Published online Oct 21, 2012. doi: 10.3748/wjg.v18.i39.5570
Revised: March 28, 2012
Accepted: April 9, 2012
Published online: October 21, 2012
AIM: To analysis the factors that predict the response to entecavir therapy in chronic hepatitis patients with hepatitis B virus (HBV) genotype C.
METHODS: Fifty patients [hepatitis B e antigen (HBeAg)-negative:HBeAg-positive = 26:24] with HBV genotype C, who received naïve entecavir therapy for > 2 years, were analyzed. Patients who showed HBV DNA levels ≥ 3.0 log viral copies/mL after 2 years of entecavir therapy were designated as slow-responders, while those that showed < 3.0 log copies/mL were termed rapid-responders. Quantitative hepatitis B surface antigen (HBsAg) levels (qHBsAg) were determined by the Architect HBsAg QT immunoassay. Hepatitis B core-related antigen was detected by enzyme immunoassay. Pre-C and Core promoter mutations were determined using by polymerase chain reaction (PCR). Drug-resistance mutations were detected by the PCR-Invader method.
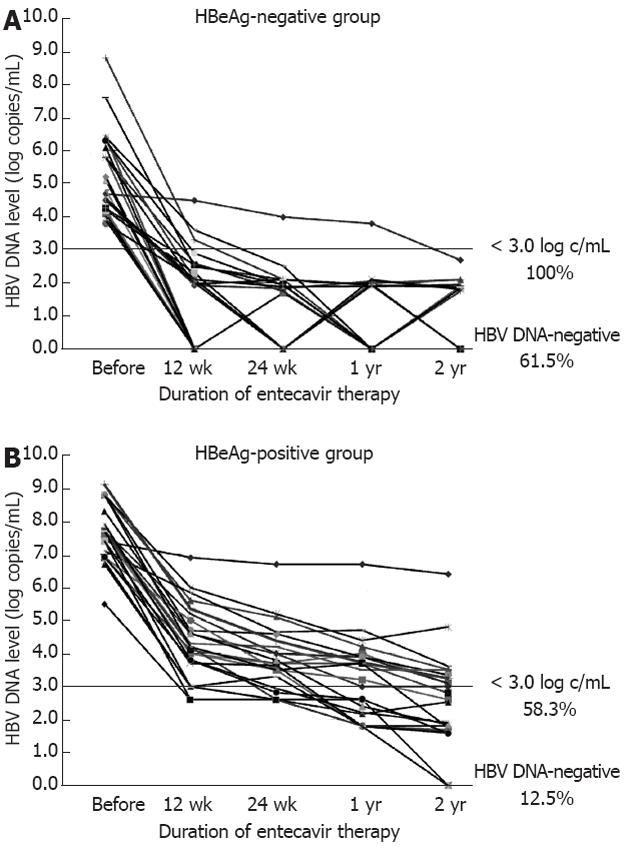
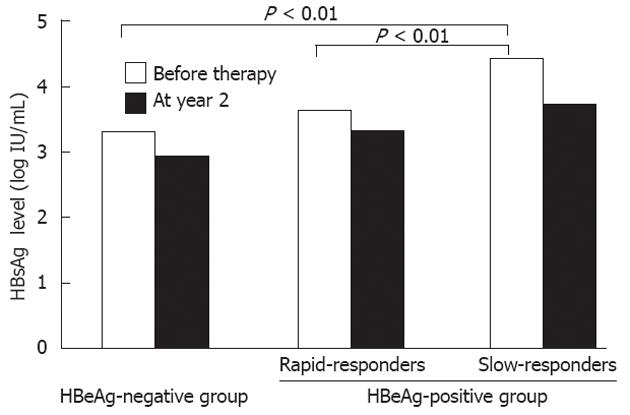
RESULTS: At year 2, HBV DNA levels in all patients in the HBeAg-negative group were < 3.0 log copies/mL. In contrast, in the HBeAg-positive group, 41.7% were slow-responders, while 58.3% were rapid-responders. No entecavir-resistant mutants were detected in the slow-responders. When the pretreatment factors were compared between the slow- and rapid-responders; the median qHBsAg in the slow-responders was 4.57 log IU/mL, compared with 3.63 log IU/mL in the rapid-responders (P < 0.01). When the pretreatment factors predictive of HBV DNA-negative status at year 2 in all 50 patients were analyzed, HBeAg-negative status, low HBV DNA levels, and low qHBsAg levels were significant (P < 0.01). Multivariate analysis revealed that the low qHBsAg level was the most significant predictive factor (P = 0.03).
