Published online Dec 25, 2023. doi: 10.5501/wjv.v12.i5.296
Peer-review started: September 11, 2023
First decision: October 9, 2023
Revised: October 19, 2023
Accepted: November 30, 2023
Article in press: November 30, 2023
Published online: December 25, 2023
Processing time: 105 Days and 2.9 Hours
Chronic hepatitis B virus (HBV) infection is often associated with increased lipid deposition in hepatocytes. However, when combined with non-alcoholic fatty liver disease or hyperlipidemia, it tends to have a lower HBV deoxyribonucleic acid (DNA) load. The relationship between lipid metabolism and HBV DNA replication and its underlying mechanisms are not well understood.
To investigate the relationship between lipid metabolism and HBV DNA repli
1603 HBsAg-seropositive patients were included in the study. We first explored the relationship between patients' lipid levels, hepatic steatosis, and HBV DNA load. Also, we constructed an HBV infection combined with a hepatic steatosis cell model in vitro by fatty acid stimulation of HepG2.2.15 cells to validate the effect of lipid metabolism on HBV DNA replication in vitro. By knocking down and overexpressing Plin2, we observed whether Plin2 regulates autophagy and HBV replication. By inhibiting both Plin2 and cellular autophagy under high lipid stimulation, we examined whether the Plin2-autophagy pathway regulates HBV replication.
The results revealed that serum triglyceride levels, high-density lipoprotein levels, and hepatic steatosis ratio were significantly lower in the HBV-DNA high load group. Logistic regression analysis indicated that hepatic steatosis and serum triglyceride levels were negatively correlated with HBV-DNA load. Stratified analysis by HBeAg showed significant negative correlations between HBV-DNA load and hepatic steatosis ratio in both HBeAg-positive and HBeAg-negative groups. An in vitro cell model was developed by stimulating HepG2.2.15 cells with palmitic acid and oleic acid to study the relationship between HBV-DNA load and lipid metabolism. The results of the in vitro experiments suggested that fatty acid treatment increased lipid droplet deposition and decreased the expression of cell supernatant HBsAg, HBeAg, and HBV DNA load. Western blot and polymerase chain reaction analysis showed that fatty acid stimulation significantly induced Plin2 protein expression and inhibited the expression of hepatocyte autophagy proteins. Inhibition of Plin2 protein expression under fatty acid stimulation reversed the reduction in HBsAg and HBeAg expression and HBV DNA load induced by fatty acid stimulation and the inhibition of cellular autophagy. Knocking down Plin2 and blocking autophagy with 3-methyladenine (3-MA) inhibited HBV DNA replication.
In conclusion, lipid metabolism is a significant factor affecting HBV load in patients with HBV infection. The in vitro experiments established that fatty acid stimulation inhibits HBV replication via the Plin2-autophagy pathway.
Core Tip: Our data suggest that fatty acid stimulation inhibits hepatitis B virus (HBV) replication by upregulating Plin2 expression, inhibiting hepatocyte autophagy. This process associates with lipid metabolism, autophagy pathway, and HBV replication. Further study of lipid metabolism-Plin2-autophagy is important to understand HBV host interactions and pathogenesis better and suggests a possible route for treating patients with chronic HBV infection combined with nonalcoholic fatty liver disease.
- Citation: Wang C, Gao XY, Han M, Jiang MC, Shi XY, Pu CW, Du X. Perilipin2 inhibits the replication of hepatitis B virus deoxyribonucleic acid by regulating autophagy under high-fat conditions. World J Virol 2023; 12(5): 296-308
- URL: https://www.wjgnet.com/2220-3249/full/v12/i5/296.htm
- DOI: https://dx.doi.org/10.5501/wjv.v12.i5.296
Hepatitis B virus (HBV) infection is a global health issue[1]. According to the World Health Organization, about one-third of the world's population will contract acute HBV at some point in their life[2]. Current main treatments include nucleoside analogs and interferon; however, they are not effective in eliminating the virus[3]. Therefore, it is imperative to better understand the underlying mechanisms behind HBV infection-induced disease to find new targets for anti-HBV therapy.
The relationship between HBV infection and lipid metabolism has received more attention in the last decade. Clinical studies have demonstrated that chronic HBV infection enhances the incidence of nonalcoholic fatty liver disease (NAFLD), with NAFLD co-infected with HBV accounting for 13.5% of all HBV patients[4]. These studies indicate a close association between HBV infection and altered lipid metabolism. This present study intends to clarify the mechanism of why increased lipid deposition in the liver can also inhibited HBV replication in hepatocytes[5].
