©The Author(s) 2021.
World J Gastroenterol. Nov 7, 2021; 27(41): 7100-7112
Published online Nov 7, 2021. doi: 10.3748/wjg.v27.i41.7100
Published online Nov 7, 2021. doi: 10.3748/wjg.v27.i41.7100
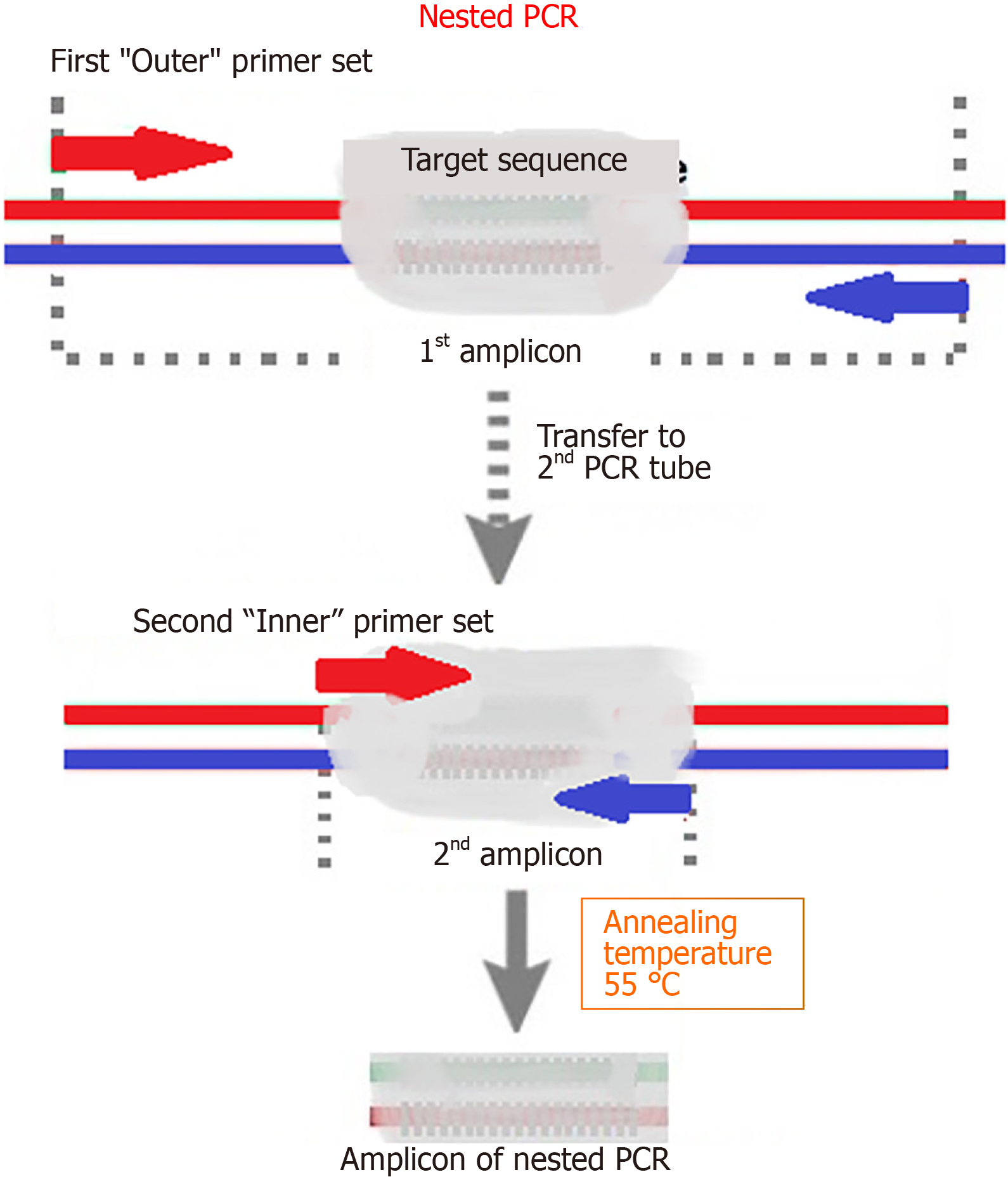
Figure 1 Nested polymerase chain reaction.