CONCLUSION: Quantitation of HBsAg could be a useful indicator to predict response to entecavir therapy.
- Citation: Orito E, Fujiwara K, Kanie H, Ban T, Yamada T, Hayashi K. Quantitation of HBsAg predicts response to entecavir therapy in HBV genotype C patients. World J Gastroenterol 2012; 18(39): 5570-5575
- URL: https://www.wjgnet.com/1007-9327/full/v18/i39/5570.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i39.5570
Hepatitis B virus (HBV) is a major causative agent of chronic liver diseases[1]. Various strains of HBV have been isolated all over the world, and have been classified as HBV genotypes from A to J[2]. In Japan, about 85% of patients have HBV genotype C, and about 12% have HBV genotype B[3]. Worldwide, HBV genotypes show specific geographical distributions. HBV genotypes A and D are prevalent in the United States and Europe, while HBV genotypes B and C are prevalent in Asia[4]. Disease progression and prevalence of hepatitis B e antigen (HBeAg)-positive status are often associated with HBV genotypes[5]. Therefore, we analyzed the clinical and virological features of patients with HBV genotype C and homogenous backgrounds, because HBV genotype C is the predominant type in Japan.
Entecavir is widely used as a first-choice nucleot(s)ide analog (NA) for chronic hepatitis B patients, because less than 1% of entecavir-naive patients developed resistant mutants after 5 years of therapy[6-8]. However, in HBeAg-positive patients, the response rate to entecavir therapy is less favorable compared with HBeAg-negative patients[6,7]. In addition, some patients show a slow-response, which indicates that serum HBV DNA levels remain high after long-term entecavir therapy. However, it is unclear which patients become slow-responders. Therefore, the aim of this study is to clarify the virological and clinical characteristics of the slow-responders before and during long-term entecavir therapy for HBV genotype C.
From July 2007, 102 consecutive hepatitis B surface antigen (HBsAg)-positive patients with chronic liver disease were enrolled in a naïve entecavir therapy in our hospital. Ten patients dropped out, 15 patients discontinued therapy, 10 patients received immunosuppressive therapy during entecavir therapy, and 10 patients received entecavir for less than 2 years. Thus, 57 patients were analyzed in this prospective, single center study. The institutional review board of the hospital approved the study. Serum samples were drawn from the patients after obtaining written informed consent.
All the patients received 0.5 mg of entecavir daily. Patients with poor adherence were excluded from the study.
All patients were positive for HBsAg for more than 6 mo, had serum HBV DNA of ≥ 3 log viral copies/mL, were negative for anti-HCV, and were negative for anti-human immunodeficiency virus before entecavir therapy. Patients with decompensated cirrhosis, acute hepatitis, or acute exacerbation were excluded. Liver biopsy was not performed in some patients; therefore, the liver disease status was diagnosed by the clinical, laboratory, and imaging tests.
HBeAg-positive patients whose serum HBV DNA levels remained ≥ 3.0 log copies/mL after 2 years of entecavir therapy were considered to be slow-responders, while patients with < 3.0 log copies/mL were designated as rapid-responders.
Quantitation of HBsAg (qHBsAg) was performed using the Architect HBsAg QT immunoassay (Abott Japan, Tokyo, Japan), in accordance with the manufacturer’s instructions[9]. The detection range was 0.05 to 250 IU/mL. If HBsAg levels were found to be higher than 250 IU/mL, samples were diluted to 1:500 to 1:20 000. In this study, the results of the quantitative HBsAg levels are shown as the logarithmic value. HBV genotypes were detected by enzyme immunoassay (EIA) (Institute of Immunology, Tokyo, Japan)[10]. The HBV core-related antigen (HBcrAg) was detected by a chemilimunescent EIA method (Fujirebio Inc., Tokyo, Japan)[11]. Pre-C mutation and Core promoter mutations were detected by polymerase chain reaction (PCR) (HBV DNA precore/core promoter mutation decision kit; Roche Diagnostics Japan, Tokyo, Japan). Drug-resistant mutations in HBV against nucleotide analogs (NAs; lamivudine, adefovir and entecavir) were detected by the PCR-Invader method (BML Inc., Tokyo, Japan)[12].