Autophagy is an evolutionarily conserved catabolic process that regulates HBV replication and is required to maintain cellular homeostasis in response to the microenvironment. It involves selective and non-selective mechanisms that cause intracellular substrate degradation[6]. HBV was found to be able to maintain its own replication by inducing hepatocyte autophagy, and when cellular autophagy was inhibited, HBV replication expression in hepatocytes was significantly reduced[7]. Perilipin2 (Plin2) is involved in the formation of lipid droplets in the liver and peripheral tissues[8]. Plin2 is highly upregulated in humans and rodents with NAFLD[5,9]. Purposeful knockdown of Plin2 protein in the mouse liver was found to significantly reduce liver weight, body weight, and adipose tissue mass[10]. In a previous study on NAFLD pathogenesis, it was found that Plin2 is not only involved in intracellular lipid deposition but also regulates intracellular autophagy, and stimulation of hepatocytes with high concentrations of fatty acids results in increased Plin2 expression and cellular autophagy inhibition[11].
Combined with the above, we speculated that the Plin2-autophagy pathway might be involved in regulating lipid deposition and HBV replication in hepatocytes. In this study, we first explored the relationship between patients' lipid levels, hepatic steatosis, and HBV deoxyribonucleic acid (DNA) load. Also, we constructed an HBV infection combined with a hepatic steatosis cell model in vitro by fatty acid stimulation of HepG2.2.15 cells to validate the effect of lipid metabolism on HBV DNA replication in vitro. By knocking down and overexpressing Plin2, we observed whether Plin2 regulates autophagy and HBV replication. By inhibiting both Plin2 and cellular autophagy under high lipid stimulation, we examined whether the Plin2-autophagy pathway regulates HBV replication. This present study intends to investigate the relationship between lipid metabolism and HBV replication through retrospective analysis and in vitro studies, providing a novel theoretical basis for the mechanism of HBV replication and a new target for searching new therapeutic sites.
The study has been approved by the Ethics Committee of Dalian Sixth People's Hospital. The privacy rights of human subjects were always respected during human experimentation, and informed consent was obtained prior to the experiment. The ethics program number is DLY/CB-IRB-026.
In this cross-sectional hospital-based study, all patients were recruited from the Dalian Sixth People's Hospital. A total of 1603 HBsAg-positive patients underwent a comprehensive health examination with no prior antiviral treatments to evaluate the effect of lipid profile on HBV viral replication. Additionally, 132 chronic hepatitis B patients were included in the study to investigate the effect of antiviral treatment on lipid profile. Patients with hepatitis C and D, autoimmune hepatitis, alcoholic fatty liver, Wilson's disease, drug-related hepatic steatosis, liver surgery, or liver transplantation were excluded from the study.
During the study, data on age, sex, alcohol consumption, and medical history were collected through face-to-face interviews. Following an overnight fast, blood samples were obtained from all participants. HBsAg, antibodies against HBsAg, HBeAg, antibodies against HBeAg, and antibodies against hepatitis B core antigen were measured using an immunoassay analyzer. The levels of serum HBV-DNA copy were measured using the COBAS Amplicor HBV monitor test (Cap/ctm, Roche, Switzerland). Clinical chemistry systems were used to evaluate the serum levels of alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), gamma-glutamyl transferase, total bile acids, total bilirubin, albumin, and bile acid.
The HBV-producing HepG2.2.15 hepatoma cell line with an integrated HBV genomic dimer was obtained from the Chinese Academy of Sciences (CAS) in Beijing, China. Trypsin-EDTA (#27250–018), fetal bovine serum (#10100147), phosphate-buffered saline (PBS) (#226013), Opti MEM medium (#22600134), and Dulbecco's modified essential medium (DMEM) (#31600083) were purchased from GIBCO BRL (Grand Island, NY, United States). Oleic acid (OA, #15724), palmitic acid (PA, #27567713), and 3-methyladenine (3MA, #M9281) were acquired from Sigma-Aldrich (Missouri, United States). Perilipin2 (Plin2, #ac219686) was obtained from Abcam Technologies (Abcam, Cambridge, United Kingdom). Light chain 3 (LC3, #A11280), glyceraldehyde-3-phosphate dehydrogenase (GAPDH, #AC002), anti-rabbit IgG (#AS014), and anti-mouse IgG (#LV-AS003) were obtained from Wuhan Abcotec Biotechnology Co Ltd (Abclonal, Wuhan, China).
