Peer-review started: May 9, 2015
First decision: June 18, 2015
Revised: November 17, 2015
Accepted: December 1, 2015
Article in press: December 2, 2015
Published online: February 12, 2016
Processing time: 269 Days and 3.7 Hours
AIM: To test the pathogenicity of pseudorabies virus (PRV) variant HN1201 and compare its pathogenicity with a classical PRV Fa strain.
METHODS: The pathogenicity of the newly-emerging PRV variant HN1201 was evaluated by different inoculating routes, virus loads, and ages of pigs. The classical PRV Fa strain was then used to compare with HN1201 to determine pathogenicity. Clinical symptoms after virus infection were recorded daily and average daily body weight was used to measure the growth performance of pigs. At necropsy, gross pathology and histopathology were used to evaluate the severity of tissue damage caused by virus infection.
RESULTS: The results showed that the efficient infection method of RPV HN1201 was via intranasal inoculation at 107 TCID50, and that the virus has high pathogenicity to 35- to 127-d old pigs. Compared with Fa strain, pigs infected with HN1201 showed more severe clinical symptoms and pathological lesions. Immunochemistry results revealed HN1201 had more abundant antigen distribution in extensive organs.
CONCLUSION: All of the above results suggest that PRV variant HN1201 was more pathogenic to pigs than the classical Fa strain.
Core tip: Pseudorabies virus (PRV) variant HN1201 has pathogenicity in 35 to 127-d old pigs via intranasal inoculation at 107 TCID50. Intranasal inoculation is more efficient than intramuscular inoculation for PRV challenge. PRV variant HN1201 showed higher pathogenic ability, as shown by the more severe clinical symptoms, pathological lesions, and abundant antigen distribution in extensive organs than the classical PRV Fa strain.
- Citation: Yang QY, Sun Z, Tan FF, Guo LH, Wang YZ, Wang J, Wang ZY, Wang LL, Li XD, Xiao Y, Tian KG. Pathogenicity of a currently circulating Chinese variant pseudorabies virus in pigs. World J Virol 2016; 5(1): 23-30
- URL: https://www.wjgnet.com/2220-3249/full/v5/i1/23.htm
- DOI: https://dx.doi.org/10.5501/wjv.v5.i1.23
Pseudorabies virus (PRV), also known as Aujeszky’s disease virus or Suid herpesvirus type 1 (SuHV-1), is the causative agent of pseudorabies (PR). Belonging to the family Herpesviridae, subfamily Alphaherpesvirinae, and genus Varicellovirus, the virus causes substantial economic losses in the pig industry worldwide[1-3]. The PRV genome is a double-stranded linear DNA which is about 143 kb in size and has about 70 ORFs[4,5]. This pathogen can infect numerous mammals, including carnivores, ruminants, and rodents, yet pigs are the only natural host for PRV as the reservoir of the virus[6,7]. PRV infection is characterized by neurologic symptoms and death in newborn piglets, respiratory disorders in elder pigs, and reproductive failure like stillbirths and abortions in sows. Like other alphaherpesviruses, PRV can establish a lifelong latent infection in the peripheral nervous system of infected pigs. Latently infected pigs can be recognized as a source of reinfection when the latent viral genome reactivates spontaneously and the infectious virus is developed[8].
Attenuated live or killed PRV vaccines have played a critical role in the control and eradication of PR. Bartha-K61, a vaccine imported from Hungary, have been widely used in China since the 1970s, and was reported to provide complete protection from field virus infection[2]. Nevertheless, since October 2011, severe PRV outbreaks have occurred on pig farms and spread rapidly to the northern parts of China[9,10]. Most of the infected farms had used Bartha-K61 vaccine according to the manufacturer’s instructions, and the serum samples obtained from the infected pigs had a considerable positive rate of gE Ab detected by ELISA (IDEXX Laboratories, Westbrook, United States)[10,11]. The affected pigs presented with multiple clinical signs, including high fever (usually ≥ 40.5 °C), depression, anorexia, respiratory distress, shivering, and systemic neurological symptoms[11,12]. Pathologic examination of viscera samples collected from dead pigs from different provinces displayed consolidation, edema, and hemorrhage in the lungs, as well as necrosis in the kidneys, indicating that newly-emerging PRV variants may have higher pathogenicity than the classical strains[13]. The PRV infection in vaccinated pig herds indicates that the traditional Bartha-K61 vaccine could not provide complete protection to the current prevalent PRV variants in China[11,14]. Accordingly, it is imperative to study the pathogenicity of the currently circulating PRV variant strains and develop newly effective vaccines to tackle the problem.
