubMed] |
| 28. | Niederau C, Fischer R, Sonnenberg A, Stremmel W, Trampisch HJ, Strohmeyer G. Survival and causes of death in cirrhotic and in noncirrhotic patients with primary hemochromatosis. N Engl J Med. 1985;313:1256-1262. [PubMed] |
| 29. | Hsing AW, McLaughlin JK, Olsen JH, Mellemkjar L, Wacholder S, Fraumeni JF. Cancer risk following primary hemochromatosis: a population-based cohort study in Denmark. Int J Cancer. 1995;60:160-162. [PubMed] |
| 30. | Purtilo DT, Gottlieb LS. Cirrhosis and hepatoma occurring at Boston City Hospital (1917-1968). Cancer. 1973;32:458-462. [PubMed] |
| 31. | MacSween RN, Scott AR. Hepatic cirrhosis: a clinico-pathological review of 520 cases. J Clin Pathol. 1973;26:936-942. [PubMed] |
| 32. | Blumberg RS, Chopra S, Ibrahim R, Crawford J, Farraye FA, Zeldis JB, Berman MD. Primary hepatocellular carcinoma in idiopathic hemochromatosis after reversal of cirrhosis. Gastroenterology. 1988;95:1399-1402. [PubMed] |
| 34. | Goh J, Callagy G, McEntee G, O’Keane JC, Bomford A, Crowe J. Hepatocellular carcinoma arising in the absence of cirrhosis in genetic haemochromatosis: three case reports and review of literature. Eur J Gastroenterol Hepatol. 1999;11:915-919. [PubMed] |
| 35. | Köhler HH, Höhler T, Küsel U, Kirkpatrick CJ, Schirmacher P. Hepatocellular carcinoma in a patient with hereditary hemochromatosis and noncirrhotic liver. A case report. Pathol Res Pract. 1999;195:509-513. [PubMed] |
| 36. | Pellisé M, González-Abraldes J, Navasa M, Miquel R, Bruguera M. [Hepatocellular carcinoma in a patient with hereditary hemochromatosis without cirrhosis]. Gastroenterol Hepatol. 2001;24:132-134. [PubMed] |
| 37. | Britto MR, Thomas LA, Balaratnam N, Griffiths AP, Duane PD. Hepatocellular carcinoma arising in non-cirrhotic liver in genetic haemochromatosis. Scand J Gastroenterol. 2000;35:889-893. [PubMed] |
| 38. | Gray SG, Crowe J, Lawless MW. Hemochromatosis: as a conformational disorder. Int J Biochem Cell Biol. 2009;41:2094-2097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 39. | Ryan JD, Ryan E, Fabre A, Lawless MW, Crowe J. Defective bone morphogenic protein signaling underlies hepcidin deficiency in HFE hereditary hemochromatosis. Hepatology. 2010;52:1266-1273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 40. | Fracanzani AL, Conte D, Fraquelli M, Taioli E, Mattioli M, Losco A, Fargion S. Increased cancer risk in a cohort of 230 patients with hereditary hemochromatosis in comparison to matched control patients with non-iron-related chronic liver disease. Hepatology. 2001;33:647-651. [PubMed] |
| 41. | Gordeuk VR, McLaren CE, MacPhail AP, Deichsel G, Bothwell TH. Associations of iron overload in Africa with hepatocellular carcinoma and tuberculosis: Strachan’s 1929 thesis revisited. Blood. 1996;87:3470-3476. [PubMed] |
| 42. | Moyo VM, Makunike R, Gangaidzo IT, Gordeuk VR, McLaren CE, Khumalo H, Saungweme T, Rouault T, Kiire CF. African iron overload and hepatocellular carcinoma (HA-7-0-080). Eur J Haematol. 1998;60:28-34. [PubMed] |
| 43. | Mandishona E, MacPhail AP, Gordeuk VR, Kedda MA, Paterson AC, Rouault TA, Kew MC. Dietary iron overload as a risk factor for hepatocellular carcinoma in Black Africans. Hepatology. 1998;27:1563-1566. [PubMed] |
| 44. | Asare GA, Mossanda KS, Kew MC, Paterson AC, Kahler-Venter CP, Siziba K. Hepatocellular carcinoma caused by iron overload: a possible mechanism of direct hepatocarcinogenicity. Toxicology. 2006;219:41-52. [PubMed] |
| 45. | Asare GA, Paterson AC, Kew MC, Khan S, Mossanda KS. Iron-free neoplastic nodules and hepatocellular carcinoma without cirrhosis in Wistar rats fed a diet high in iron. J Pathol. 2006;208:82-90. [PubMed] |
| 46. | Barry M, Scheuer PJ, Sherlock S, Ross CF, Williams R. Hereditary spherocytosis with secondary haemochromatosis. Lancet. 1968;2:481-485. [PubMed] |
| 47. | Chung H, Kudo M, Kawasaki T, Kitano M, Minami Y, Suetomi Y, Onda H. Hepatocellular carcinoma associated with secondary haemochromatosis in non-cirrhotic liver: a case report. Hepatol Res. 2003;26:254-258. [PubMed] |
| 48. | Borgna-Pignatti C, Vergine G, Lombardo T, Cappellini MD, Cianciulli P, Maggio A, Renda D, Lai ME, Mandas A, Forni G. Hepatocellular carcinoma in the thalassaemia syndromes. Br J Haematol. 2004;124:114-117. [PubMed] |