HepG2.2.15 cells were obtained from the CAS and cultured in T25 cell culture flasks with 10% heat-inactivated fetal bovine serum (GIBCO BRL) (#10100147), antibiotics, and high glucose DMEM (GIBCO) (#31600083). The flasks were pre-cultured at 37°C for 18-24 h in a 5% CO2 incubator.
Plin2 knockdown and overexpression plasmids were synthesized by Suzhou Jima Bio. Plasmid transfection was performed following the manufacturer's instructions for Lipofectamine 2000 transfection reagent (Invitrogen).
Whole-cell protein extracts were obtained by passive cell lysis using protease and phosphorylated protease inhibitors following the manufacturer's instructions, and protein concentrations were determined using the BCA method. SDS-PAGE (10% gel) was used to separate samples containing approximately 30 µg of protein per well, and the proteins were transferred to PVDF membranes, which were incubated with primary antibody at 4°C overnight after being closed in low-fat milk powder for 2 h at room temperature in TBST. The membranes were washed and placed in BLOTTO containing secondary antibodies (HRP-labeled goat anti-rabbit antibody) for 1.5 h at room temperature. Following TBST clearing, the membranes were placed in the chromogen for 30 s and exposed immediately to the exposure cassette. The method was to estimate the ratio of the brightness value of each sample strip to the brightness value of the corresponding GAPDH (internal reference) strip to get the corrected strip brightness value.
The triglyceride content of HepG2.2.15 was measured using the Triglyceride Quantification Assay Kit (Abcam) for colorimetric detection, following the manufacturer's protocol.
Several methods have been described for the detection of HBV daughter DNA in culture supernatants[12,13]. Real-time reverse transcription (RT) polymerase chain reaction (PCR) assay using primers 5'TCTTGCCTTACTTTTGGAAG 3' (forward) 5'AGTTCTTCTTCTTCTAGGGGACC3' (reverse) were used to measure HBV pgRNA and Plin2 mRNA levels in cells.
HBsAg and HBeAg levels were evaluated using a commercial ELISA kit (Hunan Shengxiang Biotechnology Co., Ltd., Hunan, China) following the manufacturer's instructions. The absorbance of each well was sequentially measured at 450 nm wavelength with zero blank air conditioning for the final assay.
After stimulating HepG2.2.15 cells with free fatty acids, the cells were rinsed three times with PBS and fixed in 10% formalin for 15 min at room temperature. Following fixation, the cells were stained with Oil Red O for 20 min at room temperature. Stained cells were observed by a fluorescent microscope (Leica DMI 4000 B) on the white light setting (magnification, ×100).
After preincubation in a complete medium at 37°C in 21% O2 and 5% CO2 for 24 h, the cells were transfected with GFP-LC3 following the manufacturer's instructions to monitor autophagy flux. After 8 h of transfection, the cells were rinsed with PBS; a complete culture medium was added to the cells. Finally, the samples were observed under a fluorescence microscope (Nikon, Tokyo, Japan)
Continuous variables were reported as mean ± standard deviation or median (interquartile range). Dependent variables were expressed as numbers or percentages. The Wilcoxon matched-pairs signed-rank test, a nonparametric statistical test, was employed to compare non-normally distributed continuous data. This test was used to analyze various variables, including age, gender, FBG, ALT, AST, ALP, γ-GGT, LDH, bile acids, total cholesterol, triglycerides, HDL-C, LDL-C, apoA, and apoB. Chi-square test was used to compare the differences in hepatic steatosis prevalence and the HBeAg sero-positive prevalence of patients with high or low HBV DNA load. Binary logistic regression analysis was conducted to identify potential factors influencing HBV DNA load, such as hepatic steatosis, triglycerides, apoA, apoB, cholesterol, HDL-C, and LDL-C. Statistical analysis was performed using IBM Corp's SPSS version 24.0 software. A two-sided P value < 0.05 was considered statistically significant. All experiments were conducted in triplicates, and western blot data were analyzed using t-tests.
