Published online Nov 12, 2016. doi: 10.5501/wjv.v5.i4.161
Peer-review started: June 27, 2016
First decision: August 5, 2016
Revised: August 9, 2016
Accepted: August 27, 2016
Article in press: August 29, 2016
Published online: November 12, 2016
Processing time: 137 Days and 2.2 Hours
To investigate the role of subgenotype specific RNA secondary structure in the compartment specific selection of hepatitis B virus (HBV) immune escape mutations.
This study was based on the analysis of the specific observation of HBV subgenotype A1 in the serum/plasma, while subgenotype A2 with G145R mutation in the peripheral blood leukocytes (PBLs). Genetic variability found among the two subgenotypes was used for prediction and comparison of the full length pregenomic RNA (pgRNA) secondary structure and base pairings. RNA secondary structures were predicted for 37 °C using the Vienna RNA fold server, using default parameters. Visualization and detailed analysis was done using RNA shapes program.
In this analysis, using similar algorithm and conditions, entirely different pgRNA secondary structures for subgenotype A1 and subgenotype A2 were predicted, suggesting different base pairing patterns within the two subgenotypes of genotype A, specifically, in the HBV genetic region encoding the major hydrophilic loop. We observed that for subgenotype A1 specific pgRNA, nucleotide 358U base paired with 1738A and nucleotide 587G base paired with 607C. However in sharp contrast, in subgenotype A2 specific pgRNA, nucleotide 358U was opposite to nucleotide 588G, while 587G was opposite to 359U, hence precluding correct base pairing and thereby lesser stability of the stem structure. When the nucleotides at 358U and 587G were replaced with 358C and 587A respectively (as observed specifically in the PBL associated A2 sequences), these nucleotides base paired correctly with 588G and 359U, respectively.
The results of this study show that compartment specific mutations are associated with HBV subgenotype specific alterations in base pairing of the pgRNA, leading to compartment specific selection and preponderance of specific HBV subgenotype with unique mutational pattern.
Core tip: We have previously shown that, in our study population, distribution of hepatitis B virus (HBV) subgenotypes A1 and A2 is highly biased in the serum/plasma and peripheral blood leukocyte (PBL) compartments respectively. Analysing the predicted base pairing patterns of pregenomic RNAs (pgRNAs), specific for HBV subgenotype A1 and A2, we demonstrate that the potent immune escape mutation G145R evolves specifically in the context of HBV subgenotype A2. The PBL compartment is exposed to strong anti-HBs immunity, and thus G145R is highly advantageous for the virus to persist. This explains the exclusive preponderance of subgenotype A2 in the PBL compartment, sharply contrasting the prevalence of subgenotype A1 in the serum/plasma.
- Citation: Datta S, Chakravarty R. Role of RNA secondary structure in emergence of compartment specific hepatitis B virus immune escape variants. World J Virol 2016; 5(4): 161-169
- URL: https://www.wjgnet.com/2220-3249/full/v5/i4/161.htm
- DOI: https://dx.doi.org/10.5501/wjv.v5.i4.161
Viral compartmentalization signify infection, persistence and replication of viruses in off-target cells/tissues or anatomical compartments of the host, and this phenomenon is now believed to be a crucial event in many important viral infections, including hepatitis B virus (HBV), human immunodeficiency virus, hepatitis C virus, etc.[1-5]. Recent molecular evolutionary studies have demonstrated that viruses evolve independently under the influence of unique immunological milieu in a given compartment, leading to the selection and emergence of specific viral variants, which endow the virus with an advantage to survive and persist in that particular compartment[2,6,7]. Such compartment specific viral evolution have extremely important implications in emergence and re-emergence of immune-escape mutants, antiviral resistant mutants, their long term persistence and transmission through different non-conventional routes[1,7].
HBV is the prototype member of the Hepadnaviridae family of enveloped viruses with a very unique partially double-stranded DNA genome[8]. Despite having a DNA genome, HBV exclusively uses an RNA intermediate (the pregenomic RNA or the pgRNA) and a virus encoded reverse transcriptase to replicate its genome through a complex mechanism of primer shifting[8]. Even though HBV is classically considered to be a hepatotropic virus, HBV related nucleic acids and proteins have long been detected in different tissues, suggesting that it replicates and propagates in various non-hepatic tissues[1]. Interestingly, some of these extrahepatic sites have been shown to act as reservoirs and also the source of reinfection after surgical and therapeutic interventions[9,10]. Recently, ours and other research groups have provided convincing evidences that the HBV strains and their mutational signature pattern present in different extra-hepatic compartments, are often characteristically distinct from the HBV strains circulating in the serum/plasma/hepatic compartments and that immune escape/drug resistance mutations are significantly more frequent in different extrahepatic compartments in HBV carriers[2,4,11].
