Published online Apr 1, 2004. doi: 10.3748/wjg.v10.i7.995
Revised: January 4, 2004
Accepted: January 8, 2004
Published online: April 1, 2004
AIM: To analyze the characterization of T-cell receptor-γ (TCR-γ) gene rearrangement in the gastrointestinal lymphomas and evaluate the value of PCR-SSCP analysis in gastrointestinal lymphomas investigation.
METHODS: TCR-γ gene rearrangement segments of gastrointestinal lymphomas were cloned and sequenced. Single clone plasmid and mixed clone plsamids were subsequently submitted to PCR-SSCP analysis to investigate the relationship between the number of amplified clones and band patterns of the amplified products. The PCR products of TCR-γ gene rearrangement of 40 gastrointestinal lymphomas were electrophoresed on agarose gels and the positive cases on agarose gels were studied by SSCP analysis.
RESULTS: The sequencing showed that TCR-γ gene rearrangement of the gastrointestinal lymphomas included functional gene and pseudogene with extensive variety in the junctional regions. In SSCP analysis, the number of the single-stranded bands was about two times of the number of amplified clones, and double-stranded band became broad with the increased number of the amplified clones. Thirteen of the 25 B- cell gastrointestinal lymphomas and 14 of the 15 gastrointestinal T-cell lymphomas were positive detected on agarose gel electrophoresis. Of the positive cases detected by SSCP analysis, 3 B-cell lymphomas and 13 T-cell lymphomas showed positive bands. The other cases showed only smears. The rearranged pattern included 13 monoallelic gene rearrangements and 3 biallelic or oligoclonal gene rearrangements.
CONCLUSION: The pattern of TCR-γ gene rearrangement in gastrointestinal lymphomas are similar to that of the nodular lymphomas. PCR-SSCP analysis for TCR-γ gene rearrangement can be applied both for adjuvant diagnosis of gastrointestinal lymphomas and analysis of the gene rearrangement pattern. The ratio of TCR-γ gene rearrangements occurred in T-cell gastrointestinal lymphomas is significantly higher than that in B-cell gastrointestinal lymphomas. The gene rearrangement pattern involves monoallelic and biallelic (or oligoclonal) gene rearrangement.
- Citation: Han XQ, He L, Shong LY, Jiang HY, Zhu MG, Zhao T. Investigation of T-cell receptor-γ gene rearrangement in gastrointestinal lymphomas by PCR-SSCP analysis. World J Gastroenterol 2004; 10(7): 995-999
- URL: https://www.wjgnet.com/1007-9327/full/v10/i7/995.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i7.995
Primary gastrointestinal lymphomas are the most common extranodal lymphomas and are almost exclusively of non-Hodgkin types[1-3]. These lymphomas are biologically different from their nodal counterparts. Distinguishing the two phenotypes is of importance as the survival rates are different between T-cell and B-cell gastrointestinal lymphomas[4-6]. As cytological features of gastrointestinal lymphomas are usually not specific, the combination with clinical, histological, immunophenotypic and molecular genetic studies is necessary to characterize these neoplasms[7].
T-cell receptor (TCR) gene rearrangement provides a convenient genetic marker for demonstrating clonality in T-cell malignant neoplasm. Among the four TCR gene types, the TCR-γ gene rearrangement occurs in the early stage of cell differentiation and the rearrangement retains in both αβ and γδ phenotypic T cells along with the cell differentiation[8]. Analysis of TCR-γ gene rearrangement is of practical value in determining the clonality of T-cell populations and for diagnosis T-cell malignant neoplasms[9-11]. But the junctional diversity in TCR-γ gene rearrangement is limited compared with the other TCR or IgH gene rearrangement because of the fewer inserted nucleotides during the rearrangement, this lead the PCR products of TCR-γ gene rearrangement amplified by a consensus primers pair to be similar in length. The similarity may result in false clonal bands, when analyzed by standard gel electrophoresis methods that separate DNA molecules based solely on size[12,13]. A high- resolution for the amplified products of TCRγ gene rearrangement is required.