In this study, 1603 HBsAg-seropositive patients were included, of which 674 (42.0%) were HBeAg-seropositive. Of the total patients, 1015 (63.3%) were male, and 815 (50.8%) had a high HBV viral load, defined as serum HBV DNA levels > 104 copies/mL. The median age was 52 years (range 43–60). Table 1 presents the characteristics of the HBsAg-seropositive patients. Patients in the high HBV DNA group had a higher levels of ALT, AST, ALP, γ-GGT, LDH, and HDL-C, and lower levels of triglyceride (TG), FBG, albumin, LDL-C, and apoB compared to those in the low HBV DNA group (P < 0.05). However, there were no significant differences between the two groups with regards to TC, total bilirubin, and apoA. The characteristics of the HBeAg-seropositive patients are shown in Table 2.
| Factors | ALL | High HBV DNA | Low HBV DNA | P value |
| Case n = 1603 | Case n = 815 | Case n = 788 | ||
| Demographic | ||||
| Age (yr) | 52 (43–60) | 51 (40–60) | 53 (45–61) | 0.010 |
| Male gender | 1015 (63.28) | 131 (69.6) | 401 (68.2) | 0.703 |
| Laboratory tests | ||||
| FBG (mmol/L) | 5.10 (4.61–5.87) | 5.00 (4.5–5.7) | 5.21 (4.77–6.00) | < 0.001 |
| ALT (U/L) | 47.95 (25.00–109.92) | 68.3 (37.1–155.83) | 31.7 (20.3–67.7) | < 0.001 |
| AST (U/L) | 41.19 (25.00–81.97) | 52.95 (33.33–106.8) | 29.86 (21.31–55.55) | < 0.001 |
| ALP (U/L) | 81.40 (63.63–109.00) | 85.30 (66.00–116.60) | 76.65 (61.9–104.00) | 0.003 |
| γ-GGT (U/L) | 51.20 (22.89–113.36) | 60.00 (29.13–135.00) | 41.60 (18.97–100.12) | 0.038 |
| LDH (U/L) | 194.8 (170.23–231.10) | 196.00 (171.56–239.00) | 194.00 (168.94–224.61) | 0.006 |
| Albumin (g/L) | 40.70 (35.33–44.70) | 39.60 (33.02–43.51) | 41.96 (37.49–45.80) | 0.045 |
| Total bilirubin (µmol/L) | 16.53 (11.63–25.24) | 17.60 (12.20–27.70) | 15.39 (11.37–23.20) | 0.165 |
| Bile acids (µg/mL) | 11.00 (5.18–30.00) | 14.30 (6.30–34.20) | 8.70 (4.26–23.88) | 0.002 |
| Total cholesterol (mmol/L) | 4.25 (3.55–5.01) | 4.19 (3.50–4.92) | 4.37 (3.61–5.10) | 0.517 |
| Triglycerides (mmol/L) | 1.06 (0.73–1.53) | 1.02 (0.73–1.44) | 1.12 (0.74–1.71) | < 0.001 |
| HDL-C (mmol/L) | 1.15 (0.90–1.41) | 1.19 (0.91–1.43) | 1.12 (0.89–1.38) | 0.029 |
| LDL-C (mmol/L) | 2.40 (1.87–2.98) | 2.27 (1.76–2.89) | 2.51 (1.95–3.10) | 0.018 |
| apoA (g/L) | 1.19 (1.01–1.38) | 1.14 (0.99–1.27) | 1.18 (1.02–1.39) | 0.849 |
| apoB (g/L) | 0.87 (0.70–1.06) | 0.84 (0.69–1.01) | 0.91 (0.73–1.10) | 0.002 |
| Steatosis | 163 (10.2) | 25 (3.1) | 138 (17.5) | < 0.001 |
| HBeAg sero-positive | 661 (41.2) | 460(56.4) | 201 (25.5) | < 0.001 |
| Factors | HBeAg sero-positive | HBeAg sero-negative | ||||
| High HBV DNA | Low HBV DNA | P value | High HBV DNA | Low HBV DNA | P value | |
| Case n = 460 | Case n = 201 | Case n = 355 | Case n = 587 | |||
| Demographic | ||||||
