Published online Oct 1, 2004. doi: 10.3748/wjg.v10.i19.2883
Revised: April 6, 2004
Accepted: April 13, 2004
Published online: October 1, 2004
AIM: To quantify the inhibition of HBV replication by targeted ribonuclease by using real-time fluorescent PCR.
METHODS: Targeted ribonuclease was introduced into 2.2.15 cells by liposome-mediated transfection or HIV-TAT mediated protein transduction. Forty-eight hours after the transfection and 24 h after the transduction, supernatants of 2.2.15 cells were collected and HBV DNA in the supernatants was quantified by real-time fluorescent PCR with a commercial kit.
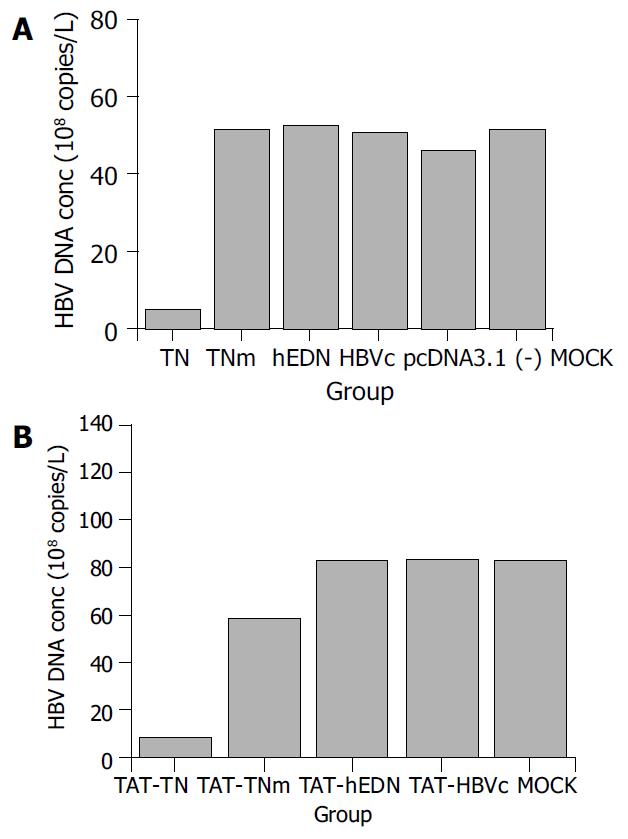
RESULTS: HBV DNA concentrations in the supernatants of 2.2.15 cells transfected or transducted with targeted ribonuclease were 4.9 ± 2.4 × 108 copies/L and 8.3 ± 4.0 × 108 copies/L, respectively. Compared with controls, transfection or transduction of targeted ribonuclease reduced HBV DNA concentration in the supernatants of 2.2.15 cells by 90.4% and 90.1%, respectively (P < 0.05).
CONCLUSION: Targeted ribonuclease can inhibit HBV replication in 2.2.15 cells.
- Citation: Liu J, Li YH, Ding J, Gong WD, Xue CF, Zhao Y, Huang YX. Quantifying anti-HBV effect of targeted ribonuclease by real-time fluorescent PCR. World J Gastroenterol 2004; 10(19): 2883-2885
- URL: https://www.wjgnet.com/1007-9327/full/v10/i19/2883.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i19.2883
Chronic hepatitis B virus (HBV) infection is a major health problem worldwide. Globally, more than 350 million people are infected with HBV, and some of them will evolve into liver cirrhosis and hepatocellular carcinoma (HCC). Current treatment regimens for chronic HBV infection, including interferon-γ , lamivudine, adefovir, or different combinations of these drugs, have only a limited long-term efficacy, but many adverse effects and drug resistance[1,2]. Therefore, the exploration of novel treatment strategies for HBV infection is both necessary and urgent. In fact, many ingenious treatment strategies have been tested for inhibition of HBV replication, such as antisense nucleotides, ribozymes, intracellular antibodies, targeted nucleases, RNA interference[3-13]. All of them can inhibit HBV replication to various degrees.