Single-strand conformational polymorphism analysis (SSCP) separates different DNA molecules based on their single-strand secondary structure conformations under certain electrophoretic condition. DNA molecules with different conformations migrate different rates on polyacrylamide gel and therefore can be separated from each other[14]. Besides gene point mutation analysis, SSCP analysis is also suitable for TCR-γ gene rearrangement study[15].
In this study, we expected to analyze the characteristics of TCR-γ gene rearrangement and evaluate the practicability of PCR-SSCP for TCR-γ gene rearrangement in the diagnosis and analysis of gastrointestinal lymphomas.
Specimens of the gastrointestinal lymphoma were obtained by biopsy and surgical resection between January 1997 to October 2002 in the department of pathology of the Nangfang Hospital of the Frist Military Medical University, the First Municipal Hospital of Guangzhou and the No. 157 Hospital of The People’s Liberation Army. Of the 40 patients, 9 had tumors located within stomach, 1 in duodenum, 1 in jejunum, 10 in ileum, 11 in ascending colon, 2 in transverse colon, 5 in descending colon and 1 in rectum. All cases were regarded gastrointestinal lymphomas because of absence of disease in other organs. The specimens were fixed in buffered formalin, embedded in paraffin and stained with haematoxylin-eosin (HE). Lineage was confirmed by staining with CD20 and CD79a for B-cells, and CD45RO and CD3 for T-cells. The 40 cases were retrospectively diagnosed according to the new WHO classification[16]. Twenty-five of the 40 cases were confirmed B-cell lymphomas and 15 cases were confirmed T-cell lymphomas. A reactive lymph node and Jurkat cell line were considered as negative and positive control, respectively.
The 7 μm thickness sections were cut with disposable blades, collected on glass slides,deparaffinized with xylene, washed with ethanol, and rehydrated in deionized water. The moist tissues of 0.5 cm × 0.5 cm containing abundant malignant cells (according to HE) were scraped off the glass slides with a sterile blade, and collected in Eppendorf tube. Jurkat cells were collected and rinsed twice in PBS. A total of 50 μL digesting buffer containing 10 mmol/L Tris, 1 mmol/L EDTA, 10 g/L Tween 20, and 200 mg/mL proteinase K were added to the tubes. The digestion was performed at 37 °C for 16 h. Samples were then heated at 96 °C for 10 min to inactivate the enzyme, centrifuged, and the supernatant was used as template for PCR amplification.
The primers design was based on those used by Benhattar et al[13], spanning the rearranged V γ 1-8 and J1/2 region with the expected products ranging from 160 to 190 bp in length. The sequences of the primers were: TVG sense, 5’AGGGTTGTGTTGGAATCAGG3’, and J1/2 antisense, 5’CCTGTGACAACAAGTGTTGT3’. PCR amplification was performed in GeneAmp PCR system 9600 (Perkin Elmer, Norwalk, CT). The reaction mixture (30 μL) contained PCR buffer (50 mmol/L KCl, 10 mmol/L Tris-HCl, pH 8.3), 200 mmol/L each of dNTP, 1.5 mmol/L MgCl2, 0.5 mmol/L of each primer, and 1.25 U of AmpliTaq (Perkin Elmer). A total of 300 ng of samples and cell line DNA (or 20 ng of plasmids) were used as templates, respectively. The PCR amplification cycles were designed to a decreasing annealing temperature similar to a touch-down PCR[17]: 5 cycles consist of 94 °C for 45 s, 60 °C for 45 s, 72 °C for 30 s; the next 5 cycles consisted of 94 °C for 45 s, 56 °C for 45 s, 72 °C for 30 s; and the last 25 cycles consisted of 94 °C for 45 s, 56 °C for 45 s, 72 °C for 30 s. The reaction mixture was first incubated at 95 °C for 5 min to denature double-stranded DNA. Finally, an additional incubation for 10 min at 72 °C was performed to ensure full extension of the products. In all experiments, monoclonal (Jurkat cell lines) and polyclonal (reactive lymph node) controls were run in parallel with the test samples. Products were analyzed by electrophoresis on 20 g/L agarose gels stained with ethidium bromide, visualized and photographed on an UV transilluminator. SSCP analysis was performed only if a single band of the expected size was detected on the gel. DNA from each sample was amplified at least twice.