In this study, we first established a PRV variant HN1201 infection model in pigs according to different inoculation routes, virus loads, and pig ages. The characterized PRV variant HN1201 was then compared with the virulent classical PRV strain Fa to determine pathogenicity.
The PRV variant HN1201 was previously isolated from the brain of infected pigs in Henan province[12]. Briefly, the infected pig brain sample was homogenized and the supernatant of homogenization was subjected to 0.22 μm filtration. The filtrated supernatant was inoculated on a PK-15 cell monolayer until the appearance of CPE after 3 d. The virus was harvested after two cycles of freeze-thaw and store at -80 °C until use. The classical PRV Fa was purchased from the Institute of China Veterinary Medicine Inspection[15]. Permissive PK-15 cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 5% fetal bovine sera.
To establish a PRV HN1201 infection model in pigs, in the first animal experiment, twenty 60-d old pigs, five 35-d old pigs, and five 127-d old pigs were used to evaluate the pathogenicity of the virus by different inoculating routes, virus loads, and ages of pigs. Twenty 60-d old pigs were randomly allocated into the first four groups (Table 1). Pigs in group 1 and group 2 were inoculated with 107 TCID50 PRV HN1201 strain via intramuscular (im) and intranasal (in) routes, respectively. Pigs in group 3 and 4 were inoculated via the intranasal route with 106 TCID50 and 105 TCID50 of PRV HN1201 strain, respectively. To test the susceptibility of pigs to the virus at different ages, five 35-d old pigs in group 5 and five 127-d old pigs in group 6 were inoculated via the intranasal route with 107 TCID50 PRV HN1201 (Table 1).
| Group | Pig age (d) | Pig No. | Virus titer | Route | Euthanized |
| 1 | 60 | 5 | 107 | im | 3/5 |
| 2 | 60 | 5 | 107 | in | 5/5 |
| 3 | 60 | 5 | 106 | in | 2/5 |
| 4 | 60 | 5 | 105 | in | 1/5 |
| 5 | 35 | 5 | 107 | in | 5/5 |
| 6 | 127 | 5 | 107 | in | 5/5 |
In the second animal study, ten 56-d old pigs were randomly divided into two groups with five pigs in each group. Pigs in group I were inoculated with 107 TCID50 PRV HN1201 via the intranasal route and group II were inoculated with classical PRV Fa strain with the same dose and route.
All pigs used in the above two animal trials were free of PRV and excluded by using gB- and gE-ELISA Kits (HerdChek PRV, IDEXX, United States) and PCR method. All pigs were also free of porcine reproductive and respiratory syndrome virus, classical swine fever virus, and porcine circovirus 2. Experimental pigs in different groups were insulated in separate rooms throughout the study. After virus inoculation, rectal temperature and clinical signs were recorded on a daily basis. At 14 d post-inoculation (dpi), all surviving pigs were humanely euthanized and necropsied, and different organ samples were collected. The collected samples were subjected to pathological examination and gently inflated with 10% neutral-buffered formalin for immunohistochemistry examination. All animal trials in this study were approved by the Animal Care and Ethics Committee of the China National Research Center for Veterinary Medicine.
Representative samples were cut from the fixed tissues and processed into paraffin blocks. Sections approximately 3-4 μm thick were cut into slides. Duplicates of the same sections were used for hematoxylin and eosin (H and E) staining and immunohistochemistry staining separately, as previously described[16]. The H and E staining was operated automatically by Leica fully automatic dyeing machine according to standard procedures. Immunohistochemistry staining was performed as below. The prepared paraffin sections were mounted on APES-treated slides and incubated overnight at 37 °C. The slides were de-waxed via routine method by Leica automatic dyeing machine. The samples were blocked with 3% peroxide-methanol for 20 min at room temperature for endogenous peroxidase ablation and rinsed by phosphate buffer solution (PBS) twice. The following steps were carried out in a moisture chamber: (1) Samples were incubated with blocking buffer containing normal horse serum (Beijing Zhongshan Jinqiao, China) with 1:20 dilution with PBS at 37 °C for 20 min; (2) The horse serum was discarded and samples were incubated in PRV monoclonal antibody 3B5 solution (Beijing Tian Tech Biotechnology, China) with 1:800 dilution in PBS (pH 7.3) at 37 °C for half an hour and then 4 °C overnight; (3) After rinsing with PBS three times, HRP goat anti-mouse IgG (BTI, United States) with 1:100 dilution in PBS (pH 7.3) was added, and the slides were incubated for 1 h at 37 °C; (4) After rinsing with PBS three times, the slides were incubated with AEC and kept at room temperature without light for 5-10 min; (5) After rinsing with PBS three times, the slides were stained with hematoxylin (freshly prepared) 1:10 dilution for 10 s; (6) The unbound hematoxylin was washed away by running water, and the slides were placed into water for 2 min; and (7) The slides were allowed to dry naturally and then mounted with water-soluble tablet seal before visualization by 200 × microscope photographs. The results were determined by negative (-) and positive (+), with positive signals interpreted as low (+), moderate (++), and intense (+++), according to the intensity of staining.