Of the 57 patients, 50 patients were genotype C, three patients were HBV genotype A, one was genotype D, and three were of indeterminate genotype on EIA. Thus, the 50 patients with HBV genotype C were analyzed. Baseline characteristics of the 50 patients are shown in Table 1. The median age of the HBeAg-negative group age was significantly higher, the median alanine aminotransferase (ALT) level was significantly lower, and the median HBV DNA level was significantly lower than those in the HBeAg-positive group.
| HBeAg-negative group | HBeAg-positive group | P value | |
| No. | 26 | 24 | NS |
| Age (yr) | 57.2 (35-80) | 44.2 (35-71) | < 0.01 |
| Gender, M:F | 15:16 | 16:11 | NS |
| Diseases, CH:LC/HCC | 21:5 | 23:1 | NS |
| ALT (IU/mL) | 38 (13-950) | 102 (812-602) | < 0.01 |
| Platelet counts (× 104/mL) | 18.6 (3.4-4.9) | 18.0 (8.4-26.8) | NS |
| Albumin (mg/dL) | 4.3 (3.4-4.9) | 4.2 (2.3-5.0) | NS |
| Serum HBV DNA level | 5.1 (3.9-8.8) | 7.6 (5.6-8.8) | < 0.01 |
| (log copies/mL) |
After 2 years of entecavir therapy, the rates of normalization (< 40 IU/L) of ALT levels were 87.0% in the HBeAg-negative group and 92.5% in the HBeAg-positive group (P = Not significant).
In contrast, at year 2, the rates of reduction in HBV DNA to < 3.0 log copies/mL were 100% in the HBeAg-negative group and 58.3% in the HBeAg-positive group (P < 0.01). Thus, in the HBeAg-positive group, 58.3% of patients were designated as rapid-responders, and 41.7% were designated as slow-responders (HBV DNA levels ≥ 3.0 log copies/mL at year 2) (Figure 1). In addition, in the HBeAg-negative group, real-time PCR indicated that 61.5% of the patients were negative for HBV DNA, compared to 12.5% of the HBeAg-positive patients (P < 0.01).
When pre-treatment factors were compared between the rapid- and slow-responders (Table 2), age, gender, disease, platelet counts, and albumin were not significantly different. The median ALT level in the rapid-responders group was 131 IU/L compared with 31 IU/L in the slow-responders (P = 0.02). The pre-treatment median HBV DNA levels were 7.4 log copies/mL in the rapid-responders, and 8.3 in the slow-responders (P = 0.06). The was no difference in the rate of Pre-C and Core promoter mutations between the responder groups. In contrast, the rate of Pre-C mutations in the HBeAg-negative group was 83.3%, compared with 0% in the HBeAg-positive group. Pre-treatment HBcrAg levels did not differ among the three groups. In contrast, the pretreatment median qHBsAg level was 3.63 log IU/mL in the rapid-responders, compared with 4.57 log IU/mL in the slow-responders (P < 0.01).