A total of 100 μL PCR products of TCR-γ gene rearrangement from five T-cell gastrointestinal lymphomas was separately purified and ligated into pGEM-T Easy cloning vector (Promega Corporation, 2800 Woods Hollow Road Madison WI 53711-5399 USA). The recombinant vector DNA was then transformed into Escherichia coli JM109. The transformed bacteria solution was spread on selective LB-agar plate with X-gal, IPTG and ampicillin. After overnight incubation at 37 °C, 2 to 3 white colonies (with DNA inserts in the plasmid) from each plate were selected randomly and cultured in LB medium containing ampicillin for 12 h at 37 °C. Plasmid DNA was isolated from each culture following a standard protocol[18] and dissolved in double distilled deionized water. Deoxyribonucleic acid sequencing of cloned PCR products was performed following the standard protocol[19] involving the universal forward-sequencing primer M13 for pGEM-T vector. The concentration of the plasmids were adjusted to 20 ng/μL. According to the sequencing results, 2, 3 and 4 plasmids with different inserted PCR products were mixed, respectively. Single and mixed plasmids were submitted as templates to PCR.
SSCP analysis was performed according to the Signoretti et al[15] with some alternation. Briefly, 6 μL of PCR products were mixed 1:1 with denaturing loading buffer (containing 95 g/L formamide, 0.5 g/L bromophenol blue, 2.5 g/L xylene cyanol and 20 mmol/L EDTA). The mixture was heated to 95 °C for 10 min, quickly chilled on ice for 1 min, and loaded on 120 g/L nondenaturing polyacrylamide (29:1 acrylamide/biasacrylamide) gel in Tris-borate- EDTA (1 × TBE) buffer. The gel was electrophoresed at 120 (12 v/cm) volts for approximately 4 to 6 h at room temperature and subsequently stained with silver and photographed.
Comparisons between the 2 groups were performed by the chi-square test using SPSS 10.0 statistical software (SPSS Company, Chicago, Illinois, USA). P values < 0.05 in two-tailed were considered statistically significant.
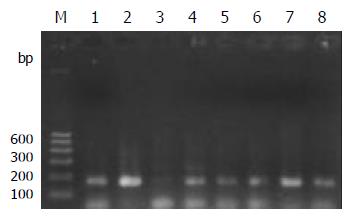
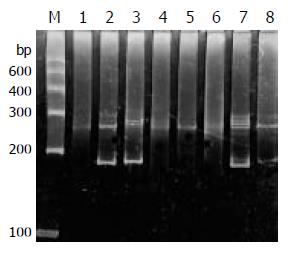
PCR amplification for the TCR-γrearranged gene showed single band of 160-190 bp in 93% (14/15) T-cell and 52% (13/25) B-cell gastrointestinal lymphomas on agarose gel electrophoresis. The positive rate was significantly higher in T-cell lymphomas (P < 0.05) than that in B-cell lymphomas. Both the reactive lymph node and the Jurkat cell line showed single band (Figure 1).
Sequence of the clones were identified to the particular Vγ (1-8) segment by internet (http://www.ncbi.nlm.nih.gov/BLAST/ Blast.cgi) assisted comparison with published TCR-γsequence (gi 28436398 ref NG_001336.2. Homo sapiens T cell receptor gamma locus (TRG@) on chromosome 7, Length = 140728). The sequenced Vγsegments were assigned to the member of the Vγ (1-8) family that, by comparison, showed the highest degree of homology (95% to 100%). Most of the Vγ (1-8) -Jγ1/2 combinations had the functional Vγ genes: V3, V4, V5, V8. Of the 14 sequenced cases, only one case was pseudogene V γ 7. In the VJ junctional N regions, the deleted nucleotides ranged from 2 to 23 nucleotides with an average length of 9.38 ± 6.42 bp, and the size of the inserted nucleotides ranged from 3 to 13 bp (average length, 7.38 ± 2.98 bp). The TCR γ junctional regions showed considerable diversity with no two clones showing the same sequence. The size of the PCR products amplified by this primers combination ranged between 160-190 bp. The letters with delete lines represent the deleted nucleotides and the capital letters represent the inserted nucleotides during the rearrangement.
