©2010 Baishideng.
World J Gastroenterol. Aug 21, 2010; 16(31): 3950-3956
Published online Aug 21, 2010. doi: 10.3748/wjg.v16.i31.3950
Published online Aug 21, 2010. doi: 10.3748/wjg.v16.i31.3950
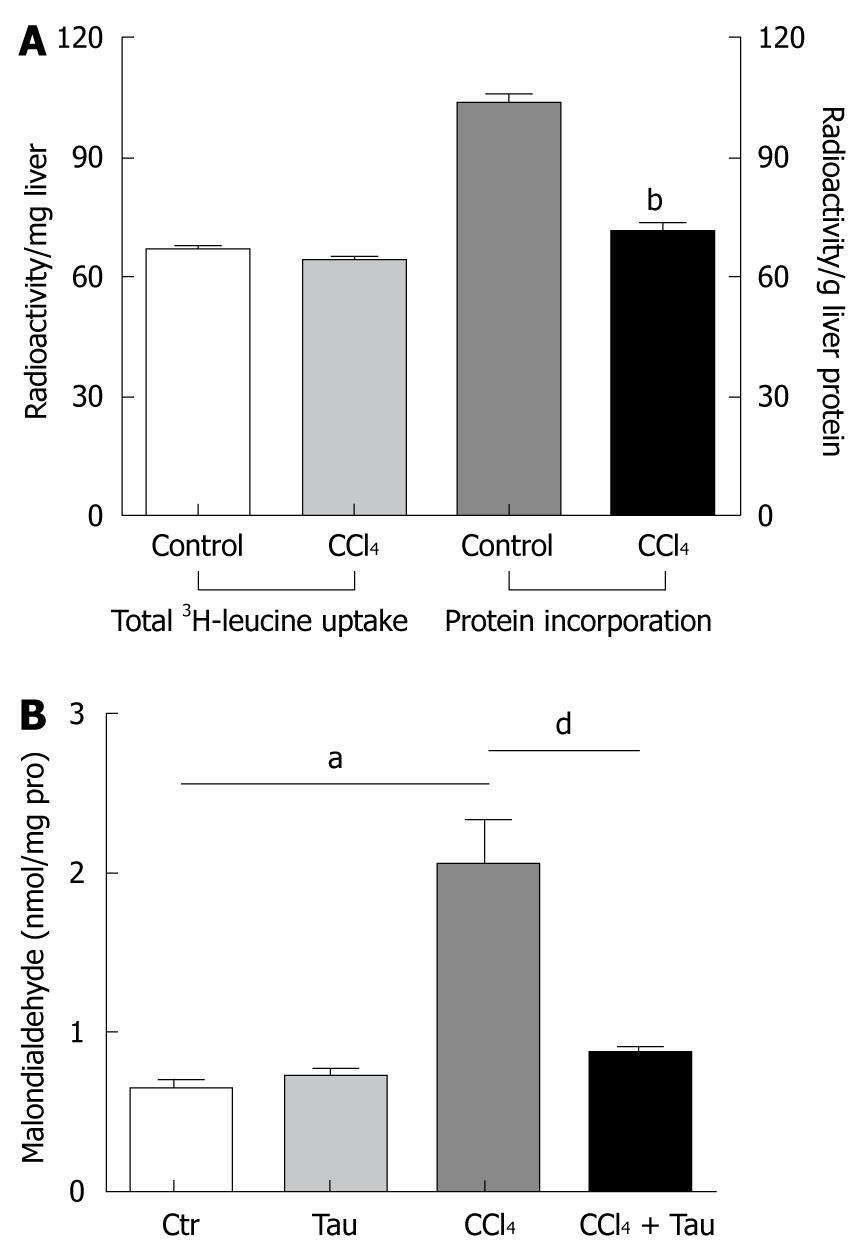
Figure 1 CCl4 inhibits protein synthesis (A) and increases malondialdehyde production (B) in liver of Sprague-Dawley rats.
Data are expressed as mean ± SE. aP < 0.05 vs control group; bP < 0.01 vs their counterpart control group; dP < 0.01 vs CCl4 treatment group.
