Published online Jul 1, 2004. doi: 10.3748/wjg.v10.i13.1964
Revised: January 22, 2004
Accepted: February 1, 2004
Published online: July 1, 2004
AIM: To investigate the association between loss of heterozygosity (LOH) on chromosome 18 and sporadic gastric cancer.
METHODS: Multiplex PCR was used to screen 14 highly polymorphic microsatellite markers on chromosome 18 in 45 cases of primary gastric cancer. PCR products were separated on polyacrylamide gels and the electrophoresis maps were analyzed with Genescan and Genotyper.
RESULTS: The LOH frequencies in gastric cancer at all 14 markers ranged from 10% to 58%. Eleven markers were found with over 20% LOH frequencies, in which 9 markers located in 18q, and 2 markers in 18p. Two overlapping deleted regions were identified: R1 between D18S61-D18S1161 at 18q22 (9 cM) with 24% LOH frequency; R2 between D18S462-D18S70 at 18q22-23 (6 cM) with 32% LOH frequency.
CONCLUSION: LOH of chromosome 18 (18q and 18p) may be involved in gastric tumorigenesis. Two overlapping deleted fragments suggested that there might be unidentified tumor suppressor genes in those two regions.
- Citation: Yu JC, Sun KL, Liu B, Fu SB. Allelotyping for loss of heterozygosity on chromosome 18 in gastric cancer. World J Gastroenterol 2004; 10(13): 1964-1966
- URL: https://www.wjgnet.com/1007-9327/full/v10/i13/1964.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i13.1964
Inactivation of tumor suppressor genes (TSGs) has been considered to be one of the most important mechanisms during the human tumorigenesis[1]. Earlier studies have shown that loss of heterozygosity (LOH) on specific chromosomal loci is related to the inactivation of TSGs. The Knudson “two-hit” hypothesis has provided the rationale for identifying TSGs by mapping regions of LOH. Analysis of LOH has been developed and fully exploited for the detection of TSGs in a variety of tumors through comparison of copy number changes in tumor DNA with matched control DNA[2]. Different chromosomal regions that are harboring putative TSGs can be found in tumors by searching for LOH markers. Furthermore, by identifying genetic distance between these markers and TSGs, the allelic loss regions can be narrowed step by step and the TSGs can be finally cloned.
Gastric cancer (GC) is one of the most frequent malignancies and remains a main cause of mortality in China[3-5]. Many regions of LOH on different chromosomes have been found in GC[6]. Allelic loss on the long arm of chromosome 18 (18q LOH) is highly related to GC[7]. Little is known about allelic loss on the short arm of chromosome 18 (18p LOH) in GC, although a significant incidence of 18p LOH (specially 18q11) has been found in tumors of the lung, brain and breast[8]. So in this paper, we chose fourteen highly polymorphic microsatellite markers spanning chromosome 18p and 18q and performed genome-wide allelotyping in 45 primary gastric cancers. We identified the chromosomal loci and overlapping regions that are frequently lost in GC for clarifying the roles of 18p LOH and 18qLOH in GC and providing evidences for finding new important TSGs.
Forty-five primary gastric tumors and corresponding non-tumorous tissue specimens were obtained at surgery from the First Hospital and the Second Hospital of Harbin Medical University. All the patients were confirmed by routine histologic examination and received no treatment before surgery. Each specimen was frozen immediately and stored at -80 °C until use. Genomic DNA was extracted using DNAzo1R reagent-genomic DNA isolation reagent (Gibcol).
