Published online Aug 14, 2013. doi: 10.3748/wjg.v19.i30.5006
Revised: June 4, 2013
Accepted: June 8, 2013
Published online: August 14, 2013
Processing time: 119 Days and 21.3 Hours
AIM: To apply a new, integrated technique for visualizing bacterial genomes to identify novel pathogenicity islands in Helicobacter pylori (H. pylori).
METHODS: A genomic barcode imaging method (converting frequency matrices to grey-scale levels) was designed to visually distinguish origin-specific genomic regions in H. pylori. The complete genome sequences of the six H. pylori strains published in the National Center for Biotechnological Information prokaryotic genome database were scanned, and compared to the genome barcodes of Escherichia coli (E. coli) O157:H7 strain EDL933 and a random nucleotide sequence. The following criteria were applied to identify potential pathogenicity islands (PAIs): (1) barcode distance distinct from that of the general background; (2) length greater than 10000 continuous base pairs; and (3) containing genes with known virulence-related functions (as determined by PfamScan and Blast2GO).
RESULTS: Comparison of the barcode images generated for the 26695, HPAG1, J99, Shi470, G27 and P12 H. pylori genomes with those for the E. coli and random sequence controls revealed that H. pylori genomes contained fewer anomalous regions. Among the H. pylori-specific continuous anomalous regions (longer than 20 kbp in each strain’s genome), two fit the criteria for identifying candidate PAIs. The bioinformatic-based functional analyses revealed that one of the two anomalous regions was the known pathogenicity island cag-PAI, this finding also served as proof-of-principle for the utility of the genomic barcoding approach for identifying PAIs, and characterized the other as a novel PAI, which was designated as tfs3-PAI. Furthermore, the cag-PAI and tfs3-PAI harbored genes encoding type IV secretion system proteins and were predicted to have potential for functional synergy.
CONCLUSION: Genomic barcode imaging represents an effective bioinformatic-based approach for scanning bacterial genomes, such as H. pylori, to identify candidate PAIs.
Core tip: The genomic barcoding technology was recently developed to increase the accuracy of genome analysis, and has facilitated the identification of origin-specific genomic regions of both eukaryotic and prokaryotic lifeforms. In this study, we applied the genomic barcode imaging approach to screen for pathogenicity islands (PAIs) in Helicobacter pylori using the six strains for which the complete genome sequences have been published and performing comparison to a common Enterobacter species. Bioinformatic-based functional analysis not only provided proof-of-principle (identifying the known cag-PAI) but also identified a novel PAI (designated as tsf3-PAI).
-
Citation: Wang GQ, Xu JT, Xu GY, Zhang Y, Li F, Suo J. Predicting a novel pathogenicity island in
Helicobacter pylori by genomic barcoding. World J Gastroenterol 2013; 19(30): 5006-5010 - URL: https://www.wjgnet.com/1007-9327/full/v19/i30/5006.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i30.5006
Helicobacter pylori (H. pylori) is a Gram-negative pathogen that colonizes the stomachs of over half the world’s population[1,2]. Despite being one of the most common chronic infections among humans, it often remains undiagnosed until an unknown trigger causes manifestation of gastric diseases (e.g., gastritis[3], ulcers[4], and gastric carcinoma[5]) with varying degrees of symptom severity and outcome. Extensive research efforts have been dedicated to understanding the molecular mechanisms of H. pylori pathogenesis, and have identified several (bona fide and putative) classes of virulence factors, including adhesins[6,7], cytotoxins[8], and lipopolysaccharide (LPS)[9]. While LPS has received the majority of research attention in the H. pylori field, due to its prevalence among pathogenic bacteria and its well-characterized interactions with the Toll-like receptor 4 of the host innate immune system, systematic investigations of the cytotoxins have also elucidated the host-pathogen signaling interactions leading to pathogenic changes in the infected tissues. For example, the vacuolating cytotoxin (VacA) has been shown to induce apoptosis in epithelial cells, and the cytotoxin-associated antigen (CagA) has been shown to counteract the VacA-induced apoptosis to promote survival of infected host cells and facilitate stomach colonization[10].