Nested polymerase chain reaction involves two amplification reactions. The first round targeted a larger DNA region, and the second targeted a narrower sub-region of the products of the first round that were used as a template. PCR: Polymerase chain reaction.
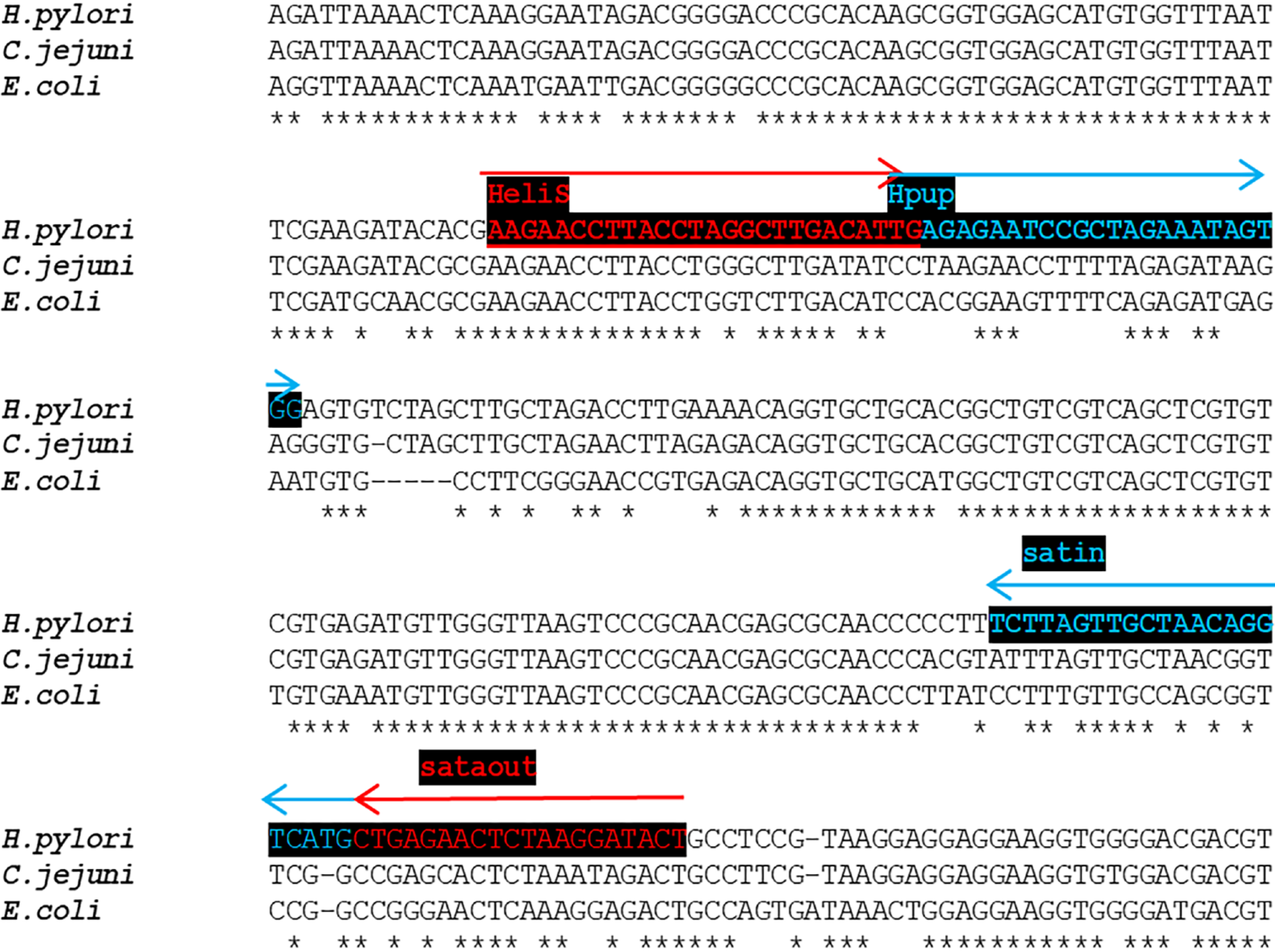
Figure 2 Design of Helicobacter pylori-specific primers for shorter 148-bp 16S rRNA amplicon.