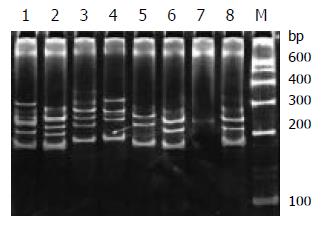
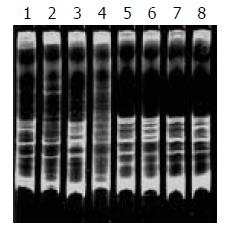
During SSCP analysis and gel electrophoresis, the PCR products of plasmids inserted with TCR-γ gene rearranged segments showed discrete bands in all lanes, except lane 7 (Figure 2) with faint band, which might be due to the insufficient DNA product in lane 7. The fastest migrating band was double-stranded DNA, located in the zone ranging from 160-190 bp. Running above the double-stranded DNA were single-stranded bands. PCR products of one cloned plasmid showed two single-stranded bands. The bands patterns of different clones were different (Figure 1 lane 5, 6 and 8). PCR products of 2 mixed cloned plasmids showed one double-stranded band and 3 to 4 sigle-stranded bands (Figure 2 lane1, 2, 3 and 4). PCR Products of 3 to 4 plasmids mixed together showed one broad double-stranded band and 6 to 8 single-stranded bands (Figure 3). Some faint single-stranded bands presented in the lanes were disregarded or interpreted with caution. Based on the results of these studies, two single-stranded bands (sometimes only one) represented the monoallelic gene rearrangement, and those with more than two single-stranded bands might be the biallelic or oligoclonal gene rearrangement.
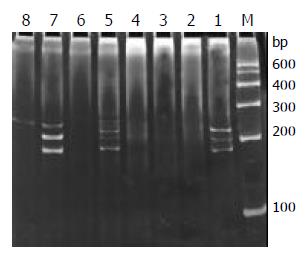
Of the 29 samples submitted to SSCP analysis, 17 showed one or more discrete bands, including the Jurkat cell line, 3/13 B-cell gastrointestinal lymphomas (Figure 4) and 13/14 T-cell gastrointestinal lymphomas (Figure 5) confirmed by the HE and immunohistochemistry. The positive rate was significantly higher in T-cell lymphomas than that in B-cell lymphomas (P < 0.01). Less than half of the cases only showed the single-stranded bands, but not double-stranded bands. This might be due to insufficient quantity of PCR products. According to the numbers of the single-stranded bands of the 17 positive cases, 13 cases were monoallelic gene rearrangement, and 4 cases might be biallelic or oligoclonal gene rearrangements. The other 10 samples and reactive lymph node which were positive on the agarose gel electrophoresis yielded only smears with no obvious band. This patterns was the characteristic of rearranged TCR-γ V-J fragments amplified from polyclonal T-cell populations without a predominant.
Most T-cells express the αβ T-cell phenotype and only a small population expresses γδ TCR. The lineage is determined by the gene rearrangement and protein expression. γ-chain gene rearrangement generally takes place in the αβ T-cell but not express a functional γδ receptor[12]. In addition, a fraction of B cells undergoes TCR-γ gene rearrangement, especially immature B cells (illegitimated gene rearrangement)[20,21]. Furthermore, a T-cell lymphoma may aberrantly express the B-cell phenotype[22]. In this study, the illegitimate TCR-γ gene rearrangement in B-cell gastrointestinal lymphomas was investigated, which showed about 12% (3/25) positive result. TCR-gamma gene rearrangement involves the recombination between one of the V region (V γ 1-8, V γ 9, V γ 10, V γ 11) and one of the J regions (J γ 1/2, J γ P1/2, J γ P)[8]. In this study, we used only V γ 1-8 and J γ 1/2 consensus primers set to analyze the most common VJ gamma recombination. It is likely that using additional V and J region primers would have further increased the yield of positive results, but our study showed that this consensus primers combination seemed to be sufficient for the diagnosis of T-cell gastrointestinal lymphomas.