Recent evidence has suggested that pathogenicity islands (PAIs) in the bacterial genome play an important role in pathogenesis[11,12]. PAIs are defined as large DNA fragments that have been acquired through horizontal transfer and which bear multiple genes encoding bacterial factors with virulence functions[13]. The genes located on each PAI serve as molecular markers for clinical testing to diagnose bacterial pathogens, estimate their pathogenic potential, and predict treatment response (i.e., antibiotic resistance)[14]. Therefore, genomic scanning to determine the PAI profile of H. pylori will not only provide insights into the molecular evolution and pathogenic mechanisms of this important human pathogen but also identify putative targets for effective molecular therapies.
The advent of high-throughput sequencing technologies has allowed for the complete genome sequences of a large number of prokaryotes; in conjunction with the rapid accumulation of such minable data in publicly available databases, various in silico methods have been developed to detect PAIs[15,16]. Most of these methods depend on finding aberrant G + C content and/or bias in codon usage[17] among various genera and species. Yet, this approach produces a high frequency of false negative results due to post-transfer changes that naturally accumulate in the transferred fragments over the course of evolution in a new environment.
In our previous studies, we addressed the limitations of the in silico methods. It was found that when genome scanning was performed using a fixed window size of at least 1000 bp, the frequency of each κ-nucleotide sequence (2 < κ < 7) was highly stable across a whole genome[18]. As a result, we represented the κ-nucleotide sequence frequency distributions across a whole genome as a 2-D barcode-like image, which was designated as a genomic barcode. By visualizing the barcodes of each genome, we were able to easily identify those sequences of foreign origin, such as horizontally transferred genes[18].
In the current study, we applied the genomic barcode imaging technique to scan the H. pylori genome for PAIs. Both known (serving as a proof-of-principle finding) and novel PAIs were detected.
Complete genomes of the 26695, HPAG1, J99, Shi470, G27 and P12 strains of H. pylori, as well as those of Escherichia coli (E. coli) O157:H7 strain EDL933 (serving as a control for comparative analysis), were downloaded from the National Center for Biotechnological Information FTP server (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/) in January 2012. In addition, a random nucleotide sequence was generated by a K-order Markov chain model for use as an additional control.
Each genome was partitioned into non-overlapping fragments of 1000 bp and a 4-nucleotide-based barcode was calculated for each genome. Specifically, the barcode for each genome is a matrix of N (4) columns and genome length/M rows, so that N (4) = 136, with the ith value being the combined frequency of the ith 4-nucleotide and its reverse complement in this fragment. The κ-nucleotide frequencies were then converted to grey-scale levels to visualize the overall barcode image profile for the whole genome. Darker grey levels represent lower frequencies.
The following criteria were applied to identify potential PAIs: (1) barcode distance distinct from that of the general background; (2) length greater than 10000 continuous base pairs; and (3) containing genes with known virulence-related functions (as determined by PfamScan[19] and Blast2GO[20]).
The distance between two barcodes was calculated as the Euclidean distance between the corresponding 136-dimensional vectors. The distance database was built using Microsoft Excel spreadsheet software, and SPSS 13.0 statistical software was employed for analysis of the data using descriptive methods and the χ2 test.
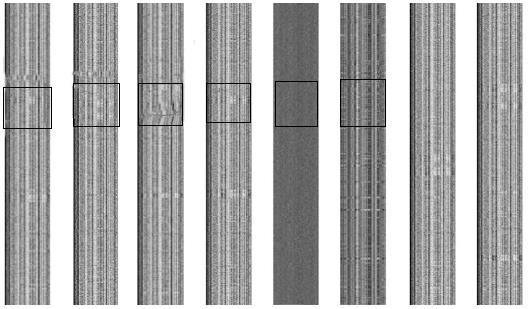
Each genome was partitioned into a series of non-overlapping fragments of 1000 bp, and the combined frequencies of each 4-nucleotide/reverse complement were calculated. The frequency matrices converted to grey-scale are shown in Figure 1. The unique barcode image for each of these microbial genomes represents the underlying base composition. The 2-D barcode images of the H. pylori strains were similar to one another but distinct from that of E. coli, demonstrating the close relationship of strains from the H. pylori species. It should be noted that no barcode structure was able to be produced for the random nucleotide sequence, indicating that the genomic barcode is an inherent property of the microbial genome.
While the genomes of different H. pylori strains possessed the conserved κ-nucleotide frequency producing the visual barcode, some regions appeared to have an abnormal structure. As shown in Figure 1, an abnormal band was apparent across the barcode image of the corresponding genome. In principle, these regions may have been acquired through horizontal gene transfer or derived from phage-mediated gene conversion.