The animal protocol was designed to minimize pain or discomfort to the animals. The animals were acclimatized to laboratory conditions (23 °C, 12 h light/12 h dark, 50% humidity, and ad libitum access to food and water) for two weeks prior to experimentation. All animals were euthanized by barbiturate overdose (intravenous injection, 150 mg/kg pentobarbital sodium) for tissue collection. All procedures involving animals were reviewed and approved by the Institutional Animal Care and Use Committee of the National Research Center for Veterinary Medicine (IACUC protocol number: 2015010402).
Differences of body temperature and body weight between two infected groups in the second animal trial were determined by using t-test in GraphPad Prism 5.0 Software (San Diego, CA). Differences were considered statistically significant when P < 0.05.
For routes of infection, all pigs in group 1 and group 2 inoculated with 107 TCID50 PRV HN1201 strain via intramuscular and intranasal routes, respectively, showed PRV-specific clinical symptoms such as fever (40.0 °C-41.5 °C), respiratory distress, excessive salvation, and neurological signs including convulsion and ataxia. All pigs in group 2 were euthanized due to moribund conditions from 5 to 7 dpi. Compared with group 2, three pigs in group 1 were euthanized from 5 to 7 dpi and the other two pigs survived until the end of the study (terminated at 14 dpi, Table 1).
All pigs in group 3 (106.0 TCID50) showed severe respiratory symptoms and neurological signs, as described above, with two being euthanized 6 dpi. Compared to pigs in group 3, respiratory symptoms such as coughing and shivering were more often observed in group 4 (105.0 TCID50). There was one pig out of the five in group 4 that showed neurological signs, and was euthanized by the end of study (terminated at 14 dpi).
Young piglets are more susceptible to PRV infection than elder pigs[6]. To determine the pathogenicity of PRV HN1201 in pigs of different ages, 35, 60, and 127-d old pigs were inoculated with 107 TCID50 of virus. After virus inoculation, pigs of different ages showed the clinical symptoms as described above. All pigs in group 5 (35-d old pigs) were euthanized from day 4 to day 6 and all pigs in group 6 (127-d old pigs) were euthanized from day 5 to day 8 due to the moribund conditions. Therefore, unlike the classical PRV strains, this PRV variant strain showed high pathogenicity in pigs of different ages.
Since the above results showed PRV variant HN1201 has high pathogenicity in pigs of different ages, a classical PRV Fa strain was used to compare pathogenicity. To exclude the bias of pathogenicity of two PRV strains due to the age of experimental pigs, ten 56-d-old healthy pigs were randomly assigned to two groups, with five pigs in each group. Pigs in groups I and II were inoculated with PRV HN1201 and Fa strain, respectively, via intranasal method at 107 TCID50.