| 49. | Wang Q, Liu Y, An D, Diao H, Xu W, He X, Sun R, Wei L, Li L. Regulation of hepatitis C virus translation initiation by iron: role of eIF3 and La protein. Virus Res. 2012;167:302-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 50. | Liu Y, An D, Sun R, Jin L, Wang Q. Inhibition of translation initiation factors might be the potential therapeutic targets for HCV patients with hepatic iron overload. Med Hypotheses. 2012;78:142-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 51. | Liu H, Trinh TL, Dong H, Keith R, Nelson D, Liu C. Iron regulator hepcidin exhibits antiviral activity against hepatitis C virus. PLoS One. 2012;7:e46631. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 52. | Valenti L, Dongiovanni P, Fargion S. Diagnostic and therapeutic implications of the association between ferritin level and severity of nonalcoholic fatty liver disease. World J Gastroenterol. 2012;18:3782-3786. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (1)] |
| 53. | Kowdley KV, Belt P, Wilson LA, Yeh MM, Neuschwander-Tetri BA, Chalasani N, Sanyal AJ, Nelson JE. Serum ferritin is an independent predictor of histologic severity and advanced fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2012;55:77-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 415] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 54. | Valenti L, Rametta R, Dongiovanni P, Motta BM, Canavesi E, Pelusi S, Pulixi EA, Fracanzani AL, Fargion S. The A736V TMPRSS6 polymorphism influences hepatic iron overload in nonalcoholic fatty liver disease. PLoS One. 2012;7:e48804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 55. | Deugnier YM, Charalambous P, Le Quilleuc D, Turlin B, Searle J, Brissot P, Powell LW, Halliday JW. Preneoplastic significance of hepatic iron-free foci in genetic hemochromatosis: a study of 185 patients. Hepatology. 1993;18:1363-1369. [PubMed] |
| 56. | Jüngst C, Cheng B, Gehrke R, Schmitz V, Nischalke HD, Ramakers J, Schramel P, Schirmacher P, Sauerbruch T, Caselmann WH. Oxidative damage is increased in human liver tissue adjacent to hepatocellular carcinoma. Hepatology. 2004;39:1663-1672. [PubMed] |
| 57. | Loeb LA, James EA, Waltersdorph AM, Klebanoff SJ. Mutagenesis by the autoxidation of iron with isolated DNA. Proc Natl Acad Sci USA. 1988;85:3918-3922. [PubMed] |
| 58. | Meneghini R. Iron homeostasis, oxidative stress, and DNA damage. Free Radic Biol Med. 1997;23:783-792. [PubMed] |
| 59. | Lawless MW, O’Byrne KJ, Gray SG. Targeting oxidative stress in cancer. Expert Opin Ther Targets. 2010;14:1225-1245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 60. | Lawless MW, Greene CM. Toll-like receptor signalling in liver disease: ER stress the missing link? Cytokine. 2012;59:195-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 61. | Romilda C, Marika P, Alessandro S, Enrico L, Marina B, Andromachi K, Umberto C, Giacomo Z, Claudia M, Massimo R. Oxidative DNA damage correlates with cell immortalization and mir-92 expression in hepatocellular carcinoma. BMC Cancer. 2012;12:177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 62. | Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81-128. [PubMed] |
| 63. | Esterbauer H. Cytotoxicity and genotoxicity of lipid-oxidation products. Am J Clin Nutr. 1993;57:779S-785S; discussion 785S-786S. [PubMed] |
| 64. | Cheeseman KH. Mechanisms and effects of lipid peroxidation. Mol Aspects Med. 1993;14:191-197. [PubMed] |
| 65. | Cadenas E. Biochemistry of oxygen toxicity. Annu Rev Biochem. 1989;58:79-110. [PubMed] |
| 66. | Benhar M, Engelberg D, Levitzki A. ROS, stress-activated kinases and stress signaling in cancer. EMBO Rep. 2002;3:420-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 472] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 67. | Matzner Y, Hershko C, Polliack A, Konijn AM, Izak G. Suppressive effect of ferritin on in vitro lymphocyte function. Br J Haematol. 1979;42:345-353. [PubMed] |
| 68. | Green R, Esparza I, Schreiber R. Iron inhibits the nonspecific tumoricidal activity of macrophages. A possible contributory mechanism for neoplasia in hemochromatosis. Ann N Y Acad Sci. 1988;526:301-309. [PubMed] |
| 69. | Asare GA, Bronz M, Naidoo V, Kew MC. Interactions between aflatoxin B1 and dietary iron overload in hepatic mutagenesis. Toxicology. 2007;234:157-166. [PubMed] |
| 70. | Vautier G, Bomford AB, Portmann BC, Metivier E, Williams R, Ryder SD. p53 mutations in british patients with hepatocellular carcinoma: clustering in genetic hemochromatosis. Gastroenterology. 1999;117:154-160. [PubMed] |