Cloning and sequencing are the precise but time and labour-consuming methods in the study of clonal gene rearrangement. Because of the admixture of reactive T-cells, at least 8 to 10 clones from each patient must be sequenced for the purpose of diagnosis, to confirm whether there are any two clones showing the same sequence[21]. In this study, we intended to know the general aspects of the TCR-γ gene rearrangement in gastrointestinal lymphomas and to obtain the clonal rearranged gene segments. So, only 2 to 3 clones from each patient were sequenced, which were obviously inadequate for clonality judgement. The sequencing result showed that the TCR-γ junctional region (N region) presented considerable diversity with no two clones showing the same sequence. The junctional region differed in both sequence and size with the deleted and inserted nucleotides. The PCR products obtained by the consensus V γ1-8/J1/2 primers ranged from 160 to 190 base pairs. All these characteristics were similar to that of nodal lymphomas[23]. The narrow size of the PCR products between different clones did not provide a means for distinguishing between monoclonal and polyclonal TCR junction by the conventional agarose gel electrophoresis[13].
PCR-SSCP has been introduced to detect DNA mutation because it can resolve DNA molecules differing as litter as a single base pairs substitution. The high sensitivity of PCR-SSCP makes it fit to tell apart the clonal from polyclonal gene rearrangement, and its simplicity makes it a good candidate for application in clinical laboratory tests[15]. In the present study, we would like to study whether the different band numbers observed on SSCP gel tell the different rearranged patterns apart in gastrointestinal lymphomas. The PCR products of single clone and mixed clones submitted to PCR-SSCP analysis showed that a consistent relationship existed between the number of single-stranded bands twice the numbers of clones amplified, which suggested that the number of single-stranded bands might estimate whether the samples were monoallelic or biallelic (oligoclonal) gene rearrangements. Two single-stranded bands (sometimes only one band) could definitely be regarded as monoallelic gene rearrangement. Three to four bands could be regarded as a clonal T-cell population with biallic gene rearrangements or two clonal T-cells populations both with monoallelic gene rearrangements. Those with more than four single-stranded bands were most probably oligoclonal gene rearrangements[24]. Owing to the high sensitivity and resolution of PCR-SSCP, multiple single-stranded bands must be interpreted with caution, and the PCR-SSCP analysis conditions must be optimized individually for a special pair of primers. The condition for the Vγ1-8/J1/2 primers we used was optimized thoroughly before introducing to the study of gastrointestinal lymphomas.
Gastrointestinal lymphomas differ from their nodal counterpart in many aspects and are more likely to be misdiagnosed before surgical resections[25]. The histological and cytological features may be more complex, with most having pleomorphic small, medium, large or anaplastic cell cytologic features and generally with a mixture of reactive cells. The immunophenotype is especially important for gastrointestinal lymphomas, as the prognosis for T-cell gastrointestinal lymphomas is poorer than that for B-cell type and more incline to perforation[3]. It should be emphasized that gastrointestinal lymphomas must be defined by a combination of factors as not a single parameter is entirely specific.
The sensitivity and validity of TCR-γ gene rearrangement investigated by PCR-SSCP were compared with conventional histology and immunohistology. Of the 40 gastrointestinal lymphomas, 13 of the 15 T-cell lymphomas and 3 of the 25 B-cell lymphomas showed positive results. The high consistency indicates that PCR-SSCP analysis for the TCR-γ gene rearrangement study is an additional powerful tool for the diagnosis of T-cell gastrointestinal lymphomas.
