©The Author(s) 2025.
World J Gastroenterol. Aug 7, 2025; 31(29): 104830
Published online Aug 7, 2025. doi: 10.3748/wjg.v31.i29.104830
Published online Aug 7, 2025. doi: 10.3748/wjg.v31.i29.104830
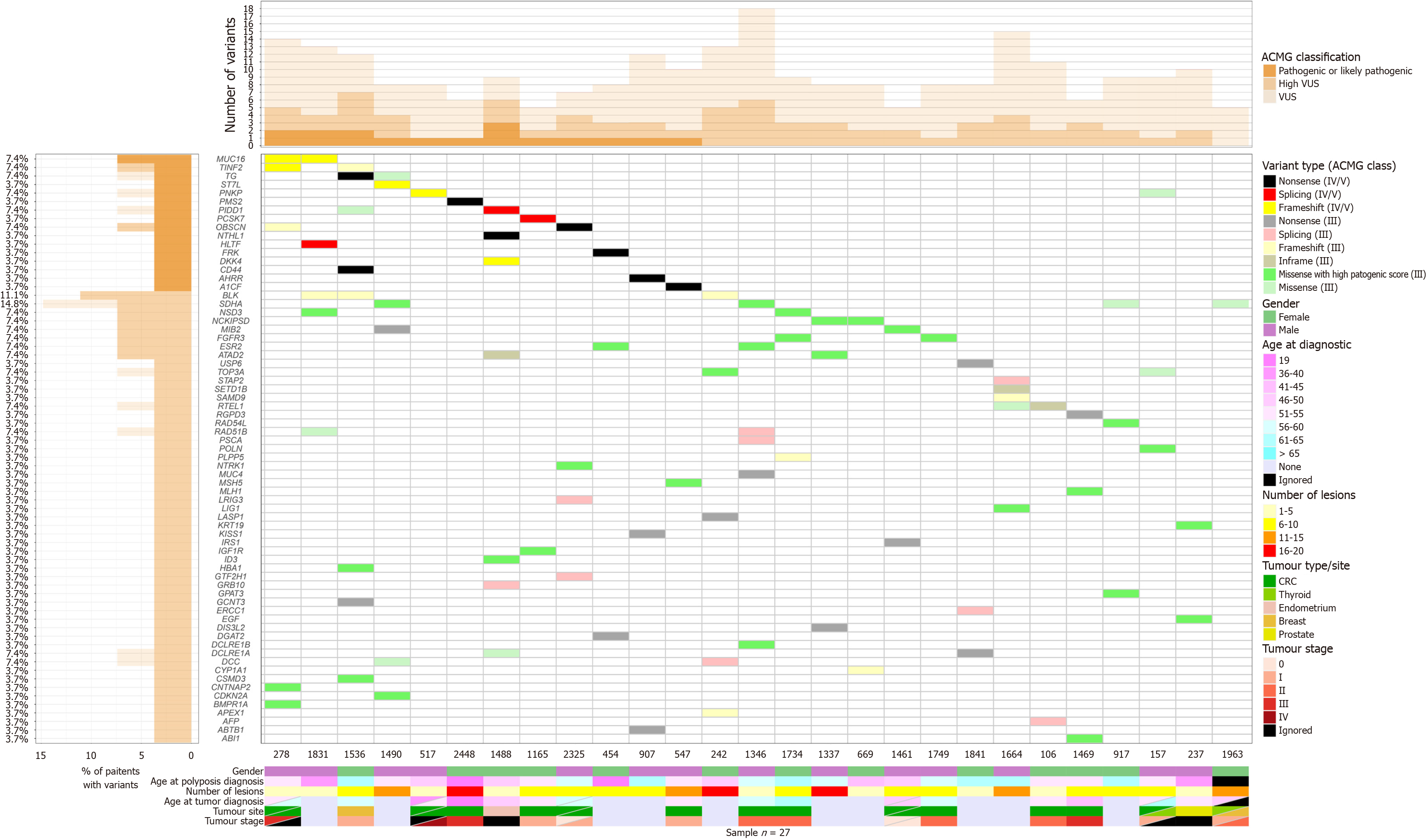
Figure 1 Representative plot of variants identified in patients with polyposis.
All genes harboring pathogenic (V) or likely pathogenic (IV) variants are shown (genes at the top). Genes harboring only variants of uncertain significance are shown if they have an indel, nonsense, or missense variant with a high pathogenic score (high variants of uncertain significance). ACMG: American College of Medical Genetics and Genomics.
- Citation: dos Santos W, Pereira AS, Laureano T, de Andrade ES, Reis MT, Garcia FA, Campacci N, Melendez ME, Reis RM, Galvão HC, Palmero EI. Whole-exome sequencing identifies new pathogenic germline variants in patients with colorectal polyposis. World J Gastroenterol 2025; 31(29): 104830
- URL: https://www.wjgnet.com/1007-9327/full/v31/i29/104830.htm
- DOI: https://dx.doi.org/10.3748/wjg.v31.i29.104830