Published online Oct 21, 2012. doi: 10.3748/wjg.v18.i39.5560
Revised: May 9, 2012
Accepted: May 26, 2012
Published online: October 21, 2012
AIM: To support probe-based confocal laser endomicroscopy (pCLE) diagnosis by designing software for the automated classification of colonic polyps.
METHODS: Intravenous fluorescein pCLE imaging of colorectal lesions was performed on patients undergoing screening and surveillance colonoscopies, followed by polypectomies. All resected specimens were reviewed by a reference gastrointestinal pathologist blinded to pCLE information. Histopathology was used as the criterion standard for the differentiation between neoplastic and non-neoplastic lesions. The pCLE video sequences, recorded for each polyp, were analyzed off-line by 2 expert endoscopists who were blinded to the endoscopic characteristics and histopathology. These pCLE videos, along with their histopathology diagnosis, were used to train the automated classification software which is a content-based image retrieval technique followed by k-nearest neighbor classification. The performance of the off-line diagnosis of pCLE videos established by the 2 expert endoscopists was compared with that of automated pCLE software classification. All evaluations were performed using leave-one-patient-out cross-validation to avoid bias.
RESULTS: Colorectal lesions (135) were imaged in 71 patients. Based on histopathology, 93 of these 135 lesions were neoplastic and 42 were non-neoplastic. The study found no statistical significance for the difference between the performance of automated pCLE software classification (accuracy 89.6%, sensitivity 92.5%, specificity 83.3%, using leave-one-patient-out cross-validation) and the performance of the off-line diagnosis of pCLE videos established by the 2 expert endoscopists (accuracy 89.6%, sensitivity 91.4%, specificity 85.7%). There was very low power (< 6%) to detect the observed differences. The 95% confidence intervals for equivalence testing were: -0.073 to 0.073 for accuracy, -0.068 to 0.089 for sensitivity and -0.18 to 0.13 for specificity. The classification software proposed in this study is not a “black box” but an informative tool based on the query by example model that produces, as intermediate results, visually similar annotated videos that are directly interpretable by the endoscopist.
CONCLUSION: The proposed software for automated classification of pCLE videos of colonic polyps achieves high performance, comparable to that of off-line diagnosis of pCLE videos established by expert endoscopists.
- Citation: André B, Vercauteren T, Buchner AM, Krishna M, Ayache N, Wallace MB. Software for automated classification of probe-based confocal laser endomicroscopy videos of colorectal polyps. World J Gastroenterol 2012; 18(39): 5560-5569
- URL: https://www.wjgnet.com/1007-9327/full/v18/i39/5560.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i39.5560
Colorectal cancer is the second leading cause of cancer-related death in the United States[1]. Its development includes several morphological stages, from benign to adenomatous polyps with low grade dysplasia to adenocarcinoma. Suspicious lesions are usually detected with standard colonoscopy by endoscopists who either perform confirmatory biopsy, or if high certainty exists, perform immediate therapy such as resection or ablation of diseased tissue. Because standard endoscopic imaging can only diagnose disease states with moderate levels of certainty[2,3], histopathology remains the criterion standard for final diagnosis[4]. However, the requirement for ex vivo histology implies a large proportion of unnecessary polypectomies and often requires a separate endoscopic procedure to be performed for treatment. It also increases the cost of colorectal cancer screening.
Probe-based confocal laser endomicroscopy (pCLE, Mauna Kea Technologies, France) enables the endoscopist to image the epithelial tissue in vivo, at the microscopic level with a confocal miniprobe, and in real-time during ongoing endoscopy. Preliminary findings by Meining et al[5] demonstrated the applicability of pCLE in diagnosing colorectal neoplasia in vivo with high sensitivity and specificity (93% and 92%, respectively) in 13 patients with colorectal lesions. Venkatesh et al[6] and De Palma[7] pointed out that confocal endomicroscopy offers the ability to target biopsies much more precisely and thus to reduce the number of random biopsies. In a recent study including a large pool of 75 patients, Buchner et al[8] compared off-line diagnosis of pCLE videos to virtual chromoendoscopy (Narrow-Band Imaging and Fujinon Intelligent Color Enhancement) and showed that off-line diagnosis of pCLE videos had higher sensitivity (91% vs 77%) with similar specificity (76%). As noted by Wallace et al[9], endoscopists now have the challenging task of performing “optical biopsies” and diagnosing pCLE video sequences in vivo.
In order to provide an objective support for pCLE diagnosis, we aimed to design a computer-based system for the automated classification of colonic polyps into neoplastic and non-neoplastic lesions. As physicians typically rely on similarity-based reasoning to establish a diagnosis from image queries, we propose a content-based image retrieval (CBIR) approach to automatically estimate the pathology of a new pCLE video. Indeed, contrary to “black box” classification systems, a CBIR-based classification system extracts, from a training database, annotated pCLE videos that are visually similar to the video of interest and directly interpretable by the endoscopist. The pathology of the video query is then estimated from the histopathological votes of these already diagnosed videos. Another advantage of CBIR-based classification is that the extracted similar videos can be presented to the endoscopist in a second reader paradigm to better support pCLE diagnosis.
The main goal of this study was to compare, using the same database of colonic polyps, the clinical performance of our automated pCLE classification software with that of off-line diagnosis of pCLE videos established by endoscopists expert in pCLE, with histopathology remaining the criterion standard reference.