| Characteristics | HBeAg-negative group | HBeAg-positive group | P value | |
| (n = 26) | RR (n = 14) | SR (n = 10) | RR vs SR | |
| <Baseline data> | ||||
| Age | 58 (35-80) | 45 (34-68) | 43 (31-71) | NS |
| Gender (male:female) | 13:13 | 9:5 | 6:4 | NS |
| Disease (CH:LC/HCC) | 21:5 | 13:1 | 6:4 | NS |
| ALT (IU/L) | 38 (13-950) | 131 (12-602) | 31 (13-108) | 0.02 |
| Platelet count (× 104/mL) | 18.6 (3.4-35.1) | 17.1 (8.4-22.4) | 20.0 (11.0-26.8) | NS |
| Albumin (mg/dL) | 4.3 (3.4-4.9) | 4.0 (2.3-5.0) | 4.4 (3.7-4.6) | NS |
| HBV genotype C | 100% | 100% | 100% | NS |
| HBV DNA (log copies/mL) | 5.1 (3.9-8.8) | 7.4 (5.6-8.8) | 8.3 (7.1-8.8) | NS |
| qHBsAg (log IU/mL) | 3.17 (0.70-4.58) | 3.63 (1.68-4.34) | 4.57 (4.35-4.76) | < 0.01 |
| HBcrAg (log U/mL) | 3.6 (3.0-> 6.8) | > 6.8 (6-> 6.8) | > 6.8 (> 6.8-> 6.8) | NS |
| Pre-C mutation (%) | 83.3 | 0 | 0 | NS |
| Core promoter mutation (%) | 58.3 | 57.1 | 50.0 | NS |
| <At year 2 during therapy> | ||||
| HBV DNA (log copies/mL) | 0.0 (0.0-2.7) | 2.1 (0.0-2.1) | 3.5 (3.1-6.9) | - |
| ALT (IU/L) | 18 (9-75) | 17.5 (10-31) | 23 (13-37) | NS |
| HBeAg seroconversion | - | 23.50% | 0% | NS |
| HBsAg seroclearance | 0% | 0% | 0% | NS |
| qHBsAg (log IU/mL) | 2.91 (0.62-3.9) | 3.25 (1.70-3.92) | 4.12 (3.23-4.47) | 0.01 |
| HBcrAg (log U/mL) | 3.0 (3.0-5.4) | 5.9 (4.0-> 6.8) | > 6.8 (5.2-> 6.8) | < 0.01 |
| Resistant mutations against entecavir | UDL | UDL | 0% | - |
At year 2 of therapy, the median qHBsAg level in the rapid-responders was 3.25 log IU/mL, compared with 4.12 log IU/mL in the slow-responders (P = 0.01). The median HBcrAg level in the rapid-responders was 5.85 log U/mL, compared with > 6.8 (the upper limit of the detection range) in the slow-responders (P < 0.01). In Figure 2, qHBsAg levels before treatment and at year 2 are shown for the HBeAg-negative group, and the rapid-responder and slow-responders in the HBeAg-positive group.
Among all the slow-responders, no entecavir-resistant mutations were found, although three patients showed M204I lamivudine-resistant mutations (Table 3).
| Patient | Age (yr) | Gender | Previous therapy | HBV genotype | Drug resistant mutations against | ||||||
| Lam | Lam | Lam/Ade | Ade | Ent | Ent | Ent | |||||
| L180 | M204 | A181 | N236 | T184 | S202 | M205 | |||||
| 1 | 52 | Male | No | C | Wild | Wild | Wild | Wild | Wild | Wild | Wild |
| 2 | 35 | Male | No | C | Wild | Wild | Wild | Wild | Wild | Wild | Wild |
| 3 | 68 | Male | No | C | Wild | Wild | Wild | Wild | Wild | Wild | Wild |
| 4 | 56 | Female | No | C | Wild | M204I | Wild | Wild | Wild | Wild | Wild |
| 5 | 36 | Female | No | C | Wild | M204I | Wild | Wild | Wild | Wild | Wild |
| 6 | 45 | Male | No | C | Wild | M204I | Wild | Wild | Wild | Wild | Wild |
| 7 | 35 | Male | No | C | Wild | Wild | Wild | Wild | Wild | Wild | Wild |
| 8 | 67 | Female | No | C | Wild | Wild | Wild | Wild | Wild | Wild | Wild |
| 9 | 39 | Male | No | C | Wild | Wild | Wild | Wild | Wild | Wild | Wild |
| 10 | 44 | Female | No | C | Wild | Wild | Wild | Wild | Wild | Wild | Wild |
At year 2 of entecavir therapy, among 50 patients, real-time PCR showed that 19 (38.0%) were negative for HBV DNA, compared with 31 (62.0%) who were still positive for HBV DNA (Table 4). The pretreatment clinical and virological characteristics between the HBV DNA-negative and -positive groups were compared by univariate analysis. In the HBV DNA-negative group, the median ALT level was significantly lower, the rate of HBeAg-negative status was significantly higher, the median HBV DNA level was lower, and the median qHBsAg level was lower, than those in the HBV DNA-positive group.