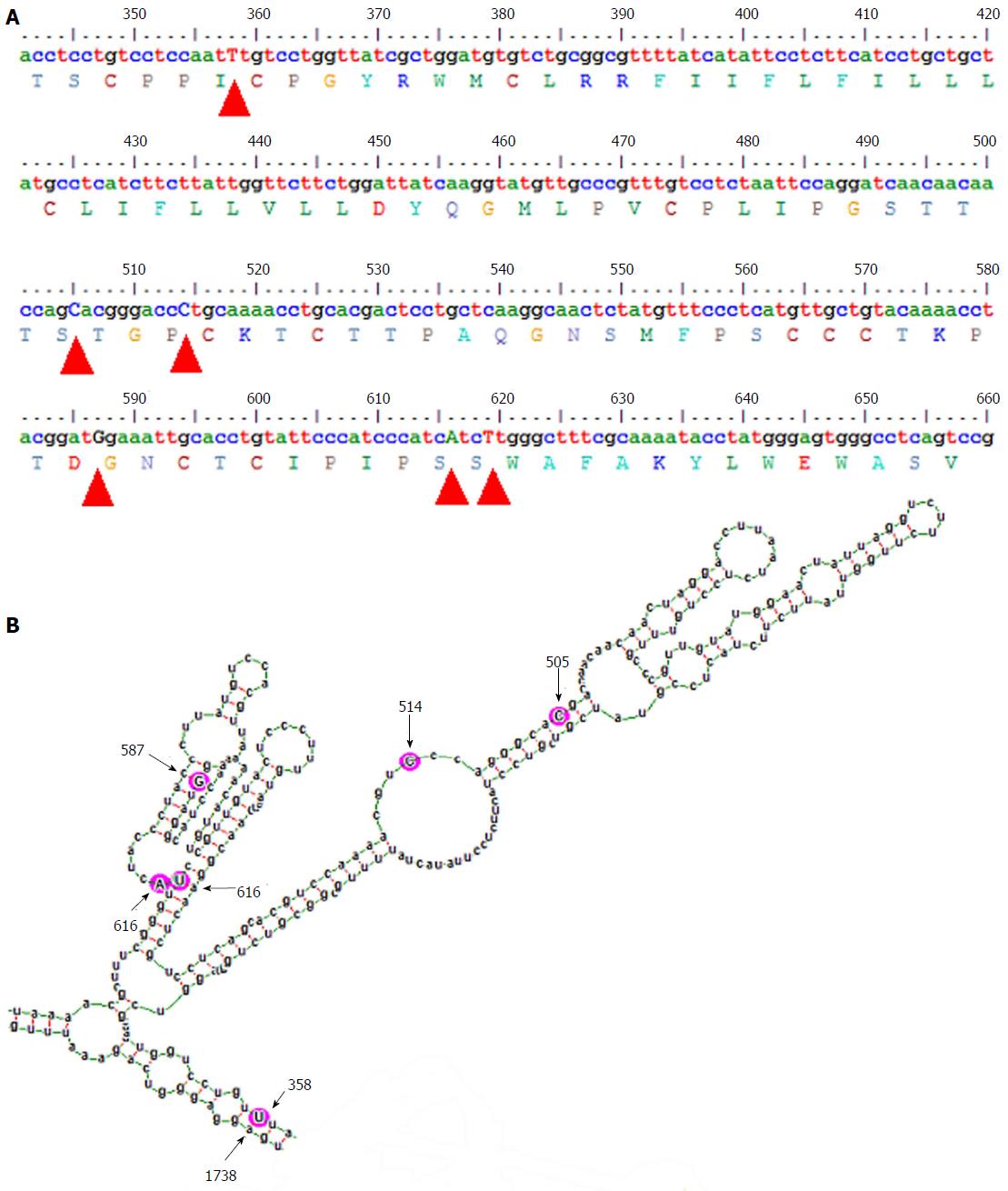
In our previous studies, we have recognized the subgenotype A1 (Afro-Asian subgenotype) as the predominant subgenotype of HBV genotype A circulating in the sera/plasma of our study population and that the occurrence of G145R mutation therein was sporadic[12-14]. In sharp contrast, we documented the confined and exclusive existence of HBV subgenotype A2 with the potent “immune escape” mutation G145R within the peripheral blood leukocytes (PBL), across the study population, irrespective of the HBV genotype/subgenotype circulating in the serum/plasma of the respective individual[2]. G145R is the mutation signifying Glycine to Arginine substitution at amino acid residue 145 in the major hydrophilic loop (MHL), a B-cell epitope of the hepatitis B surface antigen (HBsAg), which provides a strong immune escape property. These observations strongly signify that viral mutants with G145R does have an explicit replicative advantage within the PBLs, that are exposed to strong anti-HBs immunity and that this mutation emerges specifically in the perspective of subgenotype A2, but not in subgenotype A1. Moreover, all the subgenotype specific nucleotide substitutions in the MHL encoding region of A1 (505C, 514C, 616A and 619T) and A2 (505T, 514A, 616G and 619C) are synonymous in nature, which aptly rules out the possibility that the predilection of the subgenotype A2 in the PBL is due to the subgenotype specific epitopic difference in the HBsAg[2]. Taken together, the above observations led us to hypothesize that the subgenotype specific nucleotide substitutions might modulate the base pairing of the A2 specific pgRNA in a way, which favours the emergence of G145R.