To explore alternative treatment methods against HBV infection, targeted ribonuclease (TN), a fusion protein of HBVc and human eosinophil-derived neurotoxin (hEDN), was constructed and its effect on HBV replication was tested. After the targeted ribonuclease was introduced by transfection or transduction into 2.2.15 cells, a cell model of HBV infection, we found that it significantly reduced the serological markers of HBV replication, namely HBsAg and HBeAg, in the supernatants of 2.2.15 cells, suggesting that the targeted ribonuclease could inhibit HBV replication.
To further characterize the anti-HBV effect of the targeted ribonuclease, the HBV DNA was quantified in the supernatants of 2.2.15 cells treated by the targeted ribonuclease. Our results showed that the targeted ribonuclease markedly reduced HBV DNA concentration in the supernatants, which together with our previous findings, demonstrate that HBV replication can be inhibited by targeted ribonuclease.
2.2.15 cells, a human hepatoblastoma Hep G2 cell line stably transfected by HBV genome, were cultured in Dulbecco’s modified Eagle’s medium (DMEM, purchased from Gibco Life Technologies, Grand Island, NY) supplemented with 100 mL/L fetal calf serum (Sijiqing Biotech Company, Hangzhou, China).
The transfection methods were previously described[14]. Briefly, twenty-four hours before transfection, 2.2.15 cells were seeded into a culture plate at the density of 2 × 108/L. LipofectamineTM 2000 reagent (Gibco Life Technologies) was used for the transfection of 2.2.15 cells by p/TN, p/TNm, p/hEDN, p/HBVc, pcDNA3.1 (-), or mock solution (DMEM plus LipofectamineTM 2000 reagent containing no plasmid) according to the manufacturer’s protocol. p/TN, p/TNm, p/hEDN and p/HBVc are the eukaryotic expression plasmids for targeted ribonuclease, point-mutated targeted ribonuclease without ribonuclease activity, human eosinophil-derived neurotoxin, and HBV core protein, respectively.
2.2.15 cells were seeded into a culture plate at the density of 2 × 108/L. Twenty-four hours later, purified recombinant proteins with protein transduction domain, TAT-TN, TAT-TNm, TAT-hEDN, and TAT-HBVc were added into the culture medium. For mock transduction, an equal volume of DMEM instead of protein was added into the culture medium.
Forty-eight hours after transfection and 24 h after transduction, the supernatants of 2.2.15 cells were collected and HBV DNA in the supernatants was quantified by using fluorescent PCR kit for quantification of HBV DNA (Daan Gene Company, Guangzhou, China) according to the manufacturer’s protocol. PCR primers were: P1: 5’ATCCTGCTGCTATGCCTCATCTT3’, P2: 5’ACAGTGGGGAAAGCCCTACGAA3’. The probe was 5’TGGCTAGTTTACTAGTGCCATTTTG3’. PCR reaction was analyzed by PE 5700 (Perkin Elmer, USA).
All data were analyzed by SPSS 10.0 software. Differences were considered statistically significant when P < 0.05.
HBV DNA in the supernatants of 2.2.15 cells transfected or transducted with the targeted ribonuclease and controls was quantified by real-time fluorescent PCR. The results are shown in Figure 1. Compared with the controls, HBV DNA concentration in the supernatant of 2.2.15 cells was markedly reduced by the targeted ribonuclease, which was introduced into the cells by both pathways, i.e. transfection and transduction. The transfected targeted ribonuclease reduced HBV DNA concentration by 90.4% and the transducted targeted ribonuclease by 90.1% (P < 0.05). Either hEDN or HBVc alone had no such an effect, indicating that it was the fusion protein itself, i.e. the targeted ribonuclease, but not its constituent molecules that exerted the anti-HBV effect. Interestingly, TNm which was mutated at just one amino acid residue (Lys113→Arg) but had no ribonuclease activity as compared with TN, did not decrease HBV DNA concentration either, suggesting that the ribonuclease activity was needed in the anti-HBV effect of the targeted ribonuclease. To exclude the possibility that the anti-HBV effect of the targeted ribonuclease was due to nonspecific killing of 2.2.15 cells, we also detected cell viability of the transfected and transducted cells by MTT assay. The results showed that the targeted ribonuclease had no adverse effects on cell viability and proliferation as compared with controls (P > 0.05, data not shown).