| Characteristics | HBV DNA-negative group | HBV DNA-positive group | P value | |
| (n = 19) | (n = 31) | Univariate analysis | Multivariate analysis | |
| Age | 51 (31-73) | 52 (32-80) | NS | |
| Gender (male:female) | 12:7 | 20:11 | NS | |
| Disease (CH:LC/HCC) | 17:2 | 27:4 | NS | |
| ALT (IU/L) | 36 (12-366) | 108 (13-602) | 0.03 | NS |
| Platelet counts (× 104/mL) | 19.0 (8.8-35.1) | 17.8 (3.4-26.8) | NS | |
| Albumin (mg/dL) | 4.35 (3.84-4.85) | 4.14 (2.28-4.72) | NS | |
| HBV genotype (B:C:others) | 0:19:0 | 0:31: | NS | |
| HBeAg status (positive:negative) | 3:16 | 21:10 | < 0.01 | NS |
| HBV DNA (log copies/mL) | 5.1 (3.1-7.4) | 7.6 (3.7-8.8) | < 0.01 | NS |
| qHBsAg level (log IU/mL) | 3.31 (1.90-4.08) | 4.20 (3.06-4.87) | < 0.01 | 0.03 |
| HBcrAg level (log U/mL) | 3.45 (3.0-> 6.8) | > 6.8 (3.0-> 6.8) | NS | |
| Pre-C mutation (%) | 75.0 | 43.3 | NS | |
| Core promoter mutation (%) | 37.5 | 60.0 | NS | |
However, when multivariate analysis using logistic regression analysis was performed, the median qHBsAg level was the only significant factor that predicted the negative HBV DNA status at year 2 of entecavir therapy (odds ratio 8.16, 95% CI: 1.28-52.18, P = 0.03).
In this study, the clinical and virological features of patients with HBV genotype C who received naïve-entecavir therapy were analyzed. After 2 years of entecavir therapy, about 42% of the HBeAg-positive patients showed HBV DNA levels ≥ 3 log copies/mL, while all of the HBeAg-negative patients showed < 3 log copies/mL. Therefore, the factors associated with the slow response to entecavir therapy among the HBeAg-positive group were studied initially. In addition, among the 50 patients, 38% showed HBV DNA-negative status at year 2. Thus, the pretreatment factors that predict the loss of HBV DNA were analyzed in all 50 patients. According to the multivariate analysis, qHBsAg levels are the most important factor for predicting the response to entecavir therapy in patients with HBV genotype C.
In Japan, HBV genotype C is the most prevalent[3]. The response rates to interferon or NA therapy in patients with HBV genotype C, as well as D, are poor when compared to those with HBV genotype B or A[13]. Thus, in this study, only subjects with HBV genotype C were studied.
Recently, a decline in HBsAg levels during PEG-interferon therapy was reported to be significant in the evaluation of the response to therapy[14-17]. In these reports, HBsAg levels were found to be one of the best viral markers for predicting response to anti-viral therapy and viral activity levels in hepatocytes, correlating with cccDNA levels[14,18]. Although qHBsAg is a predictor of response to entecavir therapy[19], there have been no reports in a homogeneous HBV genotype setting. In this study, qHBsAg was demonstrated to be the most significant predictor of entecavir therapy in patients with HBV genotype C.
HBV DNA levels are also considered an important factor associated with response to anti-viral therapy[17]. In this study, there was a tendency for higher HBV DNA levels in the slow-responders as compared to rapid-responders. The association between HBV DNA levels and response to therapy may be clarified further in a larger number of patients.
HBcrAg levels indicate the serum HB core antigen levels plus HBeAg levels[11]. HBcrAg levels are reported to be associated with cccDNA levels in hepatocytes[20]. However, this study showed no association between HBcrAg levels and slow response. This may be explained by the narrow quantitation range of HBcrAg levels, because HBcrAg levels in most patients in the HBeAg-positive group were greater than the upper level of the detection range of the assay.
The association between Core promoter mutation and response rate to NAs is also interesting, because the replication level of HBV is thought to be high in patients with Core promoter mutations. We reported previously that, in patients with HBV genotype C, the rates of HBeAg-positive status and Core promoter mutations are higher than those in patients with HBV genotype B[5]. In this study, a higher rate of Core promoter mutations was observed in the HBeAg-positive patients with HBV genotype C. In addition, a higher rate of Pre-C mutations was observed in the HBeAg-negative group. However, no association between the mutation rate and response rate to therapy was demonstrated in this study.