In the present work, we compared the changes in the base pairing of the pgRNA due to subgenotype A1 and A2 specific substitutions in the MHL encoding region. Based on the RNA secondary structure predictions, we demonstrate that the selection and emergence of G145R within HBV subgenotype A2 sequences in the PBL compartment occurs due to the differential base pairing characteristics in the subgenotype A2 specific pgRNA.
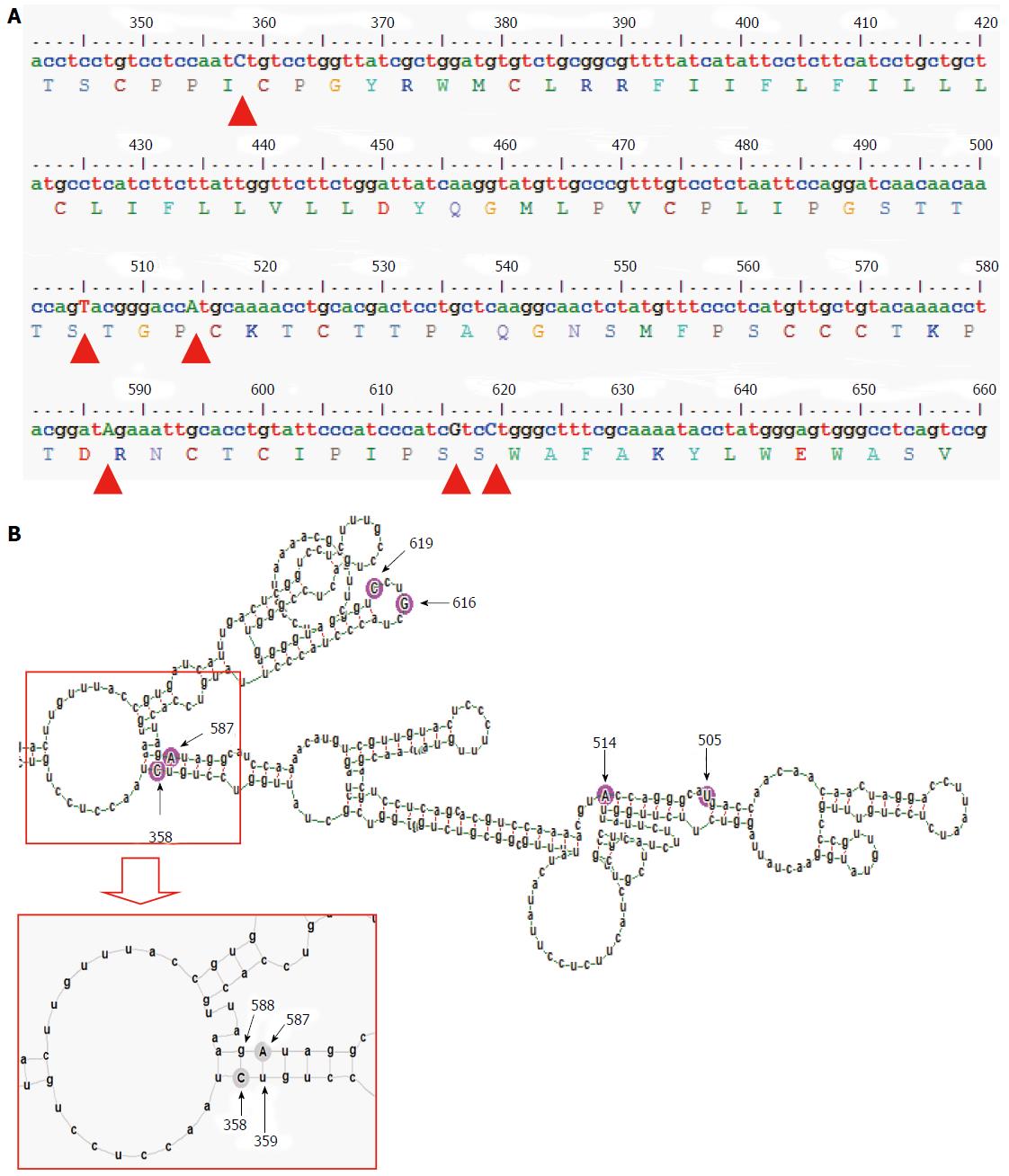
HBV surface gene sequences, corresponding to the nucleotide 341 to 660 of the HBV genome (nucleotide position counted from the unique EcoRI site in the HBV genome), that code for the epitopic MHL for both subgenotype A1 sequences (isolated from serum/plasma) and subgenotype A2 sequences (isolated from PBL), obtained during our previous studies were used in this analysis[2,14]. Using alignment of these sequences along with other reference GenBank HBV sequences[15], subgenotype A1 and A2 specific nucleotide substitutions (summarised in the Table 1) were determined earlier[2]. These nucleotide differences were consequently used for studying the alterations in the subgenotype specific base-pairing and folding of the pgRNA.
For subgenotype A1 and A2 specific pgRNA secondary structure predictions, template sequences were generated separately by editing two well defined full length sequences, namely-GenBank accession number DQ315784 (India) for subgenotype A1 and GenBank accession number AJ309370 (France) for subgenotype A2, respectively, following the method described previously for generation of full length pgRNA sequence[16]. The pgRNA templates so generated were unpolyadenylated and included the terminal