| Age (yr) | 45 (36–59) | 50 (40–59) | 0.106 | 54 (45–61) | 55 (46–61) | 0.564 |
| Male gender | 314 (68.2) | 137 (68.1) | 0.412 | 128 (56.6) | 93 (68.4) | 0.026 |
| Laboratory tests | ||||||
| FBG (mmol/L) | 4.83 (4.40–5.35) | 5.10 (4.60–5.90) | < 0.001 | 5.23 (4.73–6.20) | 5.28 (4.80–6.06) | 0.795 |
| ALT (U/L) | 83.21 (43.45–190.62) | 51.60 (26.62–120.07) | < 0.001 | 53.00 (3200–127.00) | 27.60 (19.38–53.60) | < 0.001 |
| AST (U/L) | 59.30 (38.01–130.1) | 46.85 (26.00–93.18) | < 0.001 | 46.50 (37.57–116.55) | 27.00 (20.72–46.47) | < 0.001 |
| ALP (U/L) | 87.50 (67.98–117.09) | 90.18 (67.00–127.15) | 0.489 | 83.40 (65.20–111.90) | 74.00 (60.00–96.00) | < 0.001 |
| γ-GGT (U/L) | 64.61 (33.50–139.92) | 68.25 (33.25–145.87) | 0.615 | 52.00 (21.90–115.69) | 69.6 (33.40–164.30) | < 0.001 |
| LDH (U/L) | 195.00 (171.92–238.32) | 193.40 (167.37–228.97) | 0.38 | 197.00 (170.08–239.14) | 194.00 (169.00–222.39) | 0.077 |
| Albumin (g/L) | 39.40 (32.81–43.36) | 40.30 (34.89–44.50) | 0.026 | 54.00 (45.00–61.50) | 43.90 (38.00–47.70) | < 0.001 |
| Total bilirubin (µmol/L) | 17.90 (12.40–28.35) | 17.80 (13.21–32.52) | 0.751 | 17.20 (11.9–27.00) | 14.71 (11.08–22.47) | < 0.001 |
| Bile acids (ug/mL) | 16.60 (8.01—38.77) | 16.50 (8.01–41.52) | 0.904 | 10.06 (4.98–27.50) | 7.07 (3.73–16.54) | < 0.001 |
| Total cholesterol (mmol/L) | 4.16 (3.50–4.85) | 4.00 (3.30–4.83) | 0.674 | 4.22 (3.49–5.01) | 4.46 (3.71–5.15) | 0.019 |
| Triglycerides (mmol/L) | 1.03 (0.73–1.44) | 1.08 (0.71–1.71) | 0.429 | 1.04 (0.73–1.41) | 1.14 (0.75–1.72) | 0.002 |
| HDL-C (mmol/L) | 1.19 (0.90–1.43) | 1.08 (0.81–1.40) | 0.041 | 1.20 (0.92–1.44) | 1.12 (0.91–1.38) | 0.13 |
| LDL-C (mmol/L) | 2.25 (1.76–2.90) | 2.41 (1.89–3.06) | 0.041 | 2.31 (1.72–2.87) | 2.54 (1.97–3.12) | < 0.001 |
| apoA (g/L) | 1.17 (0.97–1.35) | 1.15 (0.92–1.30) | 0.204 | 1.21 (1.00–1.40) | 1.20 (1.04–1.41) | 0.624 |
| apoB (g/L) | 0.85 (0.69–1.02) | 0.91 (0.70–1.12) | 0.01 | 0.83 (0.68–1.00) | 0.91 (0.74–1.09) | < 0.001 |
| Steatosis | 11 (2.4) | 37 (18.4) | < 0.001 | 11 (3.0%) | 101 (17.2) | < 0.001 |
Out of the 1603 HBsAg-seropositive patients, 661 were HBeAg-seropositive and 942 were HBeAg-seronegative. Among the HBeAg-seropositive group, 460 (69.6%) patients had a high viral load while in the HBeAg-seronegative group, 355 (37.7%) patients had a high viral load. Patients with high viral loads in the HBeAg-seropositive group had higher levels of ALT, AST, and HDL-C, and lower levels of albumin, FBG, LDL-C, apoB, and a lower ratio of steatosis (P < 0.05) (Table 1). In contrast, patients with high viral loads in the HBeAg-seronegative group had higher levels of ALT, AST, ALP, albumin, total bilirubin, and bile acid, and lower levels of TC, LDL-C, apoB, γ-GGT, TG, and a lower ratio of steatosis (P < 0.05) (Table 2).