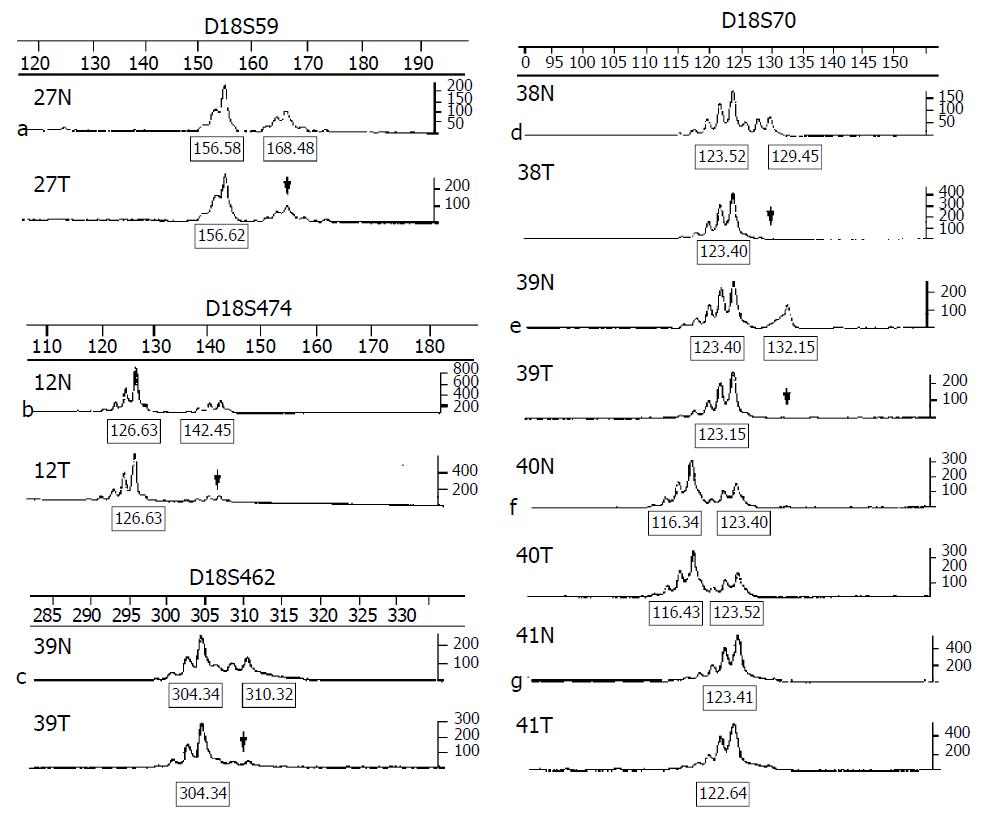
Along chromosome 18, we chose 14 polymorphic microsatellite markers (5 markers at 18p, 9 markers at 18q) at a density of approximately one marker every 9 cM (http://www.gdb.org). The oligonucleotides were labeled with FAM, HEX and NED three different fluorescent dyes for allelotyping (primers were obtained from the ABI PRISM Linkage Mapping Set v. 2, Perkin-Elmer). Multiplex PCR was carried on in a Gene AmpR PCR system 9600 (Perkin-Elmer) for amplifying matched pairs of normal and tumor DNAs. PCR reaction conditions were as follows: 5 μL final volume included 0.5 μL of 10 × PCR buffer, 0.6 μL of 25 mmol/L MgCl2, 0.1 μL of 10 mmol/L dNTP, 0.25 U of Hot-start tag polymerase, 0.04 μL of each primer and 50 ng of DNA. The following PCR run conditions were used: (a) an initial denaturation at 94 °C for 12 min; (b) 15 cycles each at 94 °C for 30 s, 63 °C for 1 min (0.5 °C decreased per cycle), 72 °C for 1 min 50 s; (c) 24 cycles each at 94 °C for 30 s, 56 °C for 1 min, 72 °C for 1 min 50 s; and (d) a final extension at 72 °C for 15 min. A portion of each PCR product (0.7 μL) was combined with 1 μL of the loading mix (ABI internal size standard and formamide loading buffer). After denaturation at 95 °C for 5 min, products were electrophoresed on 4.5% polyacrylamide gels with 7 mol/L urea on ABI prism 377 DNA sequencer (Perkin-Elmer) for 2.5 h. The data were collected automatically and analyzed using ABI prism Genescan 3.1 and Genotyper 2.1 software. Two fragments amplified from normal DNA indicated heterozygote and alleles were defined as the two highest peaks within the expected size range of heterozygote. A ratio of T1:T2/N1:N2 (less than 0.67 or greater than 1.3) was scored as a LOH (Figure 1). A single fragment amplified from normal DNA (homozygote) and these PCR reactions in which fragments were not clearly amplified were scored as non-informative. The LOH frequency of a site was equal to the percentage of the number between allelic losses and informative cases.