| 71. | Marrogi AJ, Khan MA, van Gijssel HE, Welsh JA, Rahim H, Demetris AJ, Kowdley KV, Hussain SP, Nair J, Bartsch H. Oxidative stress and p53 mutations in the carcinogenesis of iron overload-associated hepatocellular carcinoma. J Natl Cancer Inst. 2001;93:1652-1655. [PubMed] |
| 72. | Hussain SP, Raja K, Amstad PA, Sawyer M, Trudel LJ, Wogan GN, Hofseth LJ, Shields PG, Billiar TR, Trautwein C. Increased p53 mutation load in nontumorous human liver of wilson disease and hemochromatosis: oxyradical overload diseases. Proc Natl Acad Sci USA. 2000;97:12770-12775. [PubMed] |
| 73. | Gouas DA, Shi H, Hautefeuille AH, Ortiz-Cuaran SL, Legros PC, Szymanska KJ, Galy O, Egevad LA, Abedi-Ardekani B, Wiman KG. Effects of the TP53 p.R249S mutant on proliferation and clonogenic properties in human hepatocellular carcinoma cell lines: interaction with hepatitis B virus X protein. Carcinogenesis. 2010;31:1475-1482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 74. | Dautry-Varsat A, Ciechanover A, Lodish HF. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc Natl Acad Sci USA. 1983;80:2258-2262. [PubMed] |
| 75. | Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE, Fleming MD. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37:1264-1269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 580] [Cited by in RCA: 549] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 76. | Muckenthaler MU, Galy B, Hentze MW. Systemic iron homeostasis and the iron-responsive element/iron-regulatory protein (IRE/IRP) regulatory network. Annu Rev Nutr. 2008;28:197-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 440] [Cited by in RCA: 542] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 77. | Magro G, Cataldo I, Amico P, Torrisi A, Vecchio GM, Parenti R, Asioli S, Recupero D, D’Agata V, Mucignat MT. Aberrant expression of TfR1/CD71 in thyroid carcinomas identifies a novel potential diagnostic marker and therapeutic target. Thyroid. 2011;21:267-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 78. | Kwok JC, Richardson DR. The iron metabolism of neoplastic cells: alterations that facilitate proliferation? Crit Rev Oncol Hematol. 2002;42:65-78. [PubMed] |
| 79. | Rovera G, Santoli D, Damsky C. Human promyelocytic leukemia cells in culture differentiate into macrophage-like cells when treated with a phorbol diester. Proc Natl Acad Sci USA. 1979;76:2779-2783. [PubMed] |
| 80. | Castoldi M, Muckenthaler MU. Regulation of iron homeostasis by microRNAs. Cell Mol Life Sci. 2012;Jun 9; Epub ahead of print. [PubMed] |
| 81. | Schaar DG, Medina DJ, Moore DF, Strair RK, Ting Y. miR-320 targets transferrin receptor 1 (CD71) and inhibits cell proliferation. Exp Hematol. 2009;37:245-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 104] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 82. | Hubert N, Hentze MW. Previously uncharacterized isoforms of divalent metal transporter (DMT)-1: implications for regulation and cellular function. Proc Natl Acad Sci USA. 2002;99:12345-12350. [PubMed] |
| 83. | Gunshin H, Allerson CR, Polycarpou-Schwarz M, Rofts A, Rogers JT, Kishi F, Hentze MW, Rouault TA, Andrews NC, Hediger MA. Iron-dependent regulation of the divalent metal ion transporter. FEBS Lett. 2001;509:309-316. [PubMed] |
| 84. | Tchernitchko D, Bourgeois M, Martin ME, Beaumont C. Expression of the two mRNA isoforms of the iron transporter Nramp2/DMTI in mice and function of the iron responsive element. Biochem J. 2002;363:449-455. [PubMed] |
| 85. | Andolfo I, De Falco L, Asci R, Russo R, Colucci S, Gorrese M, Zollo M, Iolascon A. Regulation of divalent metal transporter 1 (DMT1) non-IRE isoform by the microRNA Let-7d in erythroid cells. Haematologica. 2010;95:1244-1252. [PubMed] |
| 86. | Faller M, Matsunaga M, Yin S, Loo JA, Guo F. Heme is involved in microRNA processing. Nat Struct Mol Biol. 2007;14:23-29. [PubMed] |