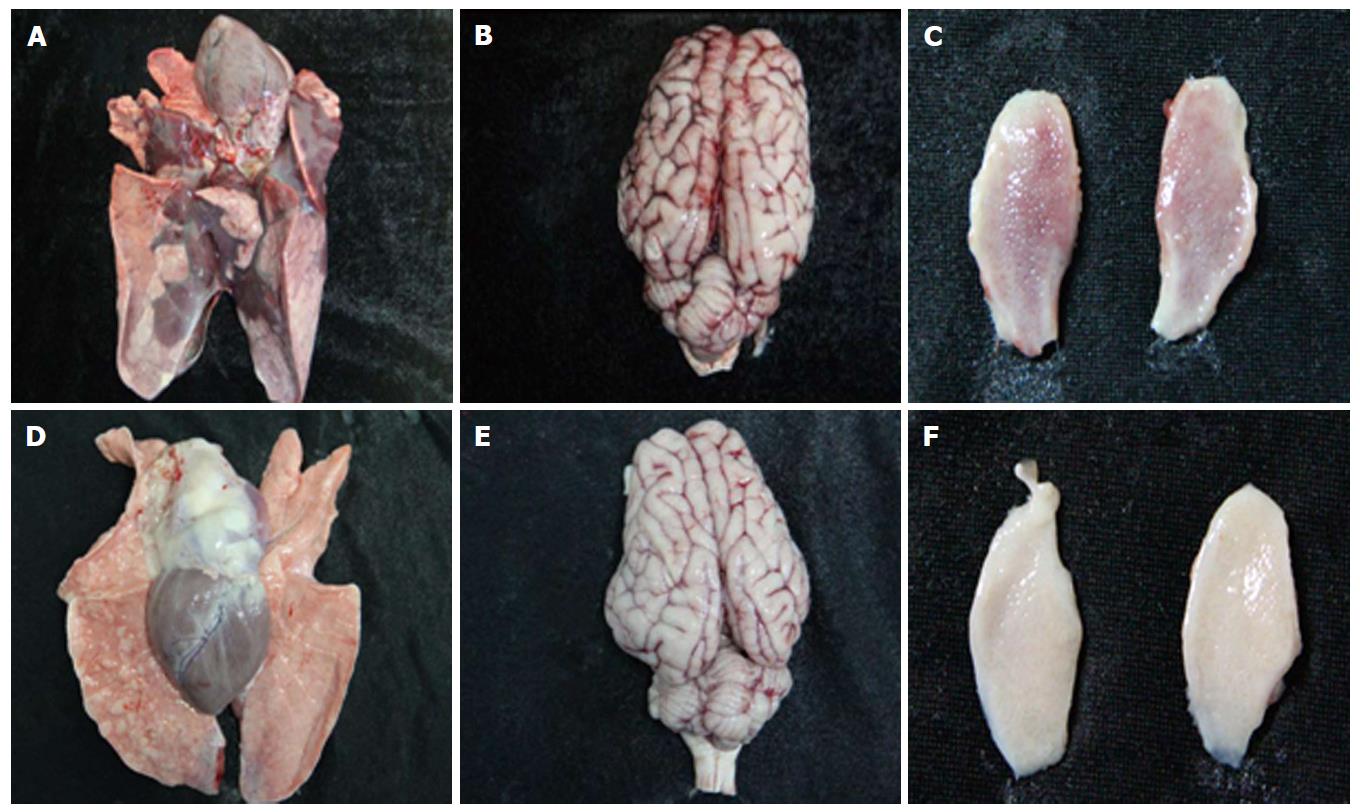
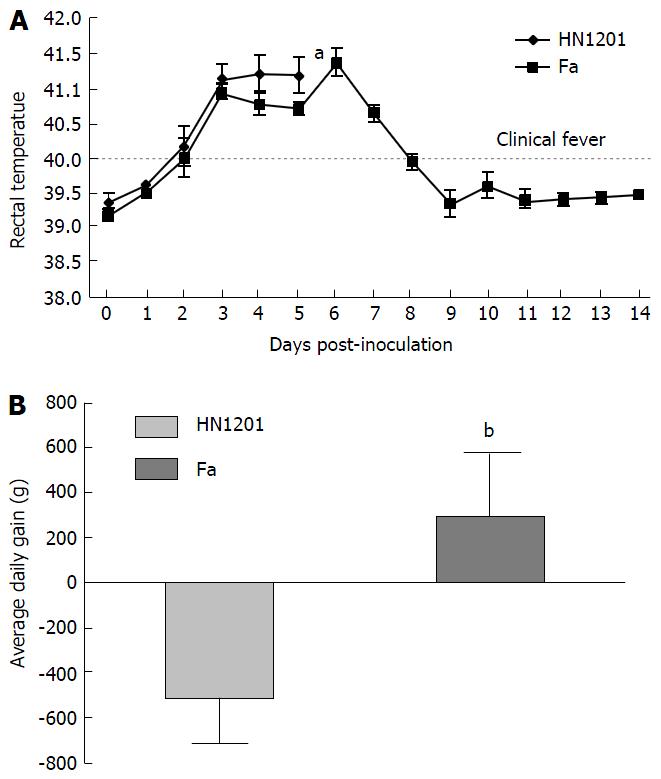
As expected, all pigs in groupI displayed high fever, anorexia, depression, respiratory symptoms, and neurological signs as described in the first animal study. In contrast, four pigs in group II had no respiratory or neurological symptoms, aside from sneezing (Table 2), while only one pig showed the same clinical signs as pigs in groupI. Gross pathology examination at necropsy showed that PRV HN1201 infection led to severe pulmonary consolidation and necrosis in the lung (Figure 1A), encephalic hemorrhage in the brain (Figure 1B), and hemorrhage and necrosis in the tonsil (Figure 1C). By contrast, pigs infected with PRV Fa showed only slight hemorrhage in the lung tissue (Figure 1D) and had no obvious changes in the brain or tonsil (Figure 1E and F). No other obvious pathologic change was found after two virus infection in heart, liver, spleen, and kidney tissues. There was no significant difference in rectal temperature between the two groups in the first 5 d of study (Figure 2A). Pigs in groupI had significant body weight losses compared to pigs in group II at 6 dpi (Figure 2B). At 5 dpi, two pigs were euthanized in group I and one pig was euthanized in group II. At 6 dpi, another three pigs were euthanized in group I and all remaining pigs in group II survived to the end of the study.
| Groups | Respiratory symptom | Neurological symptom | |||||||
| Vomit | Respiratory distress | Cough | Sneeze | Salivation | Circling | Posterior paralysis | Muscle tremors | Lay recumbent and paddle | |
| + | + | - | + | - | + | - | + | - | |
| I (HN1201) | - | + | + | + | - | + | - | + | - |
| + | + | + | + | - | - | + | + | + | |
| - | + | + | + | + | + | + | + | + | |
| + | + | + | + | + | + | + | + | + | |
| - | - | - | + | - | - | - | - | - | |
| II (Fa strain) | - | - | - | + | - | - | - | - | - |
| - | - | - | + | - | - | - | - | - | |
| - | - | - | + | - | - | - | - | - | |
| + | + | - | + | + | + | - | + | + | |
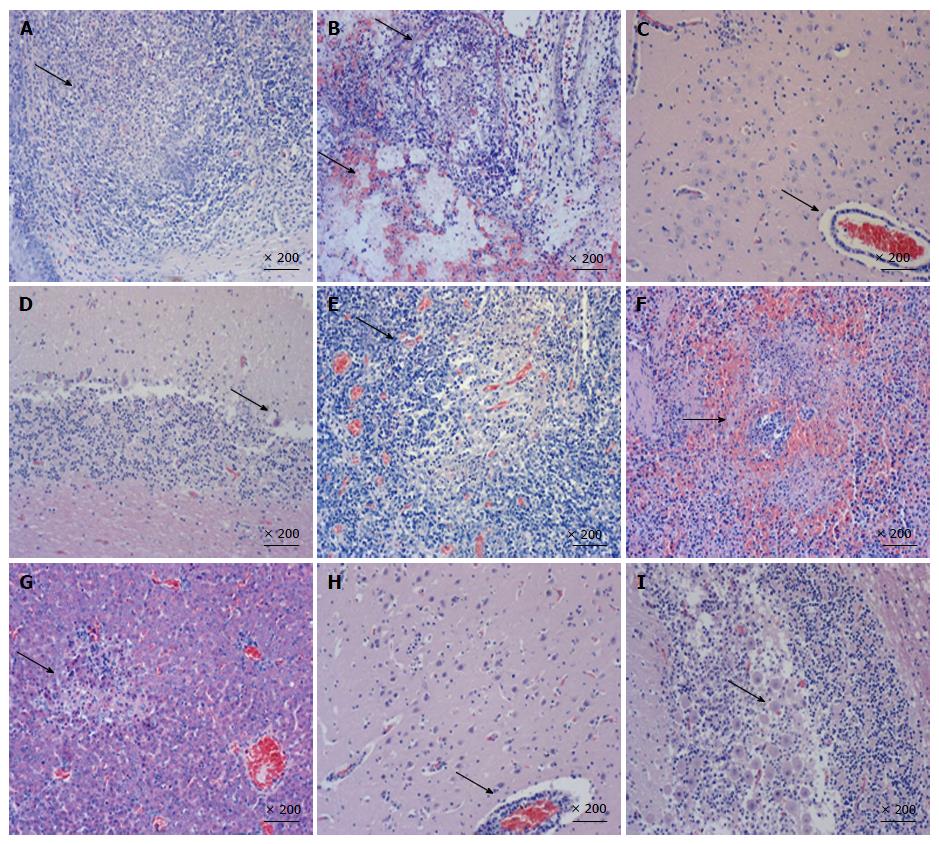