Table 3 presents the results of binary logistic regression analyses of metabolic factors associated with HBV-DNA load. The serum levels of TG (OR 0.83, 95%CI 0.70–0.98, P = 0.027), apoA (OR 0.47, 95%CI 0.26–0.83, P = 0.009), and LDL-C (OR 0.59, 95%CI 0.45–0.77, P < 0.001) were negatively associated with HBV-DNA load, while hepatic steatosis (OR 0.15, 95%CI 0.10–0.23, P < 0.001) was also negatively associated with HBV-DNA load. On the other hand, TC level (OR 1.39, 95%CI 1.09–1.77, P = 0.009) was positively associated with HBV-DNA load.
| P value | OR (95%CI) | |
| HBsAg sero-positive | ||
| Steatosis | < 0.001 | 0.15 (0.10–0.23) |
| Triglyceride | 0.027 | 0.83 (0.70–0.98) |
| apoA | 0.009 | 0.47 (0.26–0.83) |
| apoB | 0.905 | 0.99 (0.91–1.08) |
| Cholesterol | 0.009 | 1.39 (1.09–1.77) |
| HDL-C | 0.171 | 1.38 (0.87–2.18) |
| LDL-C | < 0.001 | 0.59 (0.45–0.77) |
| HBeAg sero-positive | ||
| Steatosis | < 0.001 | 0.11 (0.05–0.22) |
| Triglyceride | 0.077 | 0.74 (0.53–1.03) |
| apoA | 0.846 | 1.10 (0.41–2.96) |
| apoB | 0.5 | 0.69 (0.23–2.02) |
| Cholesterol | < 0.001 | 2.33 (1.55–3.51) |
| HDL-C | 0.204 | 0.61 (0.29–1.30) |
| LDL-C | < 0.001 | 0.38 (0.24–0.60) |
| HBeAg sero-negative | ||
| Steatosis | < 0.001 | 0.19 (0.11–0.35) |
| Triglyceride | 0.002 | 0.69 (0.55–0.87) |
| apoA | 0.209 | 0.62 (0.29–1.31) |
| apoB | 0.713 | 1.02 (0.91–1.14) |
| Cholesterol | 0.004 | 1.69 (1.19–2.40) |
| HDL-C | 0.904 | 0.96 (0.51–1.81) |
| LDL-C | < 0.001 | 0.46 (0.31–0.67) |
Table 3 provides the results of logistic regression analyses of metabolic factors associated with HBV-DNA load in HBeAg-seropositive and HBeAg-seronegative patients. In HBeAg-seropositive patients, serum LDL-C level (OR 0.38, 95%CI 0.24–0.60, P < 0.001) and hepatic steatosis (OR 0.11, 95%CI 0.05–0.22, P < 0.001) were negatively associated with HBV-DNA load, while TC level (OR 2.33, 95%CI 1.55–3.51, P < 0.001) was positively associated with