| 87. | Arosio P, Levi S. Cytosolic and mitochondrial ferritins in the regulation of cellular iron homeostasis and oxidative damage. Biochim Biophys Acta. 2010;1800:783-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 241] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 88. | Shpyleva SI, Tryndyak VP, Kovalchuk O, Starlard-Davenport A, Chekhun VF, Beland FA, Pogribny IP. Role of ferritin alterations in human breast cancer cells. Breast Cancer Res Treat. 2011;126:63-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 148] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 89. | Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592-603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 972] [Cited by in RCA: 961] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 90. | Dykxhoorn DM, Wu Y, Xie H, Yu F, Lal A, Petrocca F, Martinvalet D, Song E, Lim B, Lieberman J. miR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One. 2009;4:e7181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 242] [Cited by in RCA: 261] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 91. | Castoldi M, Vujic Spasic M, Altamura S, Elmén J, Lindow M, Kiss J, Stolte J, Sparla R, D’Alessandro LA, Klingmüller U. The liver-specific microRNA miR-122 controls systemic iron homeostasis in mice. J Clin Invest. 2011;121:1386-1396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 202] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 92. | Bolondi G, Garuti C, Corradini E, Zoller H, Vogel W, Finkenstedt A, Babitt JL, Lin HY, Pietrangelo A. Altered hepatic BMP signaling pathway in human HFE hemochromatosis. Blood Cells Mol Dis. 2010;45:308-312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 93. | Jopling CL, Norman KL, Sarnow P. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122. Cold Spring Harb Symp Quant Biol. 2006;71:369-376. [PubMed] |
| 94. | Jopling CL. Regulation of hepatitis C virus by microRNA-122. Biochem Soc Trans. 2008;36:1220-1223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 103] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 95. | Sarasin-Filipowicz M. Interferon therapy of hepatitis C: molecular insights into success and failure. Swiss Med Wkly. 2010;140:3-11. [PubMed] |
| 96. | Ryan JD, Altamura S, Devitt E, Mullins S, Lawless MW, Muckenthaler MU, Crowe J. Pegylated interferon-α induced hypoferremia is associated with the immediate response to treatment in hepatitis C. Hepatology. 2012;56:492-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 97. | Padgett KA, Lan RY, Leung PC, Lleo A, Dawson K, Pfeiff J, Mao TK, Coppel RL, Ansari AA, Gershwin ME. Primary biliary cirrhosis is associated with altered hepatic microRNA expression. J Autoimmun. 2009;32:246-253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 165] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 98. | Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671-678. [PubMed] |
| 99. | Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. miR-122, a paradigm for the role of microRNAs in the liver. J Hepatol. 2008;48:648-656. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 300] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 100. | Svasti S, Masaki S, Penglong T, Abe Y, Winichagoon P, Fucharoen S, Umemura T. Expression of microRNA-451 in normal and thalassemic erythropoiesis. Ann Hematol. 2010;89:953-958. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 101. | Geekiyanage H, Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid β, novel targets in sporadic Alzheimer’s disease. J Neurosci. 2011;31:14820-14830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 211] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 102. | Jiang X, Tsitsiou E, Herrick SE, Lindsay MA. MicroRNAs and the regulation of fibrosis. FEBS J. 2010;277:2015-2021. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 211] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 103. | Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 656] [Cited by in RCA: 666] [Article Influence: 44.4] [Reference Citation Analysis (0)] |
| 104. | Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 302] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 105. | Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125-172. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2244] [Cited by in RCA: 2260] [Article Influence: 125.6] [Reference Citation Analysis (0)] |