The patients included in the study were enrolled between November 2007 and March 2009 for previous studies approved by the Mayo Clinic Institutional Review Board, and from which we collected all available data to ensure as large a sample size as possible. These patients were enrolled into the study of Buchner et al[8] and for further studies by the same Mayo Clinic group. Only the patients with complete diagnostic data were considered in our study. All study participants gave full written consent. Patients were enrolled if they were due for surveillance or screening colonoscopies, evaluation of known or suspected polyps on other imaging modalities, and endoscopic mucosal resection of larger flat colorectal neoplasia. Exclusion criteria were patients with non-corrected coagulopathy, women who were pregnant or breast feeding, those with documented allergy to fluorescein, and patients with no colorectal lesions found during a study colonoscopy. Twenty-four hours before the procedure, patients were prepped with 2-4 L polyethylene glycol solution. Conscious sedation was performed with intravenous administration of midazolam and meperidine.
All procedures were performed by the authors (either Wallace MB or Buchner AM) using a high-definition colonoscope (Fujinon EC450HL5 or 490 ZW, Fujinon, Ft Wayne, NJ, United States; Olympus CFH180, Olympus, Center Valley, NY, United States). The system was equipped with the EPX 4400 processor (Fujinon Inc.) or CV 180 Exera (Olympus, Co.). The primary screening method was white-light high-definition colonoscopy. Then, either Fujinon Intelligent Color Enhancement mode 4 with a Fujinon colonoscope or Narrow-Band Imaging with an Olympus 180 series scope was used to characterize lesions in all patients.
The surface pit pattern of the lesion was classified according to Kudo criteria. Anatomical site and morphological class of lesions were recorded in accordance with the Paris classification[10]. Fluorescein sodium 2.5-5.0 mL 10% (AK Fluor, Akorn Pharmaceutical, Lake Forest, IL, United States) solution was administered intravenously after the first polyp was identified. Immediately after fluorescein injection, pCLE video sequences of the lesions were acquired and recorded. According to the visual examination of both endoscopic and pCLE images, real biopsies were targeted to the most suspicious parts of the polyp. Appropriate treatment procedures, ranging from simple polypectomies to complex endoscopic mucosal resection of lesions, were then performed.
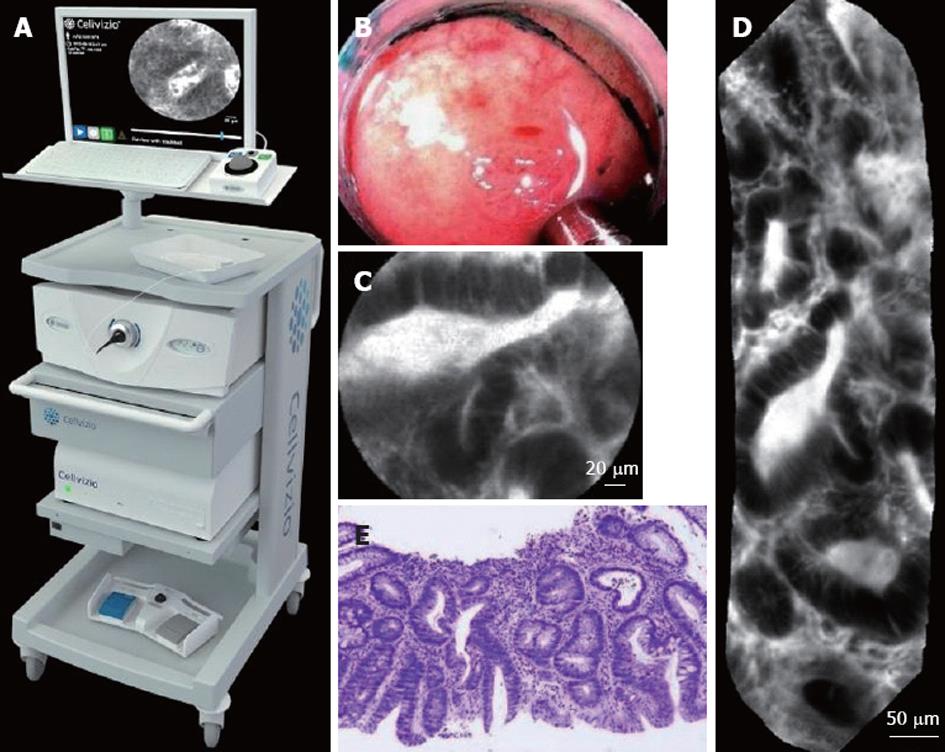
During a pCLE acquisition protocol, the endoscopist typically inserts, through the working channel of a standard endoscope, a confocal miniprobe (Coloflex UHD, Cellvizio GI) of external diameter 2.5 mm, which is made of 30 000 optical fibers bundled together. The pCLE imaging setup, shown in Figure 1, allows the acquisition of pCLE images of field-of-view 240 μm at a rate of 9 to 12 frames per second. In stable pCLE video sequences, the probe is in constant contact with the tissue. Representative endoscopic, pCLE, and histopathology images of tubular adenoma are shown in Figure 1.
Prior to pCLE evaluation of the study polyps, the 2 expert endoscopists (Wallace MB, Buchner AM) viewed extensive published material on pCLE and performed a self-calibration on training pCLE videos of 20 polyps of known pathology (10 neoplastic and 10 non-neoplastic). These “training” polyps were evaluated by a gastrointestinal pathologist (Krishna M) and came from 9 patients not included in the study. Once acquired, the pCLE videos of the study lesions were evaluated off-line and in random order by the 2 expert endoscopists, who were blinded to histology diagnosis and endoscopic appearance of the lesion. The off-line diagnosis of pCLE videos was made based on the established modified Mainz criteria[11] for diagnosis of colorectal neoplasia, and according to pit pattern and overall crypt and vessel architecture. Of the whole pCLE video imaging of a polyp, the sequence of the video containing the most malignant pCLE features was considered to represent the polyp.