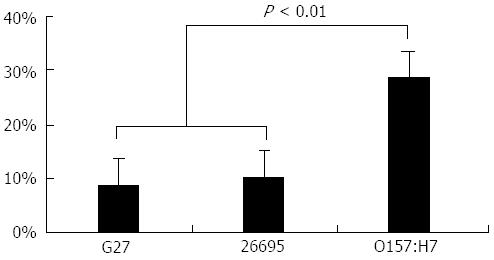
The percentage of the anomalous regions in each genome are shown in Figure 2. As expected, the H. pylori strains contain fewer anomalous regions than E. coli (P < 0.01).
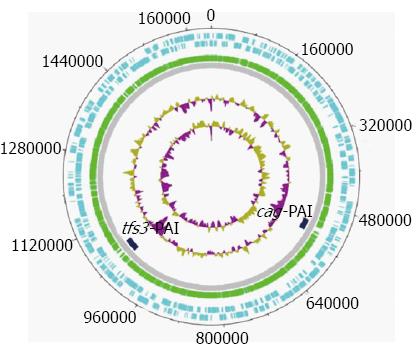
We collected continuous anomalous fragments, longer than 20 kbp in each genome, and kept only those specific for most H. pylori genomes. In addition, some anomalous fragments found only in some H. pylori genomes, but subdivided into a number of discrete smaller segments in another H. pylori genome, were excluded from further analysis since such fragments may have resulted from frequent recombination events[21,22]. As a result of this procedure, two specific genome regions were selected as potential PAI candidates. Figure 3 and Table 1 show the position of these two candidate PAIs in H. pylori.
| Size | GC | Barcodedistance | ORF | |
| Wholegenome | 1.5-1.7 Mbp | 38.0% ± 0.2% | 114.3 ± 14.9 | - |
| cag-PAI | 35 kbp | 35.4% ± 0.8% | 134.6 ± 20.1 | 20.0 ± 0.6 |
| tfs3-PAI | 30 kbp | 33.0% ± 0.8% | 138.0 ± 20.0 | 17.0 ± 3.0 |
The bioinformatic-based functional analyses revealed that one of the two anomalous regions was the known pathogenicity island cagPAI, this finding also served as proof-of-principle for the utility of the genomic barcoding approach for identifying PAIs, and characterized the other as a novel PAI, which was designated as tfs3-PAI and was located at the 3’ end of the Ser-tRNA gene.
We verified that the genes located in cag-PAI encode components of the type IV secretion system (T4SS), as characterized by previous studies[22-24].
Compared with cag-PAI, tfs3-PAI displayed some sequence variability due to rearrangements. The tfs3-PAI consisted of three distinct domains separated by mobile genetic elements. The first module contained the largest number of genes and encoded mobile sequence elements including a transposase (IS605), which is an essential element for a PAI. The second module encoded homologous genes of tfs3 gene clusters, which formed a T4SS. The function of the tfs3 gene cluster is not yet known, but it may play a role in bacterial conjugation and host cell signaling complementary to that of the cag-PAI-encoded system, which indicates a functional synergy. Most genes of the third module encoded hypothetical genes; as these genes have no orthologs in the databases, it is not clear at this point how many of them are in fact pseudogenes. It worth noting that tfs3-PAI consists of 17 open reading frames, six of which encode homologous genes of the T4SS. Therefore, this region may be related to pathogenesis in gastroduodenal diseases, and may represent a useful target for new vaccines and antibiotics.
The first potential PAI was cag-PAI, a well-known pathogenicity island in H. Pylori[25]. This approximately 35 kbp cluster of genes was acquired through horizontal transfer from an unknown extraneous source and integrated into the H. pylori chromosome. It is known that, compared to Enterobacteriaceae, H. pylori has less opportunity to obtain foreign genes by horizontal transfer since only a few bacterial species colonize human stomachs. Indeed, a previous microarray-based study of a larger strain collection suggested that up to 10% of all genes in an individual isolate may be accessory genes[26], which corroborates our finding.