| 106. | Venugopal SK, Jiang J, Kim TH, Li Y, Wang SS, Torok NJ, Wu J, Zern MA. Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am J Physiol Gastrointest Liver Physiol. 2010;298:G101-G106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 183] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 107. | Guo CJ, Pan Q, Jiang B, Chen GY, Li DG. Effects of upregulated expression of microRNA-16 on biological properties of culture-activated hepatic stellate cells. Apoptosis. 2009;14:1331-1340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 61] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 108. | Guo CJ, Pan Q, Li DG, Sun H, Liu BW. miR-15b and miR-16 are implicated in activation of the rat hepatic stellate cell: An essential role for apoptosis. J Hepatol. 2009;50:766-778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 243] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 109. | Ji J, Zhang J, Huang G, Qian J, Wang X, Mei S. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 2009;583:759-766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 252] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 110. | Mann J, Chu DC, Maxwell A, Oakley F, Zhu NL, Tsukamoto H, Mann DA. MeCP2 controls an epigenetic pathway that promotes myofibroblast transdifferentiation and fibrosis. Gastroenterology. 2010;138:705-714, 714.e1-4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 326] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 111. | Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005-1017. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1388] [Cited by in RCA: 1372] [Article Influence: 80.7] [Reference Citation Analysis (0)] |
| 112. | Fornari F, Gramantieri L, Giovannini C, Veronese A, Ferracin M, Sabbioni S, Calin GA, Grazi GL, Croce CM, Tavolari S. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761-5767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 321] [Article Influence: 18.9] [Reference Citation Analysis (1)] |
| 113. | Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092-6099. [PubMed] |
| 114. | Coulouarn C, Factor VM, Andersen JB, Durkin ME, Thorgeirsson SS. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526-3536. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 595] [Cited by in RCA: 598] [Article Influence: 35.2] [Reference Citation Analysis (10)] |
| 115. | Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015-32027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 415] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 116. | Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 320] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 117. | Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 467] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 118. | Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167-2174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 454] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 119. | le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699-3708. [PubMed] |
| 120. | Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651-5661. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 466] [Cited by in RCA: 491] [Article Influence: 27.3] [Reference Citation Analysis (0)] |
| 121. | Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, Pollutri D, Croce CM, Bolondi L, Gramantieri L. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184-5193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 338] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 122. | Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 542] [Cited by in RCA: 621] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 123. | Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647-658. [PubMed] |
| 124. | Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P. miR-221 and -222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 617] [Cited by in RCA: 651] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 125. | Lan FF, Wang H, Chen YC, Chan CY, Ng SS, Li K, Xie D, He ML, Lin MC, Kung HF. Hsa-let-7g inhibits proliferation of hepatocellular carcinoma cells by downregulation of c-Myc and upregulation of p16(INK4A). Int J Cancer. 2011;128:319-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 135] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 126. | O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839-843. [PubMed] |
| 127. | Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049-5058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 370] [Cited by in RCA: 374] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 128. | Liu AM, Poon RT, Luk JM. MicroRNA-375 targets Hippo-signaling effector YAP in liver cancer and inhibits tumor properties. Biochem Biophys Res Commun. 2010;394:623-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 194] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 129. | Liu WH, Yeh SH, Lu CC, Yu SL, Chen HY, Lin CY, Chen DS, Chen PJ. MicroRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology. 2009;136:683-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 212] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 130. | Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298-1307. [PubMed] |