All resected specimens were reviewed by a reference gastrointestinal pathologist (Krishna M) blinded to the pCLE information. Only the size and anatomic location were provided, which is the routine clinical practice at the Mayo Clinic institution. Intraepithelial neoplasia were defined using modified Vienna criteria[12,13]: benign polyps and hyperplastic polyps were classified as non-neoplastic lesions, while tubular adenoma, villous adenoma, tubulovillous adenoma and adenocarcinoma were classified as neoplastic lesions.
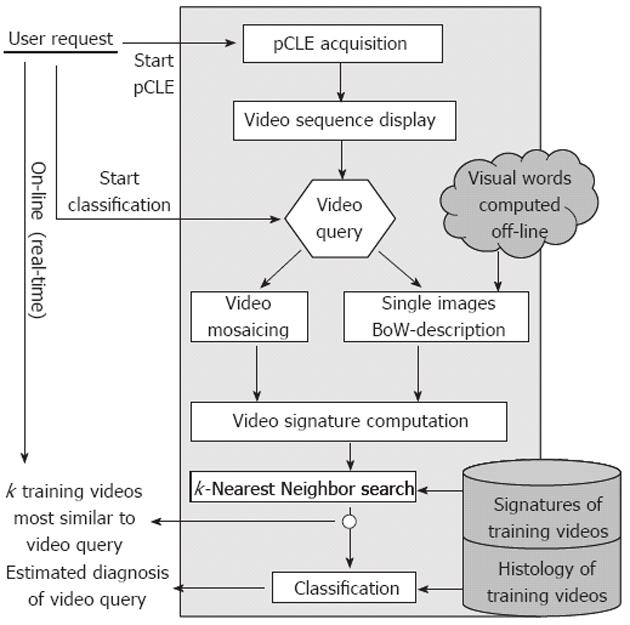
As endoscopists use perceptual similarities between pCLE videos of known diagnosis to establish a diagnosis on a new pCLE video, we propose a content-based retrieval approach to design the automated pCLE video classification software. We revisited the standard bag-of-visual-words (BoW) technique which has been successfully used in many content-based image retrieval applications in computer vision[14]. A thorough technical presentation of our methodology has been disclosed previously[15], but without detailed clinical evaluation.
The standard BoW technique for image retrieval can be divided into four steps: region detection on the image, description of the regions, discretization of the feature space and similarity measuring between images. The detection step extracts salient regions in the image using sparse detectors. During the description step, a descriptor computes for each salient region its description vector. Then, the discretization step uses the result of a clustering method that builds K clusters, i.e., K visual words, from the union of the description vector sets gathered across all the images of the training database. Each description vector counts for one visual word, so an image can be represented by a signature of size K which is the histogram of its visual words. By construction, image signatures are invariant by viewpoint changes (image translation, rotation and scaling) and affine illumination changes. Finally, the similarity measuring step defines the similarity distance between two images as an adequate distance between their signatures: the most similar training images to the image of interest are defined as being the closest ones in terms of this distance.
First, we observed that discriminative information is densely distributed in pCLE images. Second, we noticed that several pCLE image patterns have the same shape but represent different objects characterized by their different size (e.g., mesoscopic crypts and microscopic goblet cells both have a rounded shape). Therefore, pCLE image description must not be invariant by scaling. To avoid scale invariance and to extract all the image information, we decided to apply, instead of standard sparse detectors, a dense detector that was made of overlapping disks having a fixed radius and localized on a dense regular grid. We maintained the invariance by in-plane translation and rotation, because the pCLE miniprobe translates and rotates along the tissue surface. Besides, as the diffusion rate of fluorescein administered before imaging procedure decreases through time, invariance by affine illumination changes is also preserved.
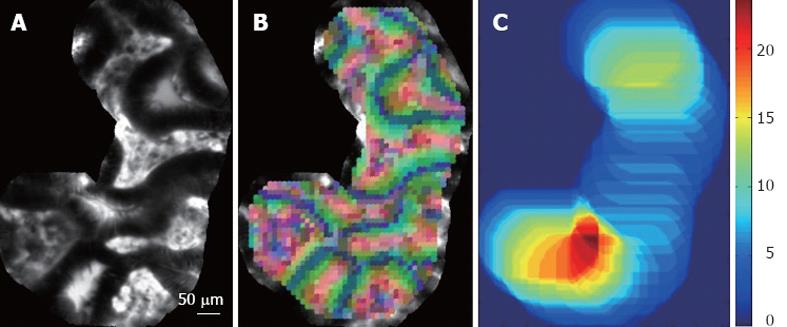
Expert endoscopists pointed out that the field-of-view of single still images may not be large enough to make a robust diagnosis. Thus, we decided to retrieve not single images but complete videos, using the video mosaicing technique[16,17] (available in the Cellvizio software) to include spatial overlap between time-related images. Examples of mosaics built with the video mosaicing tool are shown in Figure 1. To ensure on-line retrieval, we use the translation results of the real-time version of the video-mosaicing technique to weight the contribution of each local image region to its visual word, as illustrated in Figure 2. Then, we computed the video signatures with a histogram summation technique. The whole pipeline of our retrieval-based software classification framework can be run on-line during ongoing colonoscopy (Figure 3).