Real-time fluorescent PCR is a simple, sensitive, specific and precise method to quantitate nucleic acids over a vast dynamic range[15]. Due to these advantages, real-time fluorescent PCR has been widely used in both basic research and clinical diagnosis to measure the quantity of nucleic acids. As for HBV, real-time fluorescent PCR has been used in monitoring viral load during the course of antiviral treatment to evaluate the efficacy of the treatment and predict the possibility of emergence of drug-resistant variants, in the analysis of anti-HBV effect of small interfering RNA transfected into HBV-producing cell lines, and in the study of HBV pathogenesis[10,16-19]. In the current research, we also used real-time fluorescent PCR to precisely quantify HBV DNA in the supernatants of 2.2.15 cells to detect the effect of the targeted ribonuclease on HBV replication. The targeted ribonuclease reported here is a fusion protein against HBV infection constructed by us, based on the principle of capsid-targeted viral inactivation (CTVI)[5,20]. Our previous studies showed that it significantly reduced HBsAg and HBeAg in the supernatants of 2.2.15 cells, suggesting that the targeted ribonuclease could inhibit HBV replication. In consistent with our previous findings, in this study we observed the targeted ribonuclease notably reduced HBV DNA in the supernatants of 2.2.15 cells. Taken together, these results demonstrate that the targeted ribonuclease strongly inhibits HBV replication. The targeted ribonuclease may therefore be one of the promising alternative anti-HBV agents which are being studied in the pursuing of a satisfactory treatment for chronic HBV infection.
Although the precise mechanism of anti-HBV effect of targeted ribonuclease still remains unknown and awaits further study, the principle of capsid-targeted viral inactivation (CTVI) implies that it may specifically recognize pregenomic RNA (pgRNA) of HBV, the template for HBV replication, intracellularly by the HBVc domain of fusion protein, and then degrades pgRNA by the hEDN domain. This will in turn lead to a decrease in the replication of HBV DNA and synthesis of viral proteins, which finally results in a reduction of extracellular HBV DNA and viral proteins (HBsAg and HBeAg). In fact, we have found that purified targeted ribonuclease can degrade RNA in vitro (unpublished data) which supports this mechanism. Experiments are ongoing in this laboratory to clarify the mechanism and to apply the targeted ribonuclease to small animal models.