Alignment of Helicobacter pylori (amplified region) to other bacterial species. Selective primers marked in red, and blue were designed in the regions with a high divergence of Helicobacter sequence.
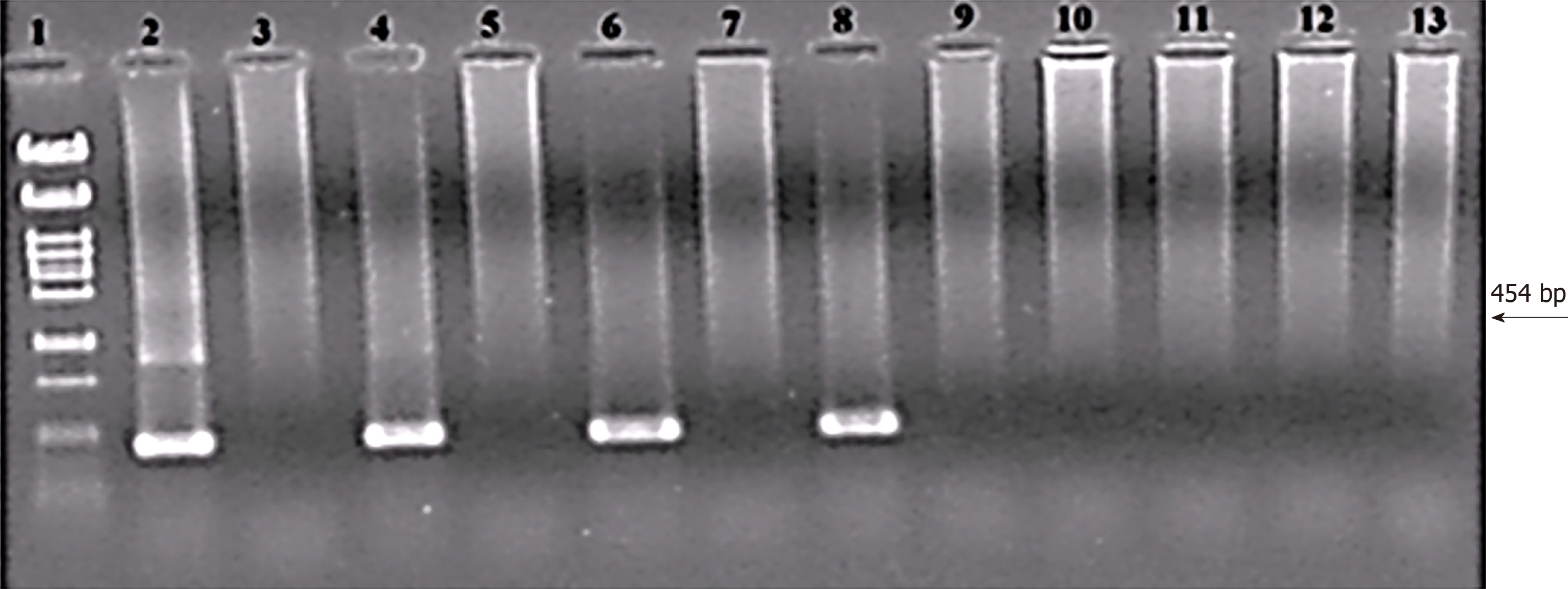
Figure 3 Threshold value of the nested polymerase chain reaction assay for Helicobacter pylori detection in cell suspension (colony polymerase chain reaction).
Lines: 1: Size marker λ/Pst1; 2: 500; 3: Negative control (NC); 4: 50; 5: NC; 6: 5; 7: NC; 8: 0.5; 9: NC; 10: 0.05; 11: NC; 12: 0.005; 13: NC. Numbers express cell counts in the polymerase chain reaction (PCR) reaction. External primers HeliS/HeliN. Internal primers Hpup/Hpdown. Size of PCR product is 454 bp[28,53]. Each sample was tested by PCR separately in two independent experiments, always with the same result. Separated on 2% agarose in TBE.
- Citation: Sulo P, Šipková B. DNA diagnostics for reliable and universal identification of Helicobacter pylori. World J Gastroenterol 2021; 27(41): 7100-7112
- URL: https://www.wjgnet.com/1007-9327/full/v27/i41/7100.htm
- DOI: https://dx.doi.org/10.3748/wjg.v27.i41.7100