Organ samples of pig tonsil, lung, cerebellum, lymph nodes, kidney, and liver were collected for histological examination and immunohistochemistry staining. Typical PRV infection is characterized by necrosis in multiple organs. As shown in Figure 3, necrosis, congestion, or hemorrhage in all above organs of PRV HN1201-infected pigs were observed after H&E staining (Figures 3A-G), with neuronal intra-nuclear inclusions also being observed in the brain. Compared to the HN1201 infection, PRV Fa-infected pigs only showed neuronal degeneration, necrosis in the brain, Purkinje cell degeneration, and necrosis in the cerebellum (Figure 3H and I). In accordance with histopathology results, immunohistochemistry staining showed significant strong positive signals in all of the above organs obtained from pigs infected with HN1201 virus, whereas only brain and cerebellum samples of one RPV Fa-infected pig revealed positive results (Table 3).
| Groups | Tonsil | Lung | Lymph nodes | Brain | Cerebellum | Spleen | Liver | ||
| Mandibular | Superficial inguinal | Mesenteric | |||||||
| HN1201 | 3+ | 3+ | 3+ | 2+ | 2+ | 2+ | 1+ | 2+ | 2+ |
| 3+ | 2+ | 2+ | 2+ | 2+ | 2+ | 1+ | 3+ | - | |
| 3+ | 3+ | 3+ | 2+ | 2+ | 1+ | 2+ | 3+ | + | |
| 3+ | 3+ | ++ | 3+ | 2+ | 1+ | 2+ | 2+ | 2+ | |
| 3+ | 3+ | 3+ | 2+ | 3+ | 2+ | 2+ | 3+ | 3+ | |
| Fa strain | - | + | - | - | - | - | - | - | - |
| - | - | - | - | - | 2+ | 2+ | - | - | |
| - | - | - | - | - | 2+ | 2+ | - | - | |
| - | - | - | - | - | 2+ | 2+ | - | - | |
| - | - | - | - | - | 2+ | 2+ | - | - | |
Since late 2011, outbreaks of PR-like diseases have occurred on numerous Bartha-K61-vaccinated pig farms and gradually spread in China, causing huge economic losses to the Chinese swine industry[10,13]. Recent studies have shown that PRV variants contributed to the recent outbreaks of PR, and the traditional Bartha-K61 vaccine could not provide complete protection against the emerging PRV strains[11,14]. Similar to classical PR, the disease is characterized by the sudden death of new born piglets, respiratory and neurological symptoms in growing pigs, and stillbirth or the birth of weak piglets from sows. However, the pathogenicity of the new emerging PRV variant was never delineated and compared with classical PRV strains. Therefore, it is necessary to determine the pathogenicity of the current PRV variants before any control measures are implemented to control the disease.