| 131. | Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745-752. [PubMed] |
| 132. | He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130-1134. [PubMed] |
| 133. | Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731-743. [PubMed] |
| 134. | Huang S, He X. The role of microRNAs in liver cancer progression. Br J Cancer. 2011;104:235-240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 176] [Cited by in RCA: 192] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 135. | Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 207] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 136. | Shimizu S, Takehara T, Hikita H, Kodama T, Miyagi T, Hosui A, Tatsumi T, Ishida H, Noda T, Nagano H. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52:698-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 274] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 137. | Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 511] [Article Influence: 30.1] [Reference Citation Analysis (0)] |
| 138. | Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Croce CM, Bolondi L, Negrini M. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009;15:5073-5081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 251] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 139. | Li Y, Tan W, Neo TW, Aung MO, Wasser S, Lim SG, Tan TM. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009;100:1234-1242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 219] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 140. | Yang L, Ma Z, Wang D, Zhao W, Chen L, Wang G. MicroRNA-602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer Biol Ther. 2010;9:803-808. [PubMed] |
| 141. | Wong QW, Ching AK, Chan AW, Choy KW, To KF, Lai PB, Wong N. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. 2010;16:867-875. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 206] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 142. | Wang B, Hsu SH, Majumder S, Kutay H, Huang W, Jacob ST, Ghoshal K. TGFbeta-mediated upregulation of hepatic miR-181b promotes hepatocarcinogenesis by targeting TIMP3. Oncogene. 2010;29:1787-1797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 287] [Cited by in RCA: 312] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 143. | Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 286] [Article Influence: 16.8] [Reference Citation Analysis (1)] |
| 144. | Yang F, Yin Y, Wang F, Wang Y, Zhang L, Tang Y, Sun S. miR-17-5p Promotes migration of human hepatocellular carcinoma cells through the p38 mitogen-activated protein kinase-heat shock protein 27 pathway. Hepatology. 2010;51:1614-1623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 142] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 145. | Ding J, Huang S, Wu S, Zhao Y, Liang L, Yan M, Ge C, Yao J, Chen T, Wan D. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol. 2010;12:390-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 257] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 146. | Yao J, Liang L, Huang S, Ding J, Tan N, Zhao Y, Yan M, Ge C, Zhang Z, Chen T. MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 2010;51:846-856. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 115] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 147. | Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW, Chen CM, Lin CD, Liao YL, Wang JL, Chau YP. MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma. Hepatology. 2009;49:1571-1582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 450] [Cited by in RCA: 479] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 148. | Ji J, Zhao L, Budhu A, Forgues M, Jia HL, Qin LX, Ye QH, Yu J, Shi X, Tang ZY. Let-7g targets collagen type I alpha2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol. 2010;52:690-697. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 176] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 149. | Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 339] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 150. | Salvi A, Sabelli C, Moncini S, Venturin M, Arici B, Riva P, Portolani N, Giulini SM, De Petro G, Barlati S. MicroRNA-23b mediates urokinase and c-met downmodulation and a decreased migration of human hepatocellular carcinoma cells. FEBS J. 2009;276:2966-2982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 116] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 151. | Li S, Fu H, Wang Y, Tie Y, Xing R, Zhu J, Sun Z, Wei L, Zheng X. MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology. 2009;49:1194-1202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 137] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 152. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. [PubMed] |