LOH at various frequencies were found in GC patients, ranging from 10% to 58%, with the highest rate at D18S70 (21/36, 58%). Of the 45 cases of GC studied, 36 (80%) exhibited allelic losses at least at one site, 25 at over two sites. Figure 1 shows allelic losses in partial patients on several markers. Figure 2 shows LOH frequencies of 14 markers and cytogenetic locations. Eleven markers were found with over 20% LOH frequencies, including 2 markers at 18p and 9 markers at 18q. They were as follows: D18S59 (18p11.3, 28%), D18S452 (18p11.3, 24%), D18S478 (18q11.1-11.2, 37%), D18S1102 (18q12, 29%), D18S474 (18q21, 22%), D18S64 (18q21, 30%), D18S68 (18q21, 25%), D18S61 (18q22, 26%), D18S1161 (18q22, 21%), D18S462 (18q22-23, 29%) and D18S70 (18q23, 58%). Two overlapping deleted regions (Figure 2) were identified at 18q22 (between D18S61 and D18S1161, 9 cM or approximately 5 Mb DNA sequence, overlapping fragment LOH 24%) and 18q22-23 (between D18S462 and D18S70, 6 cM or approximately 3 Mb DNA sequence, overlapping fragment LOH 32%), respectively.
Genetic instability is often observed in tumor cells[9]. Losses of fragments in some chromosomes may lead to allelic losses of TSGs. In the present study, more extensive genome screening was conducted on chromosome 18 in 45 cases of primary GC using 14 microsatellite markers (5 markers at 18p, 9 markers at 18q). Thirty-six out of 45 GC patients (80%) exhibited allelic losses at least one marker, allelic losses of over two markers were seen in 25 cases of GC patients. We found that two markers (D18S59 and D18S452) with over 20% LOH frequencies on chromosome band 18p11.3. DAL-1 gene (18p11.32) localizes between the two markers, which undergoes allelic losses in lung tumors and a significant proportion of ductal carcinomas in situ of the breast. It has been revealed that the DAL-1 protein suppresses the growth of MCF-7 breast cancer cell lines in part through the induction of apoptosis and that expression of DAL-1 increases attachment of these cells to a variety of extracellular matrices[10]. DAL-1 plays a critical role in the suppression of lung tumor formation and metastasis[11]. Our results support the role of DAL-1 gene in gastric carcinogenesis. Future investigations need to be carried on to understand the biological importance of the two putative loci and their clinical relevance in the pathogenesis of GC.
18qLOH is a frequent event in tumors, especially head and neck, colon, lung and gastric cancers[12-14]. All of our 9 markers on this chromosomal arm have shown allelic loss frequencies exceeding 20%. D18S70 was the highest rate of LOH (21/36, 58%) marker on which frequent allelic loss has been reported in head and neck cancers and serous ovarian carcinoma[16]. On the chromosome 18q22-23, Takebayashi et al[15] have identified three loci (D18S39, D18S61 and D18S70) which are lost in 70%-75% of head and neck primary tumor cell lines. Lassus et al observed that 59% of serous carcinomas have shown allelic loss at one or more sites and have been associated with tumor grade and poor survival. Moreover, they have identified two minimal common regions of loss at D18S465-D18S61 (18q22, 3.9 cM) and D18S462-D18S70 (18q23, 5.8 cM). Our data, together with these findings of others, suggest that D18S70 is a new putative tumor suppressor site in GC and support the existence of at least one candidate TSG near the site. Significant LOH of the two overlapping lost fragments identified by us appears novel for GC and no candidate TSGs have been reported in the two overlapping deleted regions so far. LOH frequencies of the two overlapping fragments suggest the presence of as yet unidentified TSGs. Although TSGs harboured in the two overlapping lost fragments have not been defined yet, the two regions are useful candidates for further allelotyping with higher-density microsatellite markers and may help us narrow the sites of frequent allelic loss in order to search for new critical TSGs involved in the development of GC.
The molecular genetic mechanisms of GC remain unclear. Investigators have postulated that, like other tumors, the multistep processes with a variety of factors are involved in GC. Step-wise accumulation of DNA damage from “normal” to tumor tissue serves as the basis of the tumor progression models[17]. Our findings have shown that allelic losses are widespread on chromosome 18 and gastric tumorigenesis may be involved in synthetic effect of multiple TSGs.