PRV is tropic for both the respiratory and nervous systems of swine. Viral particles enter sensory nerve endings, thereby innervating the infected mucosal epithelium. Morbidity and mortality associated with PRV infection varies with host age, the animal’s overall health status, and infectious dose[2]. In this study, we first tested the pathogenicity of PRV variant HN1201 by different routes of virus infection, virus loads for inoculation, and pig ages. Our results showed that intranasal infection is more effective than intramuscular infection when 107 TCID50 viruses were used for inoculation. Pigs infected with PRV 1201 by the intranasal route showed more severe clinical symptoms and higher mortality rates than those with intramuscular routes, and virus loads were positively correlated with mortality rates. The pathogenicity of some other PRV variant strains have been studied recently[17]. In a study by Luo et al[17] (2014), pigs infected with the 106 TCID50 PRV TJ strain by the intranasal route showed higher mortality than those with a lower dose or were infected by the intramuscular route, which is consistent with our results. Bama miniature pigs injected intramuscularly with 107.0 TCID50 PRV strain HeN1, another virulent PRV variant, the animals exhibited only transient fever for 3-5 d, and no other clinical symptoms or postmortem changes were observed[10]. Differences in pathogenicity and mortality caused by different PRV viruses could be explained by the virus load for inoculation, viral strain, and breed of pigs, although these three viruses also share a more than 99.0% similarity in their whole genome sequences. Besides routes of inoculation and virus load, PRV HN1201 could infect pigs from 35 to 127 d old with PRV-specific clinical symptoms, indicating that the PRV HN1201 strain is highly pathogenic to pigs.