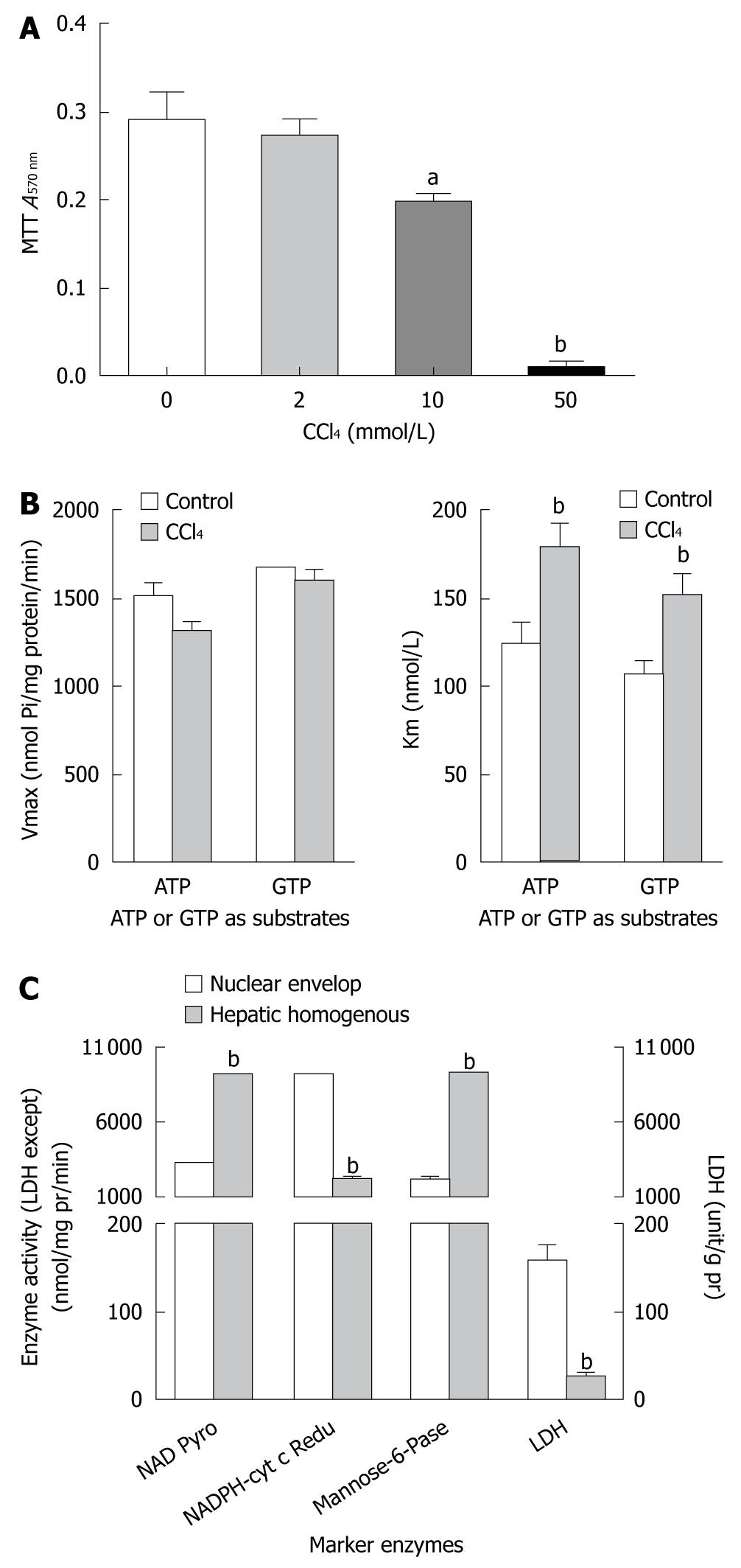
Figure 2 CC14 inhibits nucleotide triphosphatase affinity in nuclei of HepG2 cells.
A: Methyl thiazolyl tetrazolium (MTT) metabolic viability assay at a dose of 10 mmol/L showing the inhibitory effect of CCl4 on growth of HepG2 cells; B: Substrate-affinity (Km, B2) is decreased without any alterations in the substrate binding sites (Vmax,B1) for adenosine triphosphate (ATP) or guanine triphosphate (GTP) in nuclear nucleotide triphosphatase of HepG2 cells treated with CCl4 (2 mmol/L); C: Enzyme’ activities of nuclei extracts and homogenates from HepG2 cells in control and CCl4 treatment groups. aP < 0.05 vs control group, or nuclear envelop group, respectively; bP < 0.01 vs their counterpart control group, or nuclear envelop group, respectively. LDH: Lactose dehydrogenase.