Once the visual signature of the video query is computed, the k-Nearest Neighbor search step identifies the k closest training videos to the video query, by relying on the similarity distance between the video signatures. We then used the known histopathology diagnosis of these training videos to classify the query video, either as neoplastic or as non-neoplastic. Each of the k most similar training videos delivers a “histopathological” vote which is weighted by the inverse of its similarity distance to the video query.
Due to the relatively small size of our pCLE database, we needed to learn from as much data as possible. To avoid any bias while having a large training set, we employed cross-validation. As several videos were acquired for the same patient, we performed a leave-one-patient-out cross-validation[18]: all videos from a given patient were excluded from the training set before being tested as queries of our retrieval and classification software. Cross-validation also allowed us to find the optimal number of nearest neighbors, k = 9, which is the one that maximizes the accuracy of the retrieval-based software classification results.
All the reported results of the automated software classification were obtained using leave-one-patient-out cross-validation. Statistical analysis was performed by André B.
To test for statistical difference between the two methods of interest, namely automated software classification and off-line classification by expert endoscopists, we used McNemar’s tests[19] and showed the corresponding power calculations with a type I error alpha = 0.05. Two-sided P values < 0.05 were assumed to indicate statistical significance.
In order to assess statistical equivalence between the two methods, we used the two-sided Z-test between proportions[19,20] and computed 95% CI. Because the 135 pCLE videos constituted a small sample size, we used a correction for continuity for McNemar’s test.
The statistics on overall accuracy are dependent on the relative fraction of non-neoplastic and neoplastic lesions examined, which in this study were 31.1% and 68.9%, respectively. Even though observations were made for more than one polyp in some patients, for the purposes of statistical analysis, individual polyps (and their corresponding videos) were assumed to constitute independent observations. It is recognized that there was multiple testing of outcome data arising from individual polyps. Since the statistical tests were meant to highlight differences, and since the correction by Bonferroni’s method did not affect statistical significance in any of the comparisons, all P values are presented uncorrected for multiple testing.
Table 1 summarizes the demographic and general characteristics of the study population. None of the 71 patients experienced any endoscopic complications or adverse reactions to sodium fluorescein, with the exception of transient yellow discoloration of the skin and urine, which resolved by the time of discharge from the recovery room (skin) or within 24 h (urine). Histopathology and morphological classification of the 135 analyzed colorectal lesions are shown in Table 2.
| Study population | Summary (n = 71) |
| Age (yr), median (min, 25th, 75th, max) | 75 (46, 68, 79, 93) |
| Gender, % | |
| Male | 49 |
| Female | 51 |
| History of colon cancer, % | 9 |
| Family history of colon cancer, % | 10 |
| Colorectal lesions | Summary (n = 135) |
| Polyp size (mm), median (min, 25th, 75th, max) | 8 (1, 5, 20, 60) |
| Polyp location, % | |
| Cecum | 24 |
| Rectum | 20 |
| Ascending | 18 |
| Sigmoid | 14.5 |
| Transverse | 15 |
| Descending | 5.5 |
| Splenic flex | 3 |
| Histopathology diagnosis, % | |
| Hyperplastic | 31 |
| Tubular adenoma | 52 |
| Tubulovillous adenoma | 11.5 |
| Hyperplastic and adenomatous features | 2.5 |
| Adenocarcinoma | 3 |
| Neoplastic lesion, simplified histopathology, % | 69 |
| Paris classification, % | |
| 1p | 1 |
| 1s | 57 |
| 2a | 32 |
| 2b | 5 |
| 2c | 1 |
| 2a/c | 4 |
The pCLE database contained 135 pCLE videos representing each of the 135 polyps. The pCLE appearance of neoplastic lesions, compared to that of non-neoplastic lesions, included dilated irregular vessels, fluorescein leakage, cellular features of epithelial mucin depletion, and histological features of villiform crypts with increased optical density along the epithelial border.