T4SS is one of at least six specialized secretion systems characterized in bacteria. Usually consisting of 12 components, T4SS plays various functions in transporting a wide range of components, from single protein to protein-protein complexes and protein-DNA complexes. Moreover, T4SS facilitates injection of bacteria-encoded effectors into host cells during the infection process. The cag-PAI-encoded secretion systems have been implicated in modulation of bacteria-host interactions, interference with host signal-transduction pathways, and promotion of apoptosis, to name a few[22-24].
Pathogenesis of H. pylori is a multi-stage process. It is likely that multiple bacterial and host mechanisms are involved; however, a long-standing dogma of infectious biology claims that PAIs of H. pylori are stable entities and could be robustly correlated with disease progression or outcome. Screening and functional analysis of PAIs in H. pylori, as developed and demonstrated in this study, will aid in the development of more accurate and timely diagnosis and improved control of this common pathogen.
Recent evidence suggests that pathogenicity islands (PAIs) play an important role in bacterial pathogenesis. Scanning of PAIs in the Helicobacter pylori (H. pylori) genome will provide insights into the molecular evolution and pathogenic mechanisms of this important human pathogen but also identify putative targets for effective molecular therapies.
Auhtors have applied the genomic barcode imaging technique to scan PAIs in H. pylori. Bioinformatic-based functional analysis not only provided proof-of-principle (identifying the known cagPAI) but also identified a novel PAI (designated as tsf3-PAI).
A novel PAI, tsf3-PAI, was detected in H. pylori using the genomic barcode imaging technique. Bioinformatic-based functional analysis revealed that tsf3-PAI encodes a type IV secretion system (T4SS) which may functionally synergize with the T4SS encoded by cag-PAI.
The genomic barcode imaging technique is useful for identifying known and novel PAIs in bacterial genomes. The PAIs identified in this study may be related to the manifestation of H. pylori-induced gastroduodenal diseases, and may represent useful targets of new molecular therapies or vaccines.
The genomic barcode is generated by measuring the κ-nucleotide sequence frequency distributions across a whole genome using a fixed window size of at least 1000 bp. The 2-D barcode-like image is generated by converting the frequency matrices to grey-scale levels.
This manuscript applied genomic barcodes to screen for PAIs in H. Pylori, which showed that genomic barcode technique is more usefulness and accuracy tool for genome analysis so far. The proof-of-principle work showed that one known and one novel PAI could be detected using this technique.
| 1. | Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. The complete genome sequence of Helicobacter pylori strain G27. J Bacteriol. 2009;191:447-448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 174] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 2. | Greenberg ER, Chey WD. Defining the role of sequential therapy for H pylori infection. Lancet. 2013;381:180-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 3. | Essawi T, Hammoudeh W, Sabri I, Sweidan W, Farraj MA. Determination of Helicobacter pylori Virulence Genes in Gastric Biopsies by PCR. ISRN Gastroenterol. 2013;2013:606258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Jafarzadeh A, Akbarpoor V, Nabizadeh M, Nemati M, Rezayati MT. Total leukocyte counts and neutrophil-lymphocyte count ratios among Helicobacter pylori-infected patients with peptic ulcers: independent of bacterial CagA status. Southeast Asian J Trop Med Public Health. 2013;44:82-88. [PubMed] |
| 5. | Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1848] [Cited by in RCA: 1935] [Article Influence: 80.6] [Reference Citation Analysis (3)] |
| 6. | Niehues M, Hensel A. In-vitro interaction of L-dopa with bacterial adhesins of Helicobacter pylori: an explanation for clinicial differences in bioavailability. J Pharm Pharmacol. 2009;61:1303-1307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 54] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 7. | Niehues M, Stark T, Keller D, Hofmann T, Hensel A. Antiadhesion as a functional concept for prevention of pathogens: N-Phenylpropenoyl-L-amino acid amides as inhibitors of the Helicobacter pylori BabA outer membrane protein. Mol Nutr Food Res. 2011;55:1104-1117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 8. | Terebiznik MR, Raju D, Vázquez CL, Torbricki K, Kulkarni R, Blanke SR, Yoshimori T, Colombo MI, Jones NL. Effect of Helicobacter pylori’s vacuolating cytotoxin on the autophagy pathway in gastric epithelial cells. Autophagy. 2009;5:370-379. [PubMed] |