| 153. | Li J, Wang Y, Yu W, Chen J, Luo J. Expression of serum miR-221 in human hepatocellular carcinoma and its prognostic significance. Biochem Biophys Res Commun. 2011;406:70-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 170] [Article Influence: 11.3] [Reference Citation Analysis (1)] |
| 154. | Marrero JA, Feng Z, Wang Y, Nguyen MH, Befeler AS, Roberts LR, Reddy KR, Harnois D, Llovet JM, Normolle D. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology. 2009;137:110-118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 583] [Cited by in RCA: 593] [Article Influence: 34.9] [Reference Citation Analysis (9)] |
| 155. | Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol. 2011;45:355-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 191] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 156. | Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437-1447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 665] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 157. | Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 543] [Cited by in RCA: 569] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 158. | Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955-1963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 527] [Cited by in RCA: 543] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 159. | Toffanin S, Hoshida Y, Lachenmayer A, Villanueva A, Cabellos L, Minguez B, Savic R, Ward SC, Thung S, Chiang DY. MicroRNA-based classification of hepatocellular carcinoma and oncogenic role of miR-517a. Gastroenterology. 2011;140:1618-1628.e16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 180] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 160. | Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1352] [Cited by in RCA: 1294] [Article Influence: 80.9] [Reference Citation Analysis (0)] |
| 161. | Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5’ UTR. Proc Natl Acad Sci USA. 2011;108:4991-4996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 148] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 162. | Hou W, Tian Q, Zheng J, Bonkovsky HL. MicroRNA-196 represses Bach1 protein and hepatitis C virus gene expression in human hepatoma cells expressing hepatitis C viral proteins. Hepatology. 2010;51:1494-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 165] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 163. | Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233-1247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 393] [Article Influence: 28.1] [Reference Citation Analysis (0)] |
| 164. | Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703-719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1254] [Cited by in RCA: 1519] [Article Influence: 108.5] [Reference Citation Analysis (0)] |
| 165. | Yuan SX, Yang F, Yang Y, Tao QF, Zhang J, Huang G, Yang Y, Wang RY, Yang S, Huo XS. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231-2241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 285] [Cited by in RCA: 333] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 166. | Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J, Zhang Y, Chen J, Shen H, Hu Z. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One. 2012;7:e35145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 140] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 167. | Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679-1689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 556] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 168. | Hauptman N, Glavač D. Long non-coding RNA in cancer. Int J Mol Sci. 2013;14:4655-4669. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 244] [Cited by in RCA: 294] [Article Influence: 22.6] [Reference Citation Analysis (1)] |
P- Reviewers Cui GL, Ma L, Maruyama H, Tiribelli C S- Editor Wen LL L- Editor A E- Editor Ma S