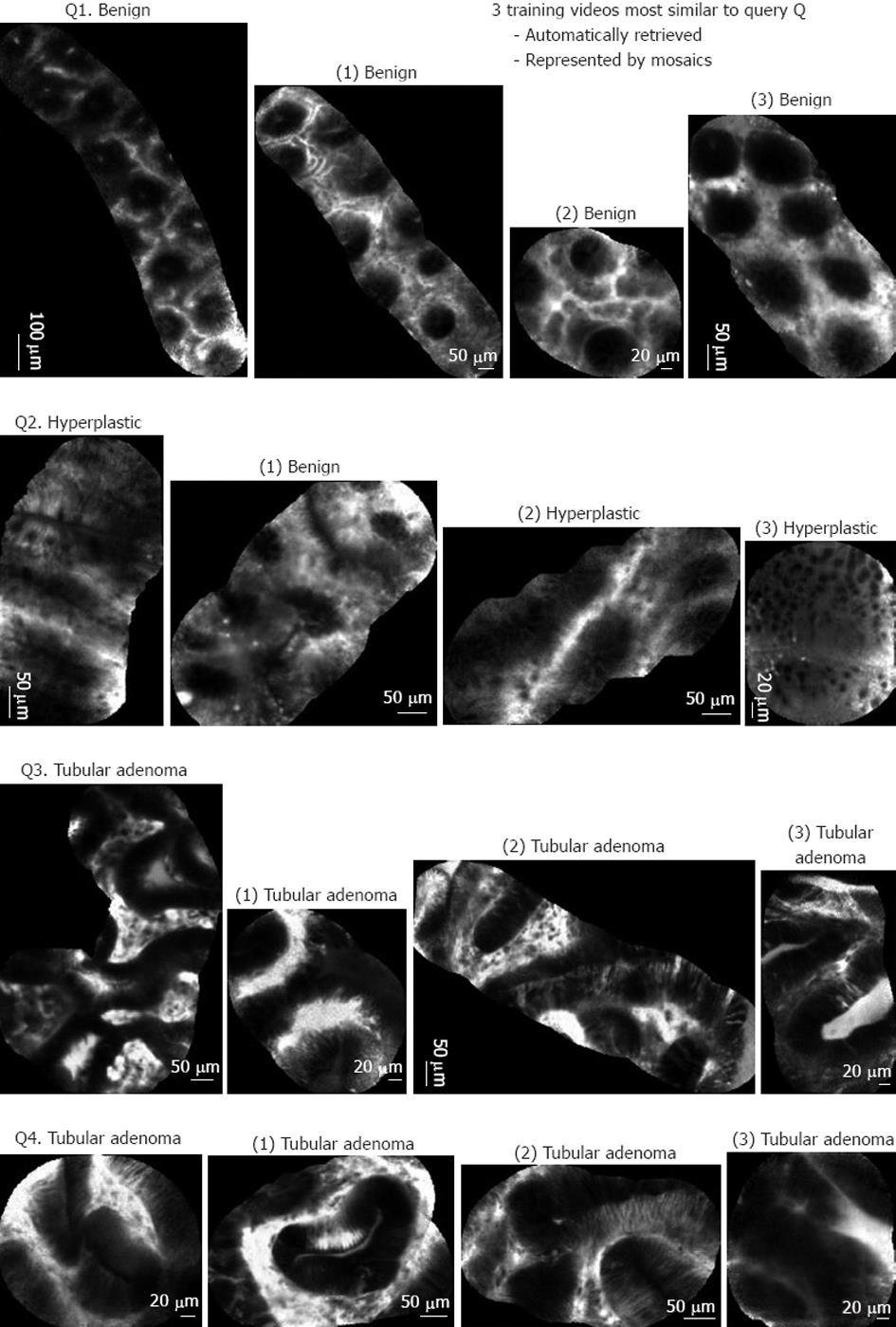
As the automated pCLE classification software is a similarity-based system that classified pCLE videos based on the votes of visually similar videos, its clinical relevance can be qualitatively evaluated by examining the intermediate results of video retrieval. Examples of mosaics built with the video mosaicing tool are shown in Figures 4 and 5. Figure 4 shows 5 typical results of the automated pCLE retrieval software. We observed that, despite the high variability in appearance of a given histopathological class (neoplastic or non-neoplastic), the automatically retrieved videos called “neighbors” looked quite similar to the video queries (Q1, Q2, Q3 and Q4, respectively). In addition, we noticed that the closer the neighbor was to the query, the more similar it was to it.
In terms of classification, the pathological class was estimated by the weighted votes of the 3 retrieved neighbors. In Figure 4, video queries Q1, Q2, Q3 and Q4 have been correctly classified with respect to histopathology, both by automated software classification and by expert endoscopists.
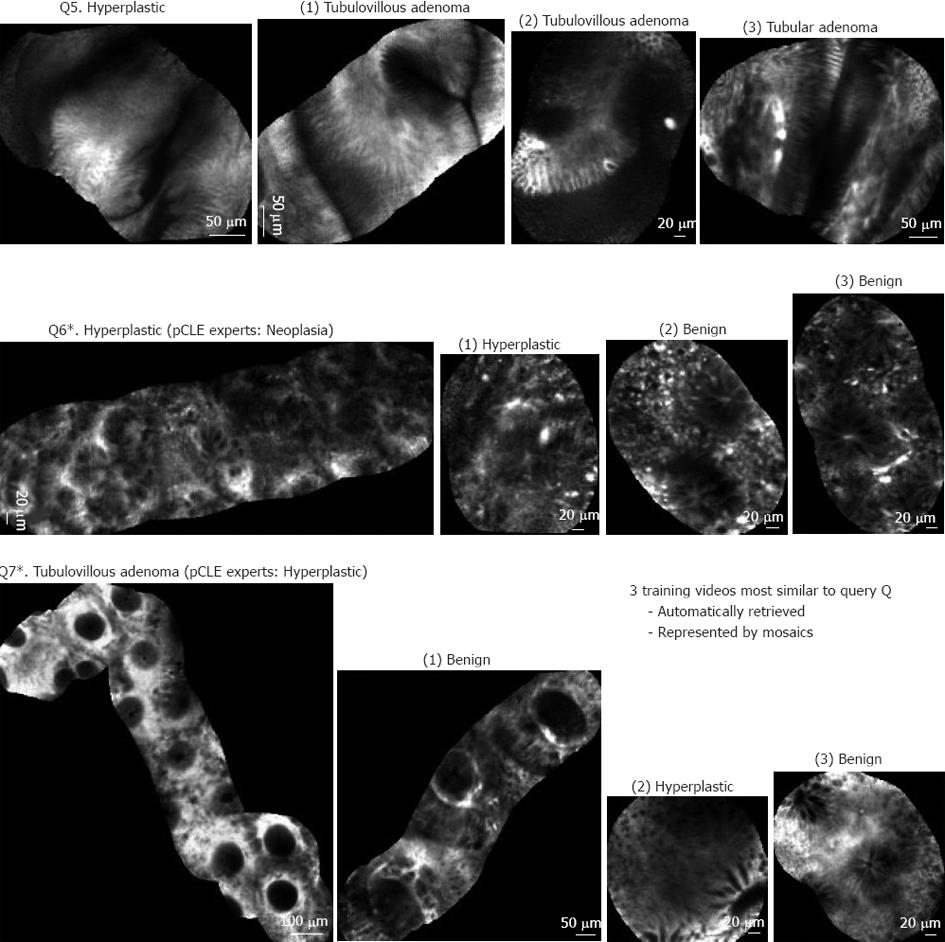
Figure 5 shows 3 other results that revealed some limitations of the automated pCLE retrieval software. Video query Q5 corresponds to a rare variety of hyperplastic polyp correctly classified as non-neoplastic by the expert endoscopists, but misclassified by the automated software classification because it was not represented in the training database for retrieval. Video query Q6 corresponds to the ambiguous serrated adenoma case, correctly classified as non-neoplastic by the automated software classification, but misclassified by the expert endoscopists who considered serrated adenomas as malignant. Video query Q7 corresponds to a tubulovillous adenoma misclassified as non-neoplastic both by the expert endoscopists and by the automated software classification (this may be explained if a sampling error occurred and the corresponding biopsy was not performed exactly on the imaging spot).