Furthermore, the gene rearrangement pattern, which may be a prognostic factor, can be revealed by PCR-SSCP[26]. Our study showed that 23.5% (4/17) of the positive cases were biallelic or oligoclonal gene rearrangements. PCR-SSCP gives reproducible band migration patterns for the same DNA molecules, therefore, this technique potentially can be used to compare multifocal lesion from a single patient or to compare a recurrent lesion with a primary one in a patient previously diagnosed with T-cell gastrointestinal lymphomas[27].
| 1. | Morton JE, Leyland MJ, Vaughan Hudson G, Vaughan Hudson B, Anderson L, Bennett MH, MacLennan KA. Primary gastrointestinal non-Hodgkin's lymphoma: a review of 175 British National Lymphoma Investigation cases. Br J Cancer. 1993;67:776-782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 95] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 2. | Kohno S, Ohshima K, Yoneda S, Kodama T, Shirakusa T, Kikuchi M. Clinicopathological analysis of 143 primary malignant lymphomas in the small and large intestines based on the new WHO classification. Histopathology. 2003;43:135-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 68] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 3. | Lee SS, Cho KJ, Kim CW, Kang YK. Clinicopathological analysis of 501 non-Hodgkin's lymphomas in Korea according to the revised European-American classification of lymphoid neoplasms. Histopathology. 1999;35:345-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 1.6] [Reference Citation Analysis (2)] |
| 4. | Nakamura S, Matsumoto T, Takeshita M, Kurahara K, Yao T, Tsuneyoshi M, Iida M, Fujishima M. A clinicopathologic study of primary small intestine lymphoma: prognostic significance of mucosa-associated lymphoid tissue-derived lymphoma. Cancer. 2000;88:286-294. [RCA] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (1)] |
| 5. | Zhang W, Li G, Liu W, Ren X, Xu H. [Analyzing of prognosis of intestinal T-cell lymphoma]. Zhonghua Binglixue Zazhi. 2002;31:295-299. [PubMed] |
| 6. | Sanchez-Bueno F, Garcia-Marcilla JA, Alonso JD, Acosta J, Carrasco L, Piñero A, Parrilla P. Prognostic factors in primary gastrointestinal non-Hodgkin's lymphoma: a multivariate analysis of 76 cases. Eur J Surg. 1998;164:385-392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 7. | Kinney MC. The role of morphologic features, phenotype, genotype, and anatomic site in defining extranodal T-cell or NK-cell neoplasms. Am J Clin Pathol. 1999;111:S104-S118. [PubMed] |
| 8. | Hodges E, Krishna MT, Pickard C, Smith JL. Diagnostic role of tests for T cell receptor (TCR) genes. J Clin Pathol. 2003;56:1-11. [RCA] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 92] [Article Influence: 4.0] [Reference Citation Analysis (3)] |
| 9. | Diss TC, Watts M, Pan LX, Burke M, Linch D, Isaacson PG. The polymerase chain reaction in the demonstration of monoclonality in T cell lymphomas. J Clin Pathol. 1995;48:1045-1050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 115] [Article Influence: 3.7] [Reference Citation Analysis (1)] |
| 10. | Sprouse JT, Werling R, Hanke D, Lakey C, McDonnel L, Wood BL, Sabath DE. T-cell clonality determination using polymerase chain reaction (PCR) amplification of the T-cell receptor gamma-chain gene and capillary electrophoresis of fluorescently labeled PCR products. Am J Clin Pathol. 2000;113:838-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 36] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 11. | Algara P, Soria C, Martinez P, Sanchez L, Villuendas R, Garcia P, Lopez C, Orradre JL, Piris MA. Value of PCR detection of TCR gamma gene rearrangement in the diagnosis of cutaneous lymphocytic infiltrates. Diagn Mol Pathol. 1994;3:275-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 0.9] [Reference Citation Analysis (1)] |