To compare the pathogenicity of newly-emerging PRV variants with the classical PRV strain, PRV HN1201 and Fa strains were used to infect pigs. Our results showed that HN1201-infected pigs showed more severe clinical signs and higher mortality rates than Fa-infected pigs (5/5 vs 1/5). Pigs in the PRV HN1201-infected group displayed high fever, anorexia, depression, respiratory symptoms, and neurological signs. In comparison, four pigs in the PRV Fa-infected group had no respiratory or neurological symptoms, aside from sneezing. Meanwhile, pigs infected with HN1201 had steady body weight loss as compared with pigs infected with the Fa strain (Figure 2B). Retarded growth was more often observed in young piglets after PRV infection. However, the loss of body weight of 56-d old pigs after PRV infection was seldom observed, which proves the high virulence of PRV HN1201.
Gross pathological examination at necropsy revealed more severe damage to the lung, tonsil, brain, cerebellum, and lymph nodes in pigs infected with HN1201 strain than in the Fa strain group. In line with pathological results, histopathology examination showed remarkably obvious necrosis in multiple tissues, such as the tonsil, lung, brain, spleen, and liver in HN1201-infected pigs; in contrast, necrosis caused by PRV Fa infection was only limited to the brain and cerebellum. Immunochemistry results also showed that PRV HN1201 infection lead to more extensive virus antigen distribution in different organs with more intense staining, while Fa infection only had one cerebellum sample from one pig that showed positive. Previous studies reported that inoculation of PRV through the nasal cavity resulted in virally-induced neuropathological lesions[2]. The kinetics and locations of lesion appearance were consistent with a transneuronal spread of PRV from the nasal epithelium to synaptically-connected higher-order structures in the nervous system. The intense PRV antigen location and severe lesions of the brain, tonsil, and lung coincided with the typical respiratory and neurological symptoms, and may be due to intranasal infection. Therefore, the above results further suggest the higher pathogenicity of PRV HN1201 when compared to the classical Fa strain.
In conclusion, PRV HN1201 infection is more effective through the intranasal route than the intramuscular inoculation route, and the virus is highly pathogenic to different ages of pig. Compared with classical PRV Fa strain, HN1201 causes more severe clinical symptoms and pathological lesions, with extensive antigen distribution in different organs.
Highly virulent pseudorabies virus (PRV) variants are circulating in most Chinese pig farms, causing huge economic losses. The pathogenicity of these PRV variants have not been previously compared with classical PRV strains.
The authors aimed to test the pathogenicity of a newly-emerging PRV variant in pigs of different inoculation routes, virus loads, and ages. Differences in pathogenicity between the newly-emerging PRV variant and the classical PRV strain were also compared.
This study demonstrates that the currently-circulating PRV HN1201 variant has higher pathogenicity in pigs than the classical PRV Fa strain via the manifestation of more severe clinical symptoms and pathological lesions, with extensive antigen distribution in different organs.
The authors proved the PRV variant to be more pathogenic in pigs as compared to the classical Fa strain, which may partially explain the inefficacy of current commercial PRV vaccines. Thus, a better understanding of the differences of pathogenicity between variant and classical PRV may facilitate the development of more effective vaccines.
Pathogenicity of pseudorabies virus is the potential capacity of PRV to cause PR-like syndrome in pigs. Pathogenicity of viruses may change due to virus mutation and/or recombination. Study into the pathogenesis of currently-circulating field viruses may provide first-hand data for disease control.