| 1. | Conjeevaram HS, Lok AS. Management of chronic hepatitis B. J Hepatol. 2003;38 Suppl 1:S90-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 133] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 2. | Jaboli MF, Fabbri C, Liva S, Azzaroli F, Nigro G, Giovanelli S, Ferrara F, Miracolo A, Marchetto S, Montagnani M. Long-term alpha interferon and lamivudine combination therapy in non-responder patients with anti-HBe-positive chronic hepatitis B: results of an open, controlled trial. World J Gastroenterol. 2003;9:1491-1495. [PubMed] |
| 3. | Li JG, Lian JQ, Jia ZS, Feng ZH, Nie QH, Wang JP, Huang CX, Bai XF. Effect of ribozymes on inhibiting expression of HBV mRNA in HepG2 cells. Shijie Huaren Xiaohua Zazhi. 2003;11:161-164. |
| 4. | Lu YY, Wang L, Liu Y, Yu M, Li K, Wang YD, Zhang LX, Cheng J. Inhibitory effect of HBcAg ScFv on hepatitis B virus. Shijie Huaren Xiaohua Zazhi. 2002;10:765-769. |
| 5. | Beterams G, Nassal M. Significant interference with hepatitis B virus replication by a core-nuclease fusion protein. J Biol Chem. 2001;276:8875-8883. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.0] [Reference Citation Analysis (1)] |
| 6. | Giladi H, Ketzinel-Gilad M, Rivkin L, Felig Y, Nussbaum O, Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol Ther. 2003;8:769-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 237] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 7. | McCaffrey AP, Nakai H, Pandey K, Huang Z, Salazar FH, Xu H, Wieland SF, Marion PL, Kay MA. Inhibition of hepatitis B virus in mice by RNA interference. Nat Biotechnol. 2003;21:639-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 458] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 8. | Shlomai A, Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 201] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 9. | Hamasaki K, Nakao K, Matsumoto K, Ichikawa T, Ishikawa H, Eguchi K. Short interfering RNA-directed inhibition of hepatitis B virus replication. FEBS Lett. 2003;543:51-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 110] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 10. | Ying C, De Clercq E, Neyts J. Selective inhibition of hepatitis B virus replication by RNA interference. Biochem Biophys Res Commun. 2003;309:482-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 67] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 11. | Chen Y, Du D, Wu J, Chan CP, Tan Y, Kung HF, He ML. Inhibition of hepatitis B virus replication by stably expressed shRNA. Biochem Biophys Res Commun. 2003;311:398-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 12. | Konishi M, Wu CH, Wu GY. Inhibition of HBV replication by siRNA in a stable HBV-producing cell line. Hepatology. 2003;38:842-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 56] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 13. | Klein C, Bock CT, Wedemeyer H, Wüstefeld T, Locarnini S, Dienes HP, Kubicka S, Manns MP, Trautwein C. Inhibition of hepatitis B virus replication in vivo by nucleoside analogues and siRNA. Gastroenterology. 2003;125:9-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 137] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 14. | Liu J, Li YH, Xue CF, Ding J, Gong WD, Zhao Y, Huang YX. Targeted ribonuclease can inhibit replication of hepatitis B virus. World J Gastroenterol. 2003;9:295-299. [PubMed] |
| 15. | Klein D. Quantification using real-time PCR technology: applications and limitations. Trends Mol Med. 2002;8:257-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 490] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 16. | Werle B, Cinquin K, Marcellin P, Pol S, Maynard M, Trépo C, Zoulim F. Evolution of hepatitis B viral load and viral genome sequence during adefovir dipivoxil therapy. J Viral Hepat. 2004;11:74-83. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 17. | Kohmoto M, Enomoto M, Yano Y, Otani S, Minamitani S, Tamori A, Habu D, Takeda T, Shiomi S, Seki S. Detection of serum hepatitis B virus DNA by real-time quantitative polymerase chain reaction (TaqMan PCR) during lamivudine treatment: comparison with three other assays. Hepatol Res. 2003;26:125-133. [RCA] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 18. | Ide T, Kumashiro R, Koga Y, Tanaka E, Hino T, Hisamochi A, Murashima S, Ogata K, Tanaka K, Kuwahara R. A real-time quantitative polymerase chain reaction method for hepatitis B virus in patients with chronic hepatitis B treated with lamivudine. Am J Gastroenterol. 2003;98:2048-2051. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 19. | Wieland SF, Spangenberg HC, Thimme R, Purcell RH, Chisari FV. Expansion and contraction of the hepatitis B virus transcriptional template in infected chimpanzees. Proc Natl Acad Sci USA. 2004;101:2129-2134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 176] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 20. | Natsoulis G, Boeke JD. New antiviral strategy using capsid-nuclease fusion proteins. Nature. 1991;352:632-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 53] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
Co-correspondents: Cai-Fang Xue
Edited by Zhang JZ and Wang XL Proofread by Xu FM