Classification accuracy, sensitivity and specificity of the two methods, automated pCLE software classification (first method) and off-line diagnosis of pCLE videos established by the 2 expert endoscopists (second method), are listed in Table 3. Automated software classification reached a sensitivity of 92.5%, a specificity of 83.3% for a resulting accuracy of 89.6%. Expert review reached a sensitivity of 91.4%, a specificity of 85.7% and the same accuracy of 89.6%.
| Automated pCLE classification | Off-line expert diagnosis of pCLE | |
| Accuracy | ||
| % | 89.6 | 89.6 |
| Fraction | 121/135 | 121/135 |
| Sensitivity | ||
| % | 92.5 | 91.4 |
| Fraction | 86/93 | 85/93 |
| Specificity | ||
| % | 83.3 | 85.7 |
| Fraction | 35/42 | 36/42 |
| Statistical significance between (1) and (2) | ||
| McNemar’s test, alpha = 0.05 | ||
| Accuracy: (P, power) | (Not significant, 2.5%) | |
| Sensitivity: (P, power) | (Not significant, 6.5%) | |
| Specificity: (P, power) | (Not significant, 5.2%) | |
| Statistical equivalence between (1) and (2) | ||
| Two-sided Z test | ||
| 95% CI for accuracy | -0.073-0.073 | |
| 95% CI for sensitivity | -0.068-0.089 | |
| 95% CI for specificity | -0.18-0.13 | |
When testing for statistical difference, the P values provided by McNemar’s tests showed that the differences between the 2 methods were not statistically significant and that there was very low power (< 6%) to detect the observed differences.
When testing for statistical equivalence, the 95% confidence intervals provided by two-sided Z-tests between proportions were: -0.073 to 0.073 for accuracy, -0.068 to 0.089 for sensitivity and -0.18 to 0.13 for specificity. These intervals included zero and were sufficiently small to suggest that the methods were equivalent. In particular, the -0.18 lower bound for specificity was acceptable if the automated pCLE classification software was only taken as a second-reader tool to support pCLE diagnosis.
The present study demonstrates that, using a fairly representative database of colonic polyps, our automated software for the pCLE video classification has overall high accuracy, sensitivity and specificity, that are comparable to those of the off-line diagnosis of pCLE videos established by two endoscopists expert in pCLE. As the automated classification software can be run on-line during ongoing colonoscopy, it could be used as a second-reader tool to support and improve not only off-line but also on-line diagnosis of pCLE established by endoscopists with various levels of expertise. In the majority of cases, the second reader would agree with a moderately experienced endoscopist, who would thus be comforted in his/her diagnosis. For cases when they disagree, the endoscopist would have the opportunity to rethink his/her diagnosis and have more accurate in vivo interpretation. Besides, especially for small polyps, this second-reader tool could assist the endoscopist in adopting the “Diagnose, Resect and Discard Strategy” that dispenses with histopathological examination.
Gomez et al[21] analyzed in vivo pCLE interpretation in distinguishing between neoplastic and non-neoplastic lesions among 3 expert endoscopists and estimated an average accuracy of 75% (sensitivity 76%, specificity 72%) with good to moderate interobserver agreement. Buchner et al[22] demonstrated that accurate interpretation of pCLE images by 11 endoscopists, considered as non-expert in pCLE, can be learned rapidly with a short 2-h training session. The learning curve pattern of pCLE in predicting neoplastic lesions was demonstrated with improved accuracies in time from 63% to 86% as observers’ experience increased. Thus, prospectively, the automated classification software could be valuable not only for in vivo diagnosis support, but also for training support to improve the learning curve of new endoscopists. Indeed, we have shown in a preliminary study[23] how interpretation difficulty can be automatically estimated by the software, in order to develop a self-training simulator for pCLE diagnosis with adjustable level of difficulty. For surgical skills, evidence of the learning effect from the use of training tools have been provided in the thesis by Brydges[24], however, further investigation is needed for the extension of learning effect analysis to diagnostic skills.