This manuscript reports the analysis of the pathogenicity of a new PRV variant that the commonly-used vaccine cannot protect against, and is therefore causing massive economic losses in China. The pathogenicity of this variant and the classical PRV Fa stain is also compared. The experiment design and results were clear and convincing. It will be interesting to see if the authors can further explore the mechanisms of the enhanced pathogenicity of the PRV variant behind these phenomena.
| 1. | Mettenleiter TC. Pseudorabies (Aujeszky’s disease) virus: state of the art. August 1993. Acta Vet Hung. 1994;42:153-177. [PubMed] |
| 2. | Pomeranz LE, Reynolds AE, Hengartner CJ. Molecular biology of pseudorabies virus: impact on neurovirology and veterinary medicine. Microbiol Mol Biol Rev. 2005;69:462-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 517] [Cited by in RCA: 685] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 3. | Roizman B, Baines J. The diversity and unity of Herpesviridae. Comp Immunol Microbiol Infect Dis. 1991;14:63-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 4. | Klupp BG, Hengartner CJ, Mettenleiter TC, Enquist LW. Complete, annotated sequence of the pseudorabies virus genome. J Virol. 2004;78:424-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 257] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 5. | Szpara ML, Tafuri YR, Parsons L, Shamim SR, Verstrepen KJ, Legendre M, Enquist LW. A wide extent of inter-strain diversity in virulent and vaccine strains of alphaherpesviruses. PLoS Pathog. 2011;7:e1002282. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 128] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 6. | Mulder WA, Pol JM, Gruys E, Jacobs L, De Jong MC, Peeters BP, Kimman TG. Pseudorabies virus infections in pigs. Role of viral proteins in virulence, pathogenesis and transmission. Vet Res. 1997;28:1-17. [PubMed] |
| 7. | Müller T, Hahn EC, Tottewitz F, Kramer M, Klupp BG, Mettenleiter TC, Freuling C. Pseudorabies virus in wild swine: a global perspective. Arch Virol. 2011;156:1691-1705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 229] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 8. | Rziha HJ, Mettenleiter TC, Ohlinger V, Wittmann G. Herpesvirus (pseudorabies virus) latency in swine: occurrence and physical state of viral DNA in neural tissues. Virology. 1986;155:600-613. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 9. | Wang TY, Xiao Y, Yang QY, Wang YZ, Sun Z, Zhang CL, Yan SJ, Wang J, Guo LH, Yan H. Construction of a gE-deleted pseudorabies virus and its efficacy to the new-emerging variant PRV challenge in the form of killed vaccine. Biomed Int Res. 2015;In press. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 10. | An TQ, Peng JM, Tian ZJ, Zhao HY, Li N, Liu YM, Chen JZ, Leng CL, Sun Y, Chang D. Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg Infect Dis. 2013;19:1749-1755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 299] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 11. | Gu Z, Dong J, Wang J, Hou C, Sun H, Yang W, Bai J, Jiang P. A novel inactivated gE/gI deleted pseudorabies virus (PRV) vaccine completely protects pigs from an emerged variant PRV challenge. Virus Res. 2015;195:57-63. [PubMed] |
| 12. | Wu R, Bai C, Sun J, Chang S, Zhang X. Emergence of virulent pseudorabies virus infection in northern China. J Vet Sci. 2013;14:363-365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 13. | Yu X, Zhou Z, Hu D, Zhang Q, Han T, Li X, Gu X, Yuan L, Zhang S, Wang B. Pathogenic pseudorabies virus, China, 2012. Emerg Infect Dis. 2014;20:102-104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 180] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 14. | Wang CH, Yuan J, Qin HY, Luo Y, Cong X, Li Y, Chen J, Li S, Sun Y, Qiu HJ. A novel gE-deleted pseudorabies virus (PRV) provides rapid and complete protection from lethal challenge with the PRV variant emerging in Bartha-K61-vaccinated swine population in China. Vaccine. 2014;32:3379-3385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 114] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 15. | Zhu L, Yi Y, Xu Z, Cheng L, Tang S, Guo W. Growth, physicochemical properties, and morphogenesis of Chinese wild-type PRV Fa and its gene-deleted mutant strain PRV SA215. Virol J. 2011;8:272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 16. | Zhang C, Guo L, Jia X, Wang T, Wang J, Sun Z, Wang L, Li X, Tan F, Tian K. Construction of a triple gene-deleted Chinese Pseudorabies virus variant and its efficacy study as a vaccine candidate on suckling piglets. Vaccine. 2015;33:2432-2437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 17. | Luo Y, Li N, Cong X, Wang CH, Du M, Li L, Zhao B, Yuan J, Liu DD, Li S. Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Vet Microbiol. 2014;174:107-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 167] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
P- Reviewer: Choi TG, Ghiringhelli PD, Hua XG, Shih L S- Editor: Song XX L- Editor: Rutherford A E- Editor: Li D