| 12. | Macintyre EA, Delabesse E. Molecular approaches to the diagnosis and evaluation of lymphoid malignancies. Semin Hematol. 1999;36:373-389. [PubMed] |
| 13. | Benhattar J, Delacretaz F, Martin P, Chaubert P, Costa J. Improved polymerase chain reaction detection of clonal T-cell lymphoid neoplasms. Diagn Mol Pathol. 1995;4:108-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 2.3] [Reference Citation Analysis (1)] |
| 14. | Orita M, Iwahana H, Kanazawa H, Hayashi K, Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989;86:2766-2770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2278] [Cited by in RCA: 2212] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 15. | Signoretti S, Murphy M, Cangi MG, Puddu P, Kadin ME, Loda M. Detection of clonal T-cell receptor gamma gene rear-rangements in paraffin-embedded tissue by polymerase chain reaction and nonradioactive single-strand conformational poly-morphism analysis. Am J Pathol. 1999;154:67-75. [RCA] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 68] [Article Influence: 2.5] [Reference Citation Analysis (2)] |
| 16. | Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD. World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting-Airlie House, Virginia, November 1997. J Clin Oncol. 1999;17:3835-3849. [PubMed] |
| 17. | Ranheim EA, Jones CD, Zehnder JL. Sensitive detection of clonal immunoglobulin rearrangements in frozen and paraffin embedded tissues by polymerase chain reaction heteroduplex analysis. Diagn Mol Pathol. 2000;9:177-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (1)] |
| 18. | Sambrook J, Russel DW. Molecular cloning, a laboratory manual. 3rd ed. Chinese edition: Sci Pub. 2002;27-30. |
| 19. | Sambrook J, Russel DW. Molecular cloning, a laboratory manual. 3rd ed. Chinese edition: Sci Pub. 2002;1005-1010. |
| 20. | Steenbergen EJ, Verhagen OJ, van Leeuwen EF, van den Berg H, von dem Borne AE, van der Schoot CE. Frequent ongoing T-cell receptor rearrangements in childhood B-precursor acute lymphoblastic leukemia: implications for monitoring minimal residual disease. Blood. 1995;86:692-702. [PubMed] |
| 21. | Chen Z, Le Paslier D, Dausset J, Degos L, Flandrin G, Cohen D, Sigaux F. Human T cell gamma genes are frequently rearranged in B-lineage acute lymphoblastic leukemias but not in chronic B cell proliferations. J Exp Med. 1987;165:1000-1015. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 82] [Article Influence: 2.1] [Reference Citation Analysis (1)] |
| 22. | Yao X, Teruya-Feldstein J, Raffeld M, Sorbara L, Jaffe ES. Peripheral T-cell lymphoma with aberrant expression of CD79a and CD20: a diagnostic pitfall. Mod Pathol. 2001;14:105-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 97] [Article Influence: 3.9] [Reference Citation Analysis (1)] |
| 23. | Kneba M, Bolz I, Linke B, Bertram J, Rothaupt D, Hiddemann W. Characterization of clone-specific rearrangement T-cell receptor gamma-chain genes in lymphomas and leukemias by the polymerase chain reaction and DNA sequencing. Blood. 1994;84:574-581. [PubMed] |
| 24. | Zhu P, Wu S, Xue H, Lu Y, Wang L, Zhang Y, Yu J. Analysis of clonality of lymphocytic leukemia and lymphoma by T-cell receptor gene rearrangement. Chin Med J (Engl). 1997;110:607-611. [PubMed] |
| 25. | Cheng AQ, Luo EZ, Lu XY. 68 cases of primary malignant gastrointestinal lymphomas. Shijie Huaren Xiaohua Zazhi. 2002;8:240-241. |
| 26. | Xu B, Tian H, Zhou SY. [Determination of clonal T cell receptor gene rearrangement in non-Hodgkin's lymphoma patients and its clinical significance]. Ai Zheng. 2003;22:397-400. [PubMed] |
| 27. | Crisi GM, Emanuel JR, Johnson C, Crotty P, Costa J, Tallini G. Semireannealing, single-stranded conformational polymorphism: a novel and effective tool for the diagnosis of T-cell clonality. Diagn Mol Pathol. 2002;11:67-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (1)] |
Edited by Gupta MK and Xu FM