redundancy. These two sequences served as the base sequences for prediction of secondary structures, to which nucleotide substitutions observed in the MHL encoding and flanking regions of serum associated A1 and PBL associated A2 (as mentioned in the previous section) were substituted respectively at appropriate nucleotide positions. Finally, these two template sequences (approximately 3.3 kb) were subjected to RNA secondary structure prediction and comparison.
For prediction of the secondary structures, the pgRNA sequences generated as stated above were submitted to the Vienna RNA secondary structure server[17,18]. The server predicts the minimum free energy (mfe) secondary structures for single RNA sequences using an algorithm proposed by Zuker and Stiegler, and also calculates the equilibrium base-pairing probabilities by means of partition function (pf) algorithm proposed by McCaskill[19,20]. Apart from the mfe and pf, the server also provides a centroid structure, which indicates the reliability of the predictions, while the dot-plot which provides information on base-pairing probabilities of all the possible predicted structures[17,21]. All the secondary structure predictions were performed for a temperature of 37 °C, keeping all the other parameters to default[22]. Visualization, annotation and analysis of the mfe structures were performed using the RNAshapes program[23]. As the present study was focused on the genetic variability of the HBV genome encoding the MHL region of the surface gene, we restricted our detailed analysis of base pairing pattern to the secondary structure of the part of pgRNA, corresponding to the MHL encoding sequence.