Higher ALT levels are considered an important factor in predicting good response to PEG-interferon therapy[21]. However, low ALT levels were observed in the HBV DNA-negative group at year 2 of therapy, because a high proportion of the HBeAg-negative patients had low ALT levels, compared to the HBeAg-positive patients. Thus, we consider that, during entecavir therapy, ALT levels are not associated with treatment response.
In this study, no resistant mutations against entecavir were found during 2 years therapy. As reported previously, resistant mutations against entecavir are rarely developed during 5 years of entecavir therapy[8]. Therefore, slow response was not caused by entecavir-resistant mutants.
In conclusion, we suggest that qHBsAg is a significant and convenient indicator for predicting response to entecavir therapy.
The authors would like to thank Dr. Namiki Izumi for assisting in this study.
Entecavir is a nucleot(s)ide analog that is widely used for the treatment of chronic hepatitis B patients. The efficacy of entecavir is very good for hepatitis B e antigen (HBeAg)-negative patients, but not so good for HBeAg-positive patients. The prognosis and response to anti-viral therapies depend on hepatitis B virus (HBV) genotype. The factors that affect the efficacy of entecavir therapy are still unclear, especially in patients with HBV genotype C.
As quantitation assay of serum hepatitis B surface antigen (HBsAg) has been recently developed, allowing the serum level of HBsAg to be determined over a very wide range. The upper range of HBsAg levels could be detected to 6.7 log IU/mL by the Architect HBsAg QT immunoassay when samples were diluted to 1:20 000. Thus, the authors could analyze the relationship between the efficacy of entecavir therapy and various HBV markers.
This study showed that the quantitative HBsAg level is a significant factor for predicting the efficacy of entecavir therapy in patients with HBV genotype C. Patients with low levels of HBsAg before entecavir therapy often show HBV DNA levels < 3.0 log copies/mL or are negative for HBV DNA at year 2 during therapy.
Using the quantitation of HBsAg, the efficacy of various anti-viral therapies can be predicted before treatment. The quantitation of HBsAg could be a useful tool for determining the treatment schedule for chronic hepatitis B patients.
Although small in patient numbers, the subject matter is interesting and original. This study adds to the emerging data suggesting HBsAg decline is a predictor of response.
| 1. | Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733-1745. [PubMed] |
| 2. | Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, Nakayoshi T, Wakuta M, Miyakawa Y, Mizokami M. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J Virol. 2009;83:10538-10547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 338] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 3. | Orito E, Ichida T, Sakugawa H, Sata M, Horiike N, Hino K, Okita K, Okanoue T, Iino S, Tanaka E. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology. 2001;34:590-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 310] [Cited by in RCA: 315] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 4. | Miyakawa Y, Mizokami M. Classifying hepatitis B virus genotypes. Intervirology. 2003;46:329-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 226] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 5. | Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, Okanoue T, Yotsuyanagi H, Iino S. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology. 2001;33:218-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 289] [Cited by in RCA: 315] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 6. | Chang TT, Gish RG, de Man R, Gadano A, Sollano J, Chao YC, Lok AS, Han KH, Goodman Z, Zhu J. A comparison of entecavir and lamivudine for HBeAg-positive chronic hepatitis B. N Engl J Med. 2006;354:1001-1010. [PubMed] |
| 7. | Lai CL, Shouval D, Lok AS, Chang TT, Cheinquer H, Goodman Z, DeHertogh D, Wilber R, Zink RC, Cross A. Entecavir versus lamivudine for patients with HBeAg-negative chronic hepatitis B. N Engl J Med. 2006;354:1011-1020. [PubMed] |