We are grateful to Dr. Shen Yan (Chinese National Human Genome Center, Beijing) for his helpful advice.
| 1. | Wang GS, Wang MW, Wu BY, You WD, Yang XY. A novel gene, GCRG224, is differentially expressed in human gastric mucosa. World J Gastroenterol. 2003;9:30-34. [PubMed] |
| 2. | Tomlinson IP, Lambros MB, Roylance RR. Loss of heterozygosity analysis: practically and conceptually flawed? Genes Chromosomes Cancer. 2002;34:349-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 58] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 3. | Wang GT. Progress in studies of mechanism of gastric precan-cerous lesions, carcinogenesis and their reversion. Shijie Huaren Xiaohua Zazhi. 2000;8:1-4. |
| 4. | Deng DJ, E Z. Overview on recent studies of gastric carcinogenesis: human exposure of N-nitrosamides. Shijie Huaren Xiaohua Zazhi. 2000;8:250-252. |
| 5. | Pan CJ, Liu KY. Proliferation/apoptosis and expression of P53 and Bcl-2 in gastric carcinoma. Shijie Huaren Xiaohua Zazhi. 2003;11:526-530. |
| 6. | Du JJ, Dou KF, Cao YX, Wang ZH, Wang WZ, Gao ZQ. CA11, a down-regulated gene in gastric cancer: a functional study. Shijie Huaren Xiaohua Zazhi. 2002;10:525-529. |
| 7. | Ren Q, Wang ZN, Luo Y, Ao Y, Lu C, Jiang L, Xu HM, Zhang X. Loss of heterozygosity on chromosome 18 in microdissected gas-tric cancer cells. Shijie Huaren Xiaohua Zazhi. 2003;11:310-313. |
| 8. | Tran Y, Benbatoul K, Gorse K, Rempel S, Futreal A, Green M, Newsham I. Novel regions of allelic deletion on chromosome 18p in tumors of the lung, brain and breast. Oncogene. 1998;17:3499-3505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 9. | Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1367] [Cited by in RCA: 1391] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 10. | Charboneau AL, Singh V, Yu T, Newsham IF. Suppression of growth and increased cellular attachment after expression of DAL-1 in MCF-7 breast cancer cells. Int J Cancer. 2002;100:181-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 57] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 11. | Yageta M, Kuramochi M, Masuda M, Fukami T, Fukuhara H, Maruyama T, Shibuya M, Murakami Y. Direct association of TSLC1 and DAL-1, two distinct tumor suppressor proteins in lung cancer. Cancer Res. 2002;62:5129-5133. [PubMed] |
| 12. | Wu CL, Kirley SD, Xiao H, Chuang Y, Chung DC, Zukerberg LR. Cables enhances cdk2 tyrosine 15 phosphorylation by Wee1, inhibits cell growth, and is lost in many human colon and squamous cancers. Cancer Res. 2001;61:7325-7332. [PubMed] |
| 13. | Tan D, Kirley S, Li Q, Ramnath N, Slocum HK, Brooks JS, Wu CL, Zukerberg LR. Loss of cables protein expression in human non-small cell lung cancer: a tissue microarray study. Hum Pathol. 2003;34:143-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 14. | Candusso ME, Luinetti O, Villani L, Alberizzi P, Klersy C, Fiocca R, Ranzani GN, Solcia E. Loss of heterozygosity at 18q21 region in gastric cancer involves a number of cancer-related genes and correlates with stage and histology, but lacks independent prognostic value. J Pathol. 2002;197:44-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 15. | Takebayashi S, Ogawa T, Jung KY, Muallem A, Mineta H, Fisher SG, Grenman R, Carey TE. Identification of new minimally lost regions on 18q in head and neck squamous cell carcinoma. Cancer Res. 2000;60:3397-3403. [PubMed] |
| 16. | Lassus H, Salovaara R, Aaltonen LA, Butzow R. Allelic analysis of serous ovarian carcinoma reveals two putative tumor suppressor loci at 18q22-q23 distal to SMAD4, SMAD2, and DCC. Am J Pathol. 2001;159:35-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Powell CA, Klares S, O'Connor G, Brody JS. Loss of heterozygosity in epithelial cells obtained by bronchial brushing: clinical utility in lung cancer. Clin Cancer Res. 1999;5:2025-2034. [PubMed] |
Edited by Chen WW and Kumar M Proofread by Xu FM