HBV-DNA load. In contrast, in HBeAg-seronegative patients, serum TG level (OR 0.69, 95%CI 0.55–0.87, P = 0.002), LDL-C level (OR 0.46, 95%CI 0.31–0.67, P < 0.001), and hepatic steatosis (OR 0.19, 95%CI 0.11–0.35, P < 0.001) were negatively associated with HBV-DNA load, while TC level (OR 1.69, 95%CI 1.19–2.40, P = 0.004) was positively associated with HBV-DNA load.
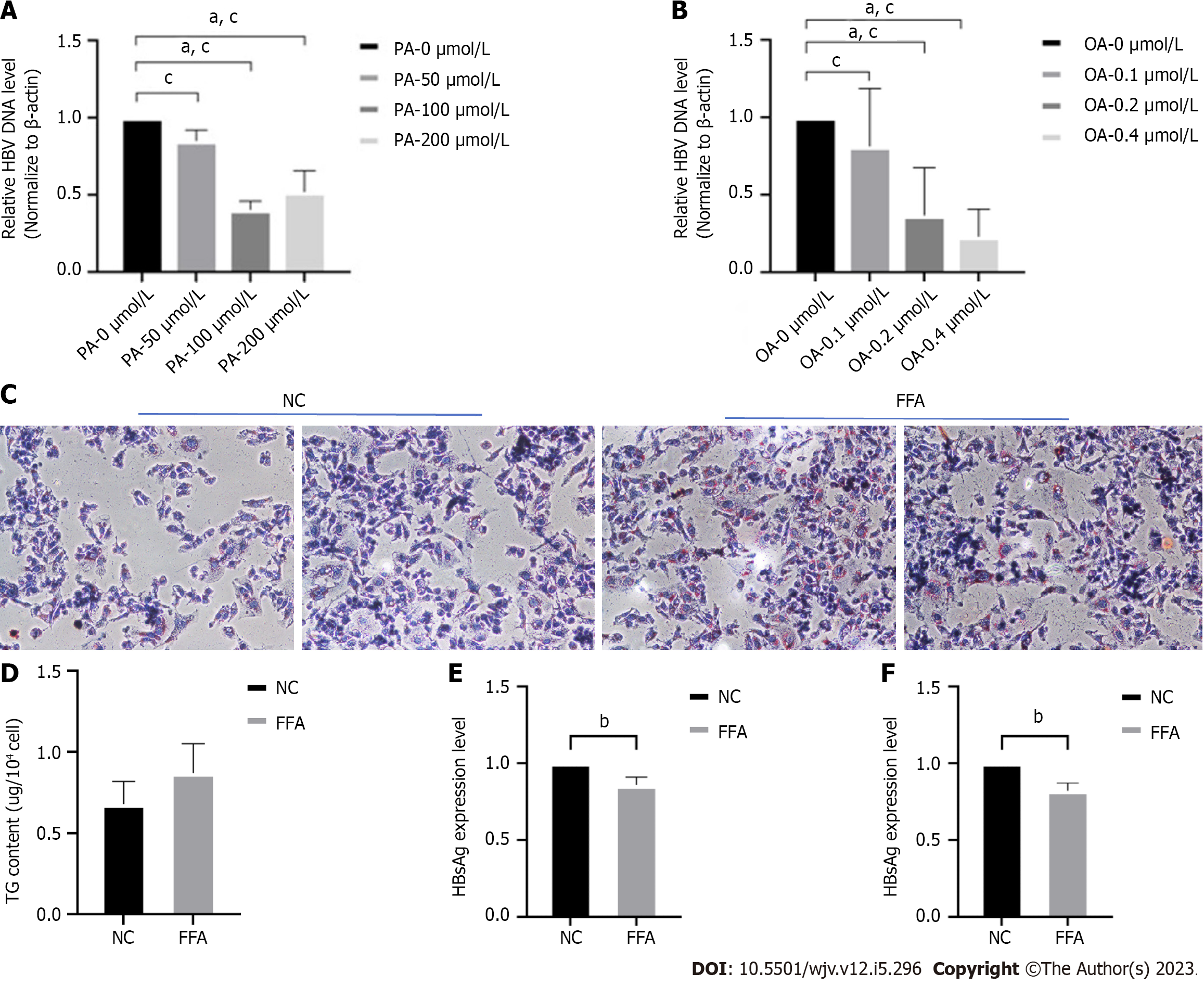
To investigate the relationship between lipid metabolism and HBV DNA replication in vitro, HepG2.2.15 cells were stimulated with varying concentrations of PA and oleic OA to create a model of HBV infection combined with hepatic steatosis. Real-time PCR was used to detect changes in HBV DNA replication levels. The results demonstrated that the expression load of HBV DNA significantly decreased in a concentration-dependent manner with the increase of PA or OA concentration (Figure 1A and B). The optimal stimulation concentration of OA was 0.2 M, while the optimal stimulation concentration of PA was 100 μmol/L. The optimal fatty acid concentrations were prepared into free fatty acids (FFA) at an OA:PA ratio of 2:1. After 72 h of FFA treatment, HepG2.2.15 cells were stained with oil red O hematoxylin, revealing a significant increase in intracellular lipid droplets in the FFA group compared to the control group, and fusion phenomena were observed (Figure 1C). The intracellular TG content was higher in the high-fat stimulation conditions than in the control group (Figure 1D). ELISA was used to measure HBsAg and HBeAg levels in cell culture supernatants, and it was found that high-fat stimulation inhibited the expression of both HBsAg (Figure 1E) and HBeAg (Figure 1F).
Based on these findings, it can be concluded that high-fat conditions inhibit the expression of HBV DNA and its serum markers in a concentration-dependent manner. However, the exact mechanism underlying this inhibition remains unclear.