One of the advantages of our classification software is that it is not a “black box” but an informative tool based on the query by example model: it produces, as intermediate results, visually similar annotated videos that are directly interpretable by the endoscopist. From the qualitative observations of visual similarities between pCLE videos, we suggest that the visually convincing results of the intermediate video retrieval step account for the relevance of the whole pCLE classification software. As few similar videos (less than 10) are necessary to classify a video query with high accuracy, this visual information should be clinically useful for the endoscopist.
Further limitations of the classification software may include three main issues. First, a large training database is needed to be sufficiently representative of non-typical pCLE cases. This is even more challenging since the practice of pCLE is evolving and that new cases with atypical pCLE features may still be encountered. Second, the definition of “criterion standard” for colorectal cancer screening is debatable because expert endoscopists and pathologists do not always agree. This can be illustrated by many examples of hyperplastic polyps redefined later as sessile serrated lesions by gastrointestinal pathologists, as in the study by Khalid et al[25]. The third limitation is that an obtained biopsy may be acquired unintentionally from the area that does not correspond with the obtained pCLE imaging.
The task of the automated pCLE classification software is not to replace the endoscopist or the pathologist but to assist the endoscopist in making an informed decision. Before using the computer-based classification tool during an ongoing endoscopy procedure, more work is needed to improve its accuracy and to develop underlying tools that are both ergonomic and complementary. In particular, the on-line display of the retrieval outputs, for instance of the 3 most similar videos to the video query, together with their histopathology and possible multimodal clinical data, may be a precious underlying indicator for diagnosis decision. Such a sophisticated “Smart Atlas” for pCLE would allow endoscopists in different centers to share and enrich their pCLE knowledge during ongoing endoscopy. Further studies are warranted to evaluate the impact of using automated pCLE retrieval and classification software on the pCLE learning curve and on the diagnostic performance of the endoscopists.
The authors thank Dr. Muhammad Waseem Shahid for his contribution to data collection and analysis. A preliminary report of this study was presented at the AGA Late Breaking Abstract Forum (Imaging and Advanced Technology-Confocal Endoscopy), Digestive Disease Week, 2010. The present study uses data from the following clinical trial: NCT 00874263.
Histopathology is the criterion standard for the diagnosis of colorectal cancers, but it implies a large proportion of unnecessary polypectomies and an inherent delay in diagnosis. Probe-based confocal laser endomicroscopy (pCLE) is a recent technology that enables, during ongoing endoscopy, in vivo imaging of the epithelium at the microscopic level.
Several studies have already demonstrated the applicability of pCLE in diagnosing colorectal neoplasia in vivo with high sensitivity and specificity. Because pCLE is a relatively recent imaging technology, the interpretation of pCLE videos of colonic polyps for diagnostic purposes is still challenging for many non-expert endoscopists.
This is believed to be the first study to propose, with the aim of supporting in vivo diagnosis of colorectal cancers, content-based image retrieval-based classification software that automatically extracts visually similar annotated videos directly interpretable by the endoscopist. The extracted annotated videos can be presented to the endoscopist in a second reader paradigm to better support pCLE diagnosis. Furthermore, this study demonstrates that this novel software achieves a high diagnostic performance, which is statistically comparable to that of off-line diagnosis of pCLE videos established by expert endoscopists.
The classification software proposed in this study is an objective tool which has the potential to support the interpretation of pCLE videos of colonic polyps for diagnostic purposes. Further studies are warranted to evaluate the impact of using the automated classification software on the pCLE learning curve and on the diagnostic performance of endoscopists.
pCLE: An imaging system that allows the endoscopist to visualize the epithelium in vivo, at the microscopic level and in real-time during ongoing endoscopy; Content-based image retrieval: A computer vision technique that automatically extracts, given a query image, several training images with the most similar appearance to the query.
This is a good descriptive study in which authors support probe-based confocal laser endomicroscopy diagnosis by designing a software for automated classification of colonic polyps. The results are interesting and suggest that the proposed software for automated classification of pCLE videos of colonic polyps achieves high performance, comparable to that of off-line diagnosis of pCLE videos established by expert endoscopists.