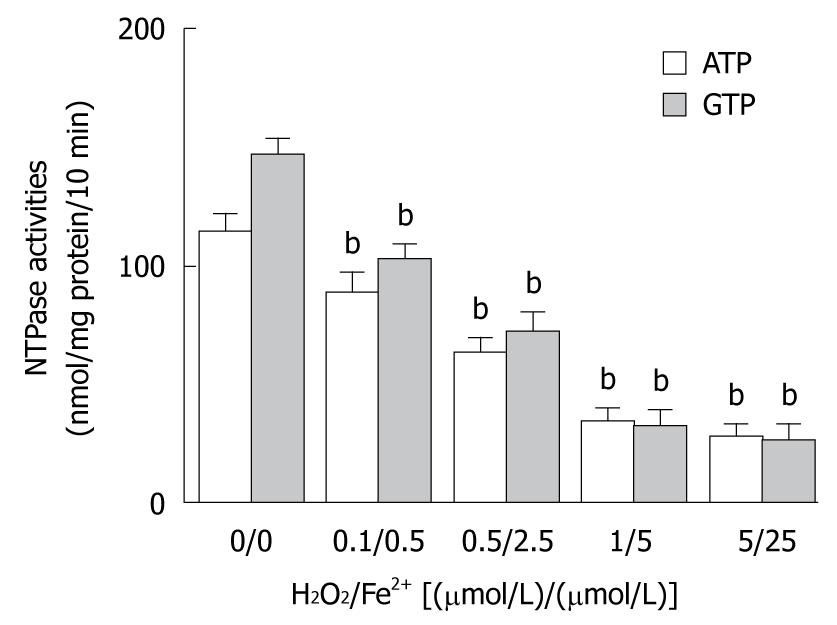
Figure 3 Involvement of reactive radicals in decreased nucleotide triphosphatase activity of nuclei.
Data are represented as mean ± SE. bP < 0.01 vs H2O2/Fe2+ group [(0 μmol/L)/(0 μmol/L)]. NTPase: Nucleotide triphosphatase.
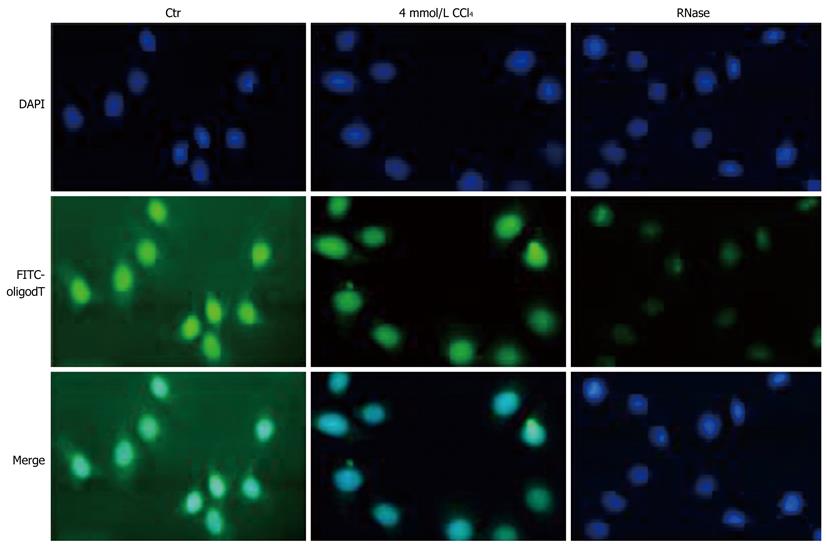
Figure 4 CCl4 decreases total mRNA nuclear export of HepG2 cells.
DAPI: 4,6-diamidino-2-phenylindole dihydrochloride; FITC: Fluorescein isothiocyanate.
- Citation: Li XW, Zhu R, Li B, Zhou M, Sheng QJ, Yang YP, Han NY, Li ZQ. Mechanism underlying carbon tetrachloride-inhibited protein synthesis in liver. World J Gastroenterol 2010; 16(31): 3950-3956
- URL: https://www.wjgnet.com/1007-9327/full/v16/i31/3950.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i31.3950