©The Author(s) 2016.
World J Gastroenterol. Feb 21, 2016; 22(7): 2284-2293
Published online Feb 21, 2016. doi: 10.3748/wjg.v22.i7.2284
Published online Feb 21, 2016. doi: 10.3748/wjg.v22.i7.2284
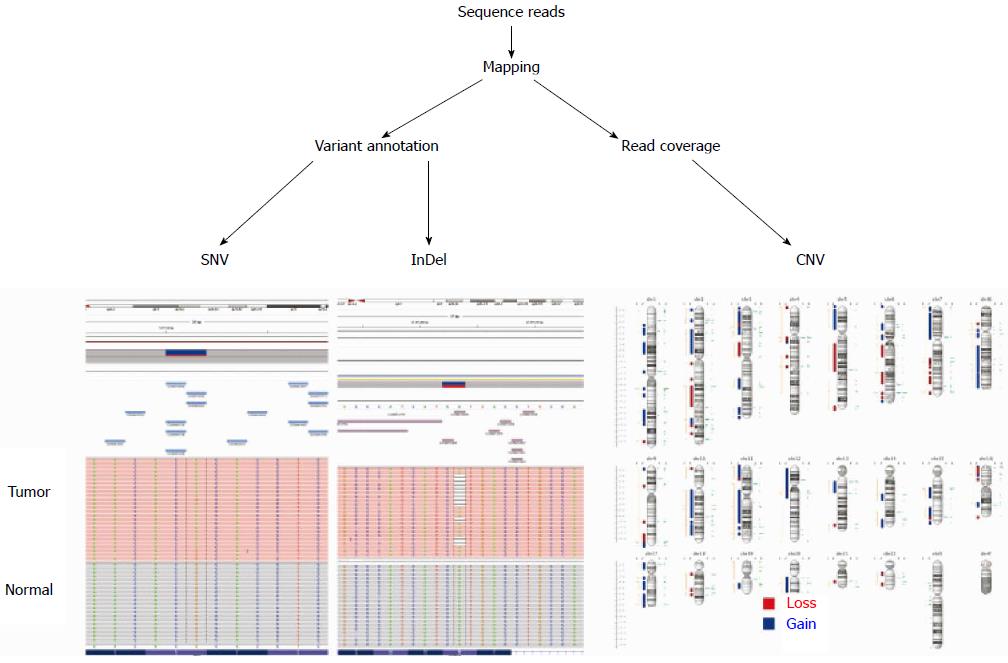
Figure 1 Cancer genome research using next-generation sequencing.
Sequence reads are quality checked and then mapped to the reference genome. Somatic DNA alterations are detected using statistical approaches in tumor and normal samples from the available software or an integrated workflow such as the GATK pipeline (Broad Institute). SNV: Single nucleotide variant; InDel: Insertion and deletion; CNV: Copy number variation.
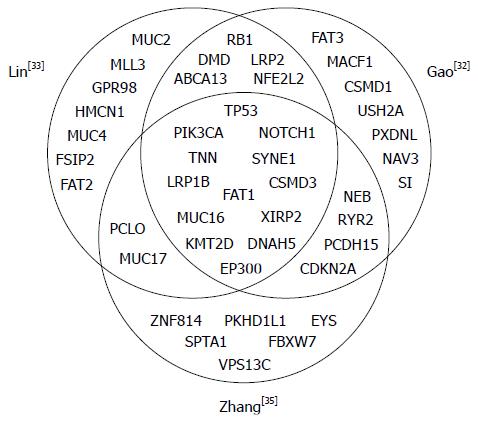
Figure 2 Comparison of most significantly mutated genes identified in the three whole genome and whole exon sequencing studies for esophageal squamous cell carcinomas.
Top significantly mutated genes (25-27 genes per study) were obtained from three recent next-generation sequencing studies of large cohorts. Only nonsynonymous mutations were considered.
- Citation: Sasaki Y, Tamura M, Koyama R, Nakagaki T, Adachi Y, Tokino T. Genomic characterization of esophageal squamous cell carcinoma: Insights from next-generation sequencing. World J Gastroenterol 2016; 22(7): 2284-2293
- URL: https://www.wjgnet.com/1007-9327/full/v22/i7/2284.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i7.2284