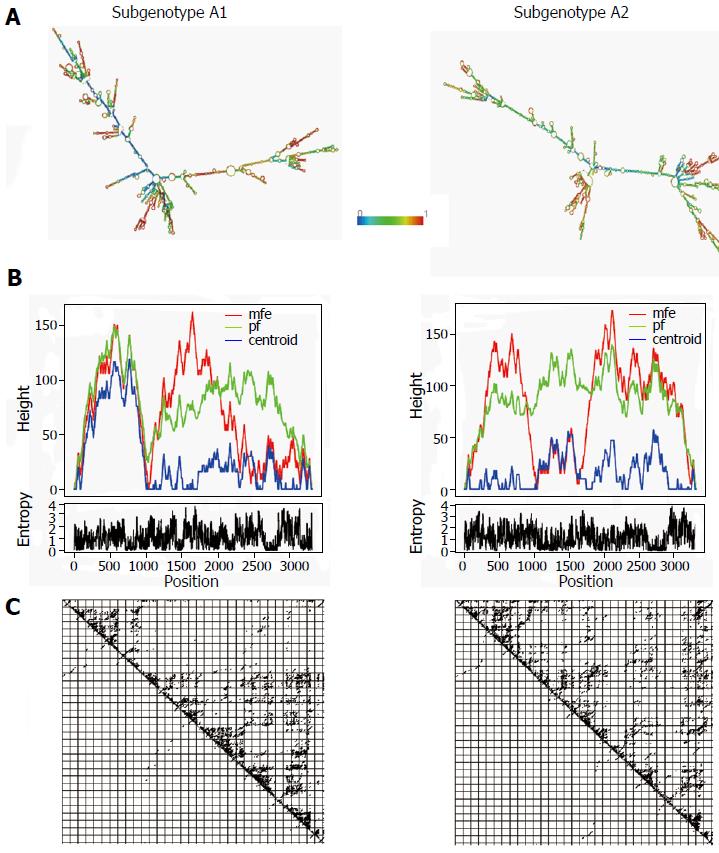
The gross structural features of the subgenotype A1 and A2 specific pgRNA were found to be entirely distinct (Figure 1). The difference in the pgRNA structure was also evident from the mountain plots showing the mfe, pf, centroid, entropy and the dot plot of the two subgenotypes. The difference in other features are summarised in the Table 2. Detailed scrutiny of the pgRNA secondary structures corresponding to the MHL encoding genetic regions, revealed entirely distinct pgRNA secondary structures with discrete intra-molecular base pairing patterns due to the subgenotype specific and variations between A1 and A2 sequences (Figures 2 and 3). Interestingly, when we focussed on the base pairing of the nucleotides encoding the MHL region, we noted that in subgenotype A1 specific pgRNA, nucleotide 358U base paired with 1738A and nucleotide 587G base paired with 607C (Figure 2). However in sharp contrast, in subgenotype A2 specific pgRNA, nucleotide 358U was opposite to nucleotide 588G, while 587G was opposite to 359U, hence precluding correct base pairing and thereby less stability of the stem structure. When the nucleotides at 358U and 587G were replaced with 358C and 587A respectively (as observed specifically in the PBL associated A2 sequences), these nucleotides base paired correctly with 588G and 359U, respectively (Figure 3), forming a correctly paired stem-loop structure, hence stabilizing the local conformation. Nevertheless, the effects of other substitutions were not as influential as these two changes. The exclusive detection of subgenotype A2 sequences with the abovementioned substitutions in the PBL clearly suggest the selective advantage of the pgRNA with 358C and 587A, and in turn the importance of G145R immune escape mutation in the PBL compartment.
| Features | Subgenotype A1 | Subgenotype A2 |
| Minimum free energy of the optimal secondary structure | -1052.10 kcal/mol | -1049.50 kcal/mol |
| Free energy of the thermodynamic ensemble | -1099.56 kcal/mol | -1098.93 kcal/mol |
| Minimum free energy of the centroid secondary structure | -722.20 kcal/mol | -679.21 kcal/mol |
| Ensemble diversity | 863.25 | 954.99 |
In this study, we present interesting observations about the possible mechanism of compartment specific selection of immune escape HBV mutants. Based on our previous studies done on serum/plasma isolated HBV genotypes, we have documented the predominance of at least three distinct HBV genotypes in our study population, namely genotype D (most abundant) followed by genotypes C and A[13,14]. However, when we investigated the paired HBV sequences isolated from serum/plasma and the PBLs, we surprisingly observed the exclusive preponderance of the genotype A in the PBL, irrespective of the HBV genotypes circulating in the serum/plasma of any given individual[2]. More interestingly, the genotype A sequences isolated from the PBL was found to be markedly distinct from that of genotype A sequences isolated from serum/plasma, in terms of subgenotype and specific nucleotide substitution patterns. More precisely, subgenotype A1 of HBV genotype A was prevalent in the serum/plasma while in sharp contrast; subgenotype A2 was solely isolated from the PBL[2].