| 8. | Chang TT, Lai CL, Kew Yoon S, Lee SS, Coelho HS, Carrilho FJ, Poordad F, Halota W, Horsmans Y, Tsai N. Entecavir treatment for up to 5 years in patients with hepatitis B e antigen-positive chronic hepatitis B. Hepatology. 2010;51:422-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 470] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 9. | Deguchi M, Yamashita N, Kagita M, Asari S, Iwatani Y, Tsuchida T, Iinuma K, Mushahwar IK. Quantitation of hepatitis B surface antigen by an automated chemiluminescent microparticle immunoassay. J Virol Methods. 2004;115:217-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 161] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 10. | Usuda S, Okamoto H, Iwanari H, Baba K, Tsuda F, Miyakawa Y, Mayumi M. Serological detection of hepatitis B virus genotypes by ELISA with monoclonal antibodies to type-specific epitopes in the preS2-region product. J Virol Methods. 1999;80:97-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 211] [Cited by in RCA: 226] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 11. | Kimura T, Rokuhara A, Sakamoto Y, Yagi S, Tanaka E, Kiyosawa K, Maki N. Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load. J Clin Microbiol. 2002;40:439-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 196] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 12. | Tadokoro K, Suzuki F, Kobayashi M, Yamaguchi T, Nagano M, Egashira T, Kumada H. Rapid detection of drug-resistant mutations in hepatitis B virus by the PCR-Invader assay. J Virol Methods. 2011;171:67-73. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Janssen HL, van Zonneveld M, Senturk H, Zeuzem S, Akarca US, Cakaloglu Y, Simon C, So TM, Gerken G, de Man RA, Niesters HG, Zondervan P, Hansen B, Schalm SW. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: a randomised trial. Lancet. 2005;365:123-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 918] [Cited by in RCA: 910] [Article Influence: 43.3] [Reference Citation Analysis (0)] |
| 14. | Wursthorn K, Lutgehetmann M, Dandri M, Volz T, Buggisch P, Zollner B, Longerich T, Schirmacher P, Metzler F, Zankel M. Peginterferon alpha-2b plus adefovir induce strong cccDNA decline and HBsAg reduction in patients with chronic hepatitis B. Hepatology. 2006;44:675-684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 354] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 15. | Brunetto MR, Moriconi F, Bonino F, Lau GK, Farci P, Yurdaydin C, Piratvisuth T, Luo K, Wang Y, Hadziyannis S. Hepatitis B virus surface antigen levels: a guide to sustained response to peginterferon alfa-2a in HBeAg-negative chronic hepatitis B. Hepatology. 2009;49:1141-1150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 364] [Cited by in RCA: 360] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 16. | Moucari R, Mackiewicz V, Lada O, Ripault MP, Castelnau C, Martinot-Peignoux M, Dauvergne A, Asselah T, Boyer N, Bedossa P. Early serum HBsAg drop: a strong predictor of sustained virological response to pegylated interferon alfa-2a in HBeAg-negative patients. Hepatology. 2009;49:1151-1157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 361] [Cited by in RCA: 362] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 17. | Rijckborst V, Hansen BE, Cakaloglu Y, Ferenci P, Tabak F, Akdogan M, Simon K, Akarca US, Flisiak R, Verhey E, Van Vuuren AJ, Boucher CA, ter Borg MJ, Janssen HL. Early on-treatment prediction of response to peginterferon alfa-2a for HBeAg-negative chronic hepatitis B using HBsAg and HBV DNA levels. Hepatology. 2010;52:454-461. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 187] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 18. | Liaw YF. Clinical utility of hepatitis B surface antigen quantitation in patients with chronic hepatitis B: a review. Hepatology. 2011;54:E1-E9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Lee JM, Ahn SH, Kim HS, Park H, Chang HY, Kim do Y, Hwang SG, Rim KS, Chon CY, Han KH. Quantitative hepatitis B surface antigen and hepatitis B e antigen titers in prediction of treatment response to entecavir. Hepatology. 2011;53:1486-1493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 114] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 20. | Wong DK, Tanaka Y, Lai CL, Mizokami M, Fung J, Yuen MF. Hepatitis B virus core-related antigens as markers for monitoring chronic hepatitis B infection. J Clin Microbiol. 2007;45:3942-3947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 141] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 21. | Buster EH, Hansen BE, Lau GK, Piratvisuth T, Zeuzem S, Steyerberg EW, Janssen HL. Factors that predict response of patients with hepatitis B e antigen-positive chronic hepatitis B to peginterferon-alfa. Gastroenterology. 2009;137:2002-2009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 301] [Cited by in RCA: 330] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
Peer reviewers: Guglielmo Borgia, MD, Professor, Public Medicine and Social Security, University of Naples “Federico II”, Malattie Infettive (Ed. 18) via S. Pansini, 5, I-80131 Naples, Italy; Ramsey Chi-man Cheung, MD, Professor, Division of GI and Hepatology, VAPAHCS (154C), 3801 Miranda Ave, Stanford University School of Medicine, Palo Alto, CA 94304, United States
S- Editor Lv S L- Editor Stewart GJ E- Editor Li JY