Copyright
©2013 Baishideng Publishing Group Co.
World J Gastroenterol. Aug 14, 2013; 19(30): 5006-5010
Published online Aug 14, 2013. doi: 10.3748/wjg.v19.i30.5006
Published online Aug 14, 2013. doi: 10.3748/wjg.v19.i30.5006
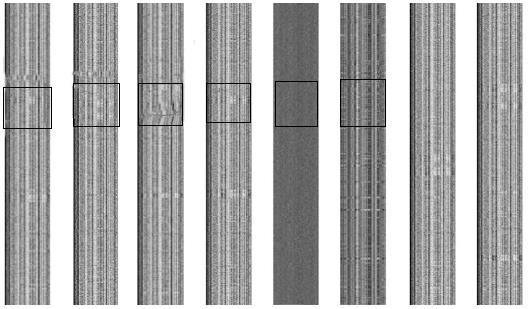
Figure 1 2-D barcode images of genomes of Helicobacter pylori strains J99, G27, 26695, HPAG1, P12, and Shi470, Escherichia coli O157:H7 strain EDL933, and a random sequence.
The y-axis represents the genome axis from top-down, with each pixel representing a fragment of n = 1000 bp; the x-axis represents the 4-nucletide frequencies. The abnormal barcode regions are demarcated by a rectangle.
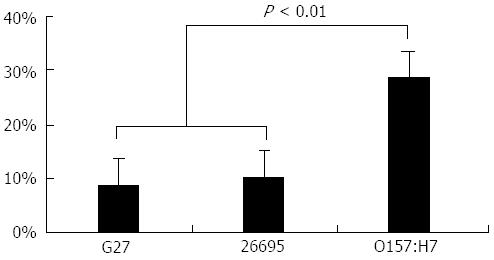
Figure 2 Fraction of anomalous fragments detected by genomic barcode imaging of Helicobacter pylori strains G27 and 26695, and Escherichia coli O157:H7 strain EDL933.
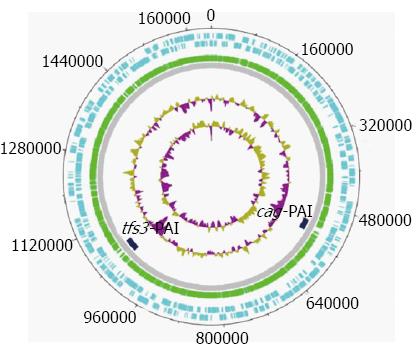
Figure 3 Circular representation of the Helicobacter pylori 26695 chromosome.
The outermost (first) concentric circle denotes the predicted coding regions on the plus strand. The second concentric circle denotes the predicted coding regions on the minus strand. The third concentric circle denotes the predicted coding regions on both strands. The fourth concentric circle denotes the buffer zone. The fifth concentric circle denotes the predicted pathogenicity island (PAI) candidates. The sixth concentric circle denotes the guanine and cytosine (GC) content. The seventh concentric circle denotes the GC content. The figure was created using GenVision from DNASTAR.
-
Citation: Wang GQ, Xu JT, Xu GY, Zhang Y, Li F, Suo J. Predicting a novel pathogenicity island in
Helicobacter pylori by genomic barcoding. World J Gastroenterol 2013; 19(30): 5006-5010 - URL: https://www.wjgnet.com/1007-9327/full/v19/i30/5006.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i30.5006