In the present study, we sought to examine the selective advantage of subgenotype A2 in the PBL compartment with the help of advance computational prediction and analysis programs. We focussed our analysis on the examination of nucleotide sequences encoding the dominant B-cell epitope (MHL) of the HBV surface antigen, since in a number of other viruses, analogous genetic regions (epitope regions of the envelope protein) have been shown to undergo faster evolution to facilitate the emergence of compartment specific immune-escape variants[2]. Interestingly, when we compared serum/plasma circulating subgenotype A1 sequences with PBL confined subgenotype A2 sequences, we found that only four subgenotype specific nucleotide substitutions differentiate both sequences[2]. However, all these four subgenotype specific nucleotide substitutions in the MHL encoding region were found to be synonymous in nature (i.e., the sequence of amino acids in the MHL remains same between subgenotypes A1 and A2), suggestive of the fact that MHL epitope diversity might not be directly relevant to the selection of subgenotype A2 over subgenotype A1 in PBL. On the other hand, in addition to these four subgenotype specific nucleotide substitutions, two additional nucleotide substitutions (358C and 587A) were evident with PBL associated A2 sequences, across the study population, which we have earlier shown to be co-evolving in the PBL[2]. Interestingly, we further noted that nucleotide substitution 358C was also synonymous, while substitution 587A was non-synonymous and translated into the potent immune escape G145R mutation of HBsAg. Earlier studies have demonstrated that by virtue of its definite advantages, G145R mutation helps HBV to dynamically evade anti-HBs specific immune response, thereby ensuring viral persistence in anatomical compartments, which are exposed to strong anti-HBs immunity[2]. The association of these five synonymous nucleotide substitutions and a potent immune escape mutation with PBL associated Ae/A2 sequences led us to hypothesize that the advantageous 587A (G145R) might be selected at the pgRNA base pairing level and to verify this hypothesis, this comparative study was undertaken.
On comparison of the pgRNA secondary structures, we observed that the invariable association of subgenotype A2 with the selection of nucleotide 587A (causing G145R) most possibly occur in the context of genotype A2 specific altered pgRNA base pairing patterns. Fascinatingly, we observed that substitution of a uracil (U, corresponding to Thymidine, T in DNA sequence) to cytosine (C) at position 358 altogether changed the local base pairing pattern of the pgRNA (358C paired with nucleotide 588G instead of the normal pairing with 1738A in subgenotype A1). In the context of this altered base pairing, a single nucleotide change (U to C) at 359 was found to stabilize the stem structures by pairing with the wild type 587G, just opposite to it. However, the nucleotide at position 359 encodes a Cysteine residue at amino acid position 69 of HBsAg, which is extremely essential for the generation of subviral 20 nm HBsAg particles, and thus, any non-synonymous substitution at this position is most likely to be detrimental for the virus persistence[24]. Therefore, based on the predicted secondary structures, we hypothesized that, instead of selecting an altered nucleotide at this exceptionally essential position (359), a compensatory alteration of a single nucleotide (G to A) at position 587 is expected to serve dual purpose, firstly it may help stabilize the stem structure (by pairing with the highly conserved 359T/U) and secondly it results in the emergence of a potent immune escape G145R mutation, both of which appears to be highly advantageous for the virus.
The HBV polymerase lacking proof reading function has been implicated in the generation of random mutations and generation of “quasi-species”, of which the genomes (viral DNA) or pregenomes (pgRNA) having mutations useful for escaping the immune response of the host are gradually selected and subsequently become the prevalent viral population[25]. Apart from viral polymerase induced random mutations, host PBL associated APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide) family of cellular cytidine deaminases have also been shown to induce hyper-mutation (G to A mutations) in the HBV genome, which has been suggested to cause genetic diversification and consequently selective evolution among the divergent genomes[26-28]. Nevertheless, the finding of selective predominance of the point mutation leading to G145R in the present study is highly significant in the context of PBL, since PBLs are exposed to strong anti-HBs humoral immune response and HBV variants with G145R are capable of strongly neutralizing this immune response, without any compromise in the replicative competence, thereby ensuring viral perseverance[29,30]. Whatever is the source of genetic diversification, in the present work we describe a probable mechanism of RNA folding, through which divergent viral genomes/pregenomes having favourable mutations are selected for propagation.
We acknowledge that the RNA folding predictions are based on statistical/mathematical algorithms and the biological relevance of these predictions are based on their corroboration with the biological data. Interestingly, the results of the present RNA folding predictions beautifully elucidate the observed co-evolution of the mutations at positions 358 and 587, which supports the biological relevance of the observed predictions. The results of the present study further implies that, certain HBV mutations are selected at the subgenomic RNA level (as they are synonymous at the protein level), which may significantly alter the base pairing of the pgRNA, which in turn may hasten the selection of mutations at other sites. Interestingly, the mechanism suggested in this work is very much similar to the mechanism described for HBV genotype specific selection of the most widely studied HBV precore mutation (1896A), which emerges to stabilize the stem-loop structure of the epsilon “ε” signal of pgRNA[31]. Altogether the present study, support the findings of Kidd-Ljunggren et al[16], that demonstrate the implications of genotype specific differences in the pgRNA secondary structures in the emergence of genotype specific variations in the HBV genome.