| 9. | Rudnicka K, Włodarczyk M, Moran AP, Rechciński T, Miszczyk E, Matusiak A, Szczęsna E, Walencka M, Rudnicka W, Chmiela M. Helicobacter pylori antigens as potential modulators of lymphocytes' cytotoxic activity. Microbiol Immunol. 2012;56:62-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 10. | Oldani A, Cormont M, Hofman V, Chiozzi V, Oregioni O, Canonici A, Sciullo A, Sommi P, Fabbri A, Ricci V. Helicobacter pylori counteracts the apoptotic action of its VacA toxin by injecting the CagA protein into gastric epithelial cells. PLoS Pathog. 2009;5:e1000603. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 109] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 11. | Ta LH, Hansen LM, Sause WE, Shiva O, Millstein A, Ottemann KM, Castillo AR, Solnick JV. Conserved transcriptional unit organization of the cag pathogenicity island among Helicobacter pylori strains. Front Cell Infect Microbiol. 2012;2:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 12. | Wang H, Han J, Chen D, Duan X, Gao X, Wang X, Shao S. Characterization of CagI in the cag pathogenicity island of Helicobacter pylori. Curr Microbiol. 2012;64:191-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 13. | Schneider G, Dobrindt U, Middendorf B, Hochhut B, Szijártó V, Emody L, Hacker J. Mobilisation and remobilisation of a large archetypal pathogenicity island of uropathogenic Escherichia coli in vitro support the role of conjugation for horizontal transfer of genomic islands. BMC Microbiol. 2011;11:210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 14. | Ostblom A, Adlerberth I, Wold AE, Nowrouzian FL. Pathogenicity island markers, virulence determinants malX and usp, and the capacity of Escherichia coli to persist in infants’ commensal microbiotas. Appl Environ Microbiol. 2011;77:2303-2308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 64] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 15. | Karlin S. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 2001;9:335-343. [PubMed] |
| 16. | Yoon SH, Park YK, Lee S, Choi D, Oh TK, Hur CG, Kim JF. Towards pathogenomics: a web-based resource for pathogenicity islands. Nucleic Acids Res. 2007;35:D395-D400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 66] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 17. | Oelschlaeger TA, Hacker J. Impact of pathogenicity islands in bacterial diagnostics. APMIS. 2004;112:930-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 18. | Wang G, Zhou F, Olman V, Li F, Xu Y. Prediction of pathogenicity islands in enterohemorrhagic Escherichia coli O157: H7 using genomic barcodes. FEBS Lett. 2010;584:194-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 19. | Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281-D288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1691] [Cited by in RCA: 1724] [Article Influence: 95.8] [Reference Citation Analysis (0)] |
| 20. | Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420-3435. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2916] [Cited by in RCA: 3156] [Article Influence: 175.3] [Reference Citation Analysis (0)] |
| 21. | Aras RA, Kang J, Tschumi AI, Harasaki Y, Blaser MJ. Extensive repetitive DNA facilitates prokaryotic genome plasticity. Proc Natl Acad Sci USA. 2003;100:13579-13584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 101] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 22. | Dobrindt U, Hochhut B, Hentschel U, Hacker J. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol. 2004;2:414-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 765] [Cited by in RCA: 801] [Article Influence: 36.4] [Reference Citation Analysis (0)] |
| 23. | Targosz A, Brzozowski T, Pierzchalski P, Szczyrk U, Ptak-Belowska A, Konturek SJ, Pawlik W. Helicobacter pylori promotes apoptosis, activates cyclooxygenase (COX)-2 and inhibits heat shock protein HSP70 in gastric cancer epithelial cells. Inflamm Res. 2012;61:955-966. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 24. | Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat Rev Gastroenterol Hepatol. 2010;7:629-641. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 500] [Cited by in RCA: 469] [Article Influence: 29.3] [Reference Citation Analysis (2)] |
| 25. | Kaplan-Türköz B, Jiménez-Soto LF, Dian C, Ertl C, Remaut H, Louche A, Tosi T, Haas R, Terradot L. Structural insights into Helicobacter pylori oncoprotein CagA interaction with β1 integrin. Proc Natl Acad Sci USA. 2012;109:14640-14645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 103] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 26. | Dong QJ, Wang Q, Xin YN, Li N, Xuan SY. Comparative genomics of Helicobacter pylori. World J Gastroenterol. 2009;15:3984-3991. [PubMed] |
P- Reviewers Balaban YH, Beales ILP, Day AS, Slomiany BL S- Editor Gou SX L- Editor A E- Editor Li JY