| 1. | Hawk ET, Levin B. Colorectal cancer prevention. J Clin Oncol. 2005;23:378-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 156] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 2. | Norfleet RG, Ryan ME, Wyman JB. Adenomatous and hyperplastic polyps cannot be reliably distinguished by their appearance through the fiberoptic sigmoidoscope. Dig Dis Sci. 1988;33:1175-1177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 30] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 3. | Rastogi A, Keighley J, Singh V, Callahan P, Bansal A, Wani S, Sharma P. High accuracy of narrow band imaging without magnification for the real-time characterization of polyp histology and its comparison with high-definition white light colonoscopy: a prospective study. Am J Gastroenterol. 2009;104:2422-2430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 140] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 4. | Winawer SJ, Zauber AG, Fletcher RH, Stillman JS, O'Brien MJ, Levin B, Smith RA, Lieberman DA, Burt RW, Levin TR. Guidelines for colonoscopy surveillance after polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer and the American Cancer Society. Gastroenterology. 2006;130:1872-1885. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 216] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 5. | Meining A, Saur D, Bajbouj M, Becker V, Peltier E, Höfler H, von Weyhern CH, Schmid RM, Prinz C. In vivo histopathology for detection of gastrointestinal neoplasia with a portable, confocal miniprobe: an examiner blinded analysis. Clin Gastroenterol Hepatol. 2007;5:1261-1267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 101] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 6. | Venkatesh K, Cohen M, Evans C, Delaney P, Thomas S, Taylor C, Abou-Taleb A, Kiesslich R, Thomson M. Feasibility of confocal endomicroscopy in the diagnosis of pediatric gastrointestinal disorders. World J Gastroenterol. 2009;15:2214-2219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 20] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 7. | De Palma GD. Confocal laser endomicroscopy in the "in vivo" histological diagnosis of the gastrointestinal tract. World J Gastroenterol. 2009;15:5770-5775. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 82] [Cited by in RCA: 91] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 8. | Buchner AM, Shahid MW, Heckman MG, Krishna M, Ghabril M, Hasan M, Crook JE, Gomez V, Raimondo M, Woodward T. Comparison of probe-based confocal laser endomicroscopy with virtual chromoendoscopy for classification of colon polyps. Gastroenterology. 2010;138:834-842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 145] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 9. | Wallace MB, Fockens P. Probe-based confocal laser endomicroscopy. Gastroenterology. 2009;136:1509-1513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 132] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 10. | The Paris endoscopic classification of superficial neoplastic lesions: esophagus, stomach, and colon: November 30 to December 1, 2002. Gastrointest Endosc. 2003;58:S3-43. [PubMed] |
| 11. | Kiesslich R, Burg J, Vieth M, Gnaendiger J, Enders M, Delaney P, Polglase A, McLaren W, Janell D, Thomas S. Confocal laser endoscopy for diagnosing intraepithelial neoplasias and colorectal cancer in vivo. Gastroenterology. 2004;127:706-713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 564] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 12. | Schlemper RJ, Riddell RH, Kato Y, Borchard F, Cooper HS, Dawsey SM, Dixon MF, Fenoglio-Preiser CM, Fléjou JF, Geboes K. The Vienna classification of gastrointestinal epithelial neoplasia. Gut. 2000;47:251-255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1463] [Cited by in RCA: 1579] [Article Influence: 60.7] [Reference Citation Analysis (0)] |
| 13. | Rubio CA, Nesi G, Messerini L, Zampi GC, Mandai K, Itabashi M, Takubo K. The Vienna classification applied to colorectal adenomas. J Gastroenterol Hepatol. 2006;21:1697-1703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 14. | Zhang J, Lazebnik S, Schmid C. Local features and kernels for classification of texture and object categories: a comprehensive study. Int J Comput Vis. 2007;73:213-238. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1266] [Cited by in RCA: 361] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 15. | André B, Vercauteren T, Buchner AM, Wallace MB, Ayache N. A smart atlas for endomicroscopy using automated video retrieval. Med Image Anal. 2011;15:460-476. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 16. | Vercauteren T, Perchant A, Malandain G, Pennec X, Ayache N. Robust mosaicing with correction of motion distortions and tissue deformations for in vivo fibered microscopy. Med Image Anal. 2006;10:673-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 92] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 17. | Becker V, Vercauteren T, von Weyhern CH, Prinz C, Schmid RM, Meining A. High-resolution miniprobe-based confocal microscopy in combination with video mosaicing (with video). Gastrointest Endosc. 2007;66:1001-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 81] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 18. | Dundar M, Fung G, Bogoni L, Macari M, Megibow A, Rao RB. A methodology for training and validating a CAD system and potential pitfalls. Int J Comput Assisted Radiol Surg. 2004;1268:1010-1014. [RCA] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 19. | Sheskin DJ. Handbook of parametric and nonparametric statistical procedures. 5th ed. Boca Raton, FL: Chapman and Hall/CRC 2011; 1-1926. |
| 20. | Jones B, Jarvis P, Lewis JA, Ebbutt AF. Trials to assess equivalence: the importance of rigorous methods. Brit Med J. 1996;313:36-39. [RCA] [DOI] [Full Text] [Cited by in Crossref: 716] [Cited by in RCA: 711] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 21. | Gomez V, Buchner AM, Dekker E, van den Broek FJ, Meining A, Shahid MW, Ghabril MS, Fockens P, Heckman MG, Wallace MB. Interobserver agreement and accuracy among international experts of probe-based confocal laser microscopy (pCLE) in predicting colorectal neoplasia. Endoscopy. 2010;42:286-291. [DOI] [Full Text] |
| 22. | Buchner AM, Gomez V, Gill KR, Ghabril M, Scimeca D, Shahid MW, Achem SR, Picco MF, Riegert-Johnson D, Raimondo M. The learning curve for in vivo probe based Confocal Laser Endomicroscopy (pCLE) for prediction of colorectal neoplasia. Gastrointest Endosc. 2009;69:AB364-AB365. [DOI] [Full Text] |
| 23. | André B, Vercauteren T, Buchner AM, Shahid MW, Wallace MB, Ayache N. An image retrieval approach to setup difficulty levels in training systems for endomicroscopy diagnosis. Med Image Comput Comput Assist Interv. 2010;13:480-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 24. | Brydges R. A critical reappraisal of self-learning in health professions education: Directed self-guided learning using simulation modalities. : University of Toronto 2010; . |
| 25. | Khalid O, Radaideh S, Cummings OW, O'Brien MJ, Goldblum JR, Rex DK. Reinterpretation of histology of proximal colon polyps called hyperplastic in 2001. World J Gastroenterol. 2009;15:3767-3770. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 111] [Cited by in RCA: 114] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
Peer reviewer: Andreas Vécsei, MD, St. Anna Children's Hospital, 1150 Vienna, Austria
S- Editor Gou SX L- Editor Webster JR E- Editor Li JY