In conclusion, our results based on the predicted RNA secondary structures suggest the role of HBV genotype/subgenotype specific base pairing patterns of the pgRNA in selection/emergence of advantageous mutations. Furthermore, the observed association of a potent immune escape mutation with a particular HBV subgenotype, confined in a specific anatomic compartment indicate the possible mechanism of genotype/subgenotype specific compartmentalization of HBV, which may have important implications in extrahepatic maintenance and transmission of HBV through hitherto unknown routes.
The authors sincerely thank the directors of their respective institute and the laboratory staff for support and cooperation.
In our previous study we have observed highly compartment specific prevalence of hepatitis B virus (HBV) genotype A. In particular, HBV subgenotype A1 was detected in serum/plasma isolates, while HBV subgenotype A2 was predominant in the peripheral blood leukocytes (PBLs). Apart from subgenotype specific differences, G145R - a potent immune escape mutation was specifically observed in the PBL related subgenotype A2 isolates. The authors undertook this study to understand the possible mechanism of the compartment specific distribution of HBV subgenotypes and immune escape mutation.
Compartment specific evolution and emergence of HBV mutations is a poorly understood area. A few recent studies, including the authors have provided evidences, indicating that HBV independently evolves in different anatomical compartments, depending upon the immune selection pressure on that compartment.
Using computational prediction methods, the authors show that the PBL specific emergence of G145R occurs in the context of HBV subgenotype A2, due to altered base-pairing patterns, as compared to the HBV subgenotype A1.
The findings of the present study are important in the long term persistence, evolution and transmission of different HBV genotypes/subgenotypes/mutations in different anatomical compartments. These findings have important implications in transmission of HBV.
Compartmentalization: Compartmentalization is the process of compartment specific infection, evolution and persistence of viral variants (genotypes/subgenotypes) in different anatomically distinct sites. Due to difference in the immune selection pressure, viral variants with alterations advantageous under the given immune pressure are gradually selected, leading to their divergence from the circulating strains. Compartmentalization has been well studied in human immunodeficiency virus, hepatitis C virus, Epstein-Barr virus, etc. in comparison, studies on HBV compartmentalization are scanty and the mechanisms of emergence of mutations is poorly understood.
This study has shown that HBV subtype A1 and A2 have entirely different pgRNA secondary structure, which may explain compartment specific selection and preponderance of specific HBV subgenotype with unique mutational pattern. This study has novel findings.
| 1. | Datta S. Compartmentalization of hepatitis B virus: Looking beyond the liver. World J Hepatol. 2015;7:2241-2244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 2. | Datta S, Panigrahi R, Biswas A, Chandra PK, Banerjee A, Mahapatra PK, Panda CK, Chakrabarti S, Bhattacharya SK, Biswas K. Genetic characterization of hepatitis B virus in peripheral blood leukocytes: evidence for selection and compartmentalization of viral variants with the immune escape G145R mutation. J Virol. 2009;83:9983-9992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 3. | Bednar MM, Sturdevant CB, Tompkins LA, Arrildt KT, Dukhovlinova E, Kincer LP, Swanstrom R. Compartmentalization, Viral Evolution, and Viral Latency of HIV in the CNS. Curr HIV/AIDS Rep. 2015;12:262-271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 4. | Ene L, Duiculescu D, Tardei G, Ruta S, Smith DM, Mehta S, Letendre S, Achim CL. Hepatitis B virus compartmentalization in the cerebrospinal fluid of HIV-infected patients. Clin Microbiol Infect. 2015;21:387.e5-387.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 5. | Ramirez S, Perez-Del-Pulgar S, Carrion JA, Costa J, Gonzalez P, Massaguer A, Fondevila C, Garcia-Valdecasas JC, Navasa M, Forns X. Hepatitis C virus compartmentalization and infection recurrence after liver transplantation. Am J Transplant. 2009;9:1591-1601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 6. | Gismondi MI, Díaz Carrasco JM, Valva P, Becker PD, Guzmán CA, Campos RH, Preciado MV. Dynamic changes in viral population structure and compartmentalization during chronic hepatitis C virus infection in children. Virology. 2013;447:187-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 7. | Blackard JT. HIV compartmentalization: a review on a clinically important phenomenon. Curr HIV Res. 2012;10:133-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 8. | Datta S, Chatterjee S, Veer V, Chakravarty R. Molecular biology of the hepatitis B virus for clinicians. J Clin Exp Hepatol. 2012;2:353-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 9. | Féray C, Zignego AL, Samuel D, Bismuth A, Reynes M, Tiollais P, Bismuth H, Brechot C. Persistent hepatitis B virus infection of mononuclear blood cells without concomitant liver infection. The liver transplantation model. Transplantation. 1990;49:1155-1158. [PubMed] |
| 10. | Brind A, Jiang J, Samuel D, Gigou M, Feray C, Bréchot C, Kremsdorf D. Evidence for selection of hepatitis B mutants after liver transplantation through peripheral blood mononuclear cell infection. J Hepatol. 1997;26:228-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 80] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 11. | Coffin CS, Osiowy C, Gao S, Nishikawa S, van der Meer F, van Marle G. Hepatitis B virus (HBV) variants fluctuate in paired plasma and peripheral blood mononuclear cells among patient cohorts during different chronic hepatitis B (CHB) disease phases. J Viral Hepat. 2015;22:416-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 12. | Datta S. An overview of molecular epidemiology of hepatitis B virus (HBV) in India. Virol J. 2008;5:156. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 91] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 13. | Banerjee A, Chandra PK, Datta S, Biswas A, Bhattacharya P, Chakraborty S, Chakrabarti S, Bhattacharya SK, Chakravarty R. Frequency and significance of hepatitis B virus surface gene variant circulating among ‘antiHBc only’ individuals in Eastern India. J Clin Virol. 2007;40:312-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 37] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 14. | Banerjee A, Kurbanov F, Datta S, Chandra PK, Tanaka Y, Mizokami M, Chakravarty R. Phylogenetic relatedness and genetic diversity of hepatitis B virus isolates in Eastern India. J Med Virol. 2006;78:1164-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 77] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 15. | Kramvis A. Genotypes and genetic variability of hepatitis B virus. Intervirology. 2014;57:141-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 265] [Cited by in RCA: 338] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 16. | Kidd-Ljunggren K, Zuker M, Hofacker IL, Kidd AH. The hepatitis B virus pregenome: prediction of RNA structure and implications for the emergence of deletions. Intervirology. 2000;43:154-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 17. | Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA websuite. Nucleic Acids Res. 2008;36:W70-W74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1446] [Cited by in RCA: 1976] [Article Influence: 109.8] [Reference Citation Analysis (0)] |
| 18. | Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429-3431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1701] [Cited by in RCA: 1652] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 19. | Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2379] [Cited by in RCA: 2494] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 20. | McCaskill JS. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105-1119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 897] [Cited by in RCA: 840] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 21. | Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157-1166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 246] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 22. | Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911-940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2839] [Cited by in RCA: 2811] [Article Influence: 104.1] [Reference Citation Analysis (0)] |
| 23. | Steffen P, Voss B, Rehmsmeier M, Reeder J, Giegerich R. RNAshapes: an integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006;22:500-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 222] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 24. | Mangold CM, Streeck RE. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J Virol. 1993;67:4588-4597. [PubMed] |
| 25. | Caligiuri P, Cerruti R, Icardi G, Bruzzone B. Overview of hepatitis B virus mutations and their implications in the management of infection. World J Gastroenterol. 2016;22:145-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 102] [Cited by in RCA: 112] [Article Influence: 11.2] [Reference Citation Analysis (2)] |
| 26. | Vieira VC, Soares MA. The role of cytidine deaminases on innate immune responses against human viral infections. Biomed Res Int. 2013;2013:683095. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 97] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 27. | Mous K, Jennes W, De Roo A, Pintelon I, Kestens L, Van Ostade X. Intracellular detection of differential APOBEC3G, TRIM5alpha, and LEDGF/p75 protein expression in peripheral blood by flow cytometry. J Immunol Methods. 2011;372:52-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 28. | Janahi EM, McGarvey MJ. The inhibition of hepatitis B virus by APOBEC cytidine deaminases. J Viral Hepat. 2013;20:821-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 29. | Jammeh S, Thomas HC, Karayiannis P. Replicative competence of the T131I, K141E, and G145R surface variants of hepatitis B Virus. J Infect Dis. 2007;196:1010-1013. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 30. | Schilling R, Ijaz S, Davidoff M, Lee JY, Locarnini S, Williams R, Naoumov NV. Endocytosis of hepatitis B immune globulin into hepatocytes inhibits the secretion of hepatitis B virus surface antigen and virions. J Virol. 2003;77:8882-8892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 79] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 31. | Croagh CM, Desmond PV, Bell SJ. Genotypes and viral variants in chronic hepatitis B: A review of epidemiology and clinical relevance. World J Hepatol. 2015;7:289-303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 103] [Article Influence: 9.4] [Reference Citation Analysis (1)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Manuscript source: Invited manuscript
Specialty type: Virology
Country of origin: India
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Chung YH, Shimizu Y, Toyoda T S- Editor: Ji FF L- Editor: A E- Editor: Wu HL