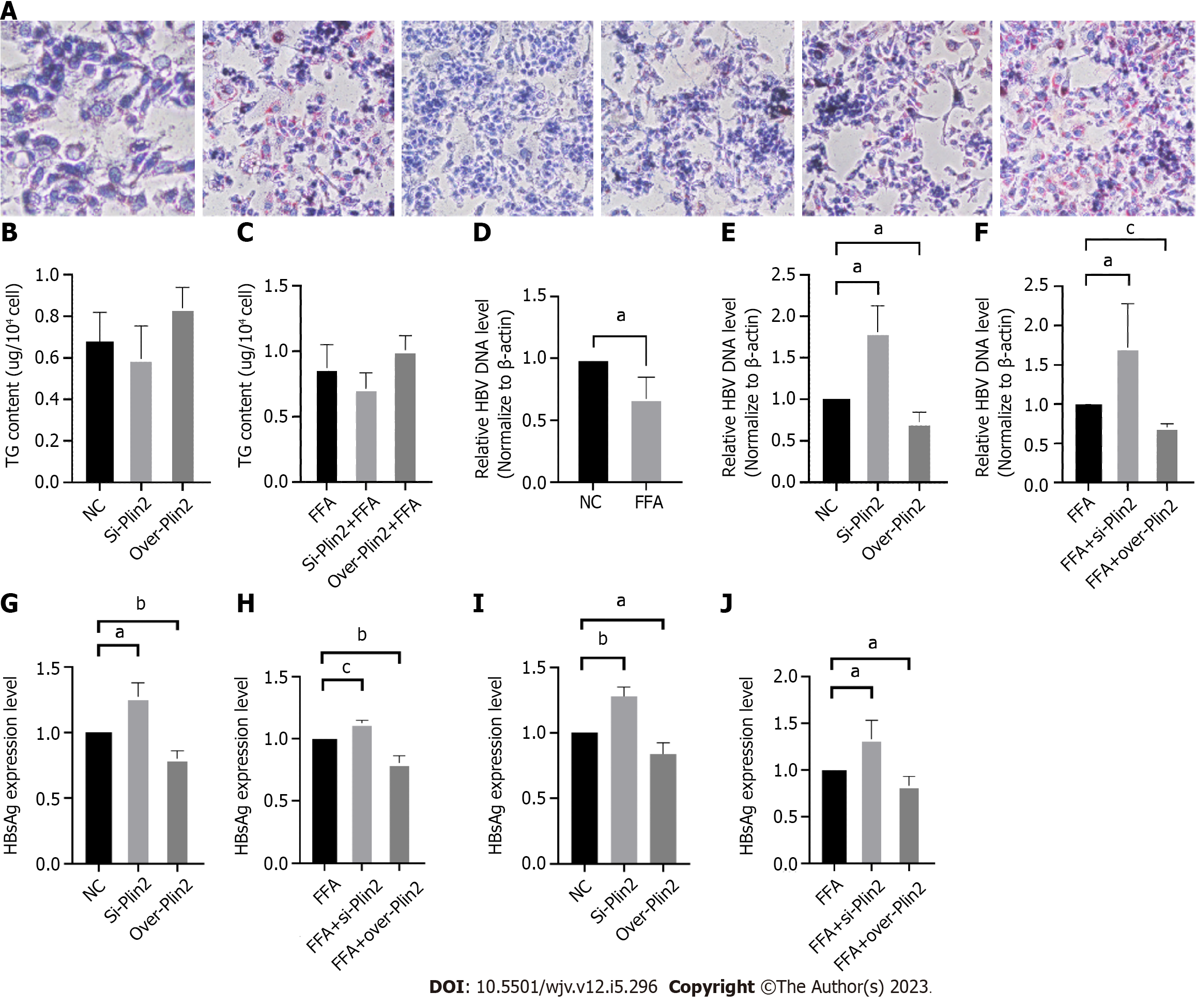
To investigate the role of Plin2 in inhibiting HBV replication under high lipid conditions, we performed both Plin2 knockdown and overexpression experiments in HepG2.2.15 cells after fatty acid stimulation. After downregulating Plin2 protein, the number and volume of intracellular lipid droplets significantly decreased in both the control group and FFA-stimulated group under microscopy, whereas the number of lipid droplets increased in the Plin2 overexpression group (Figure 2A). Additionally, TG content was observed to increase in the Plin2 overexpression group (Figure 2B and C). HBV DNA load increased significantly after knockdown of Plin2 (Figure 2D-F). The expression of HBsAg and HBeAg was also significantly upregulated after transfection with siPlin2 plasmid, whereas the expression of both markers was downregulated after overexpression of Plin2 (Figure 2G-J).
These findings suggest that high lipid conditions upregulate Plin2 expression and that Plin2 plays a role in counteracting the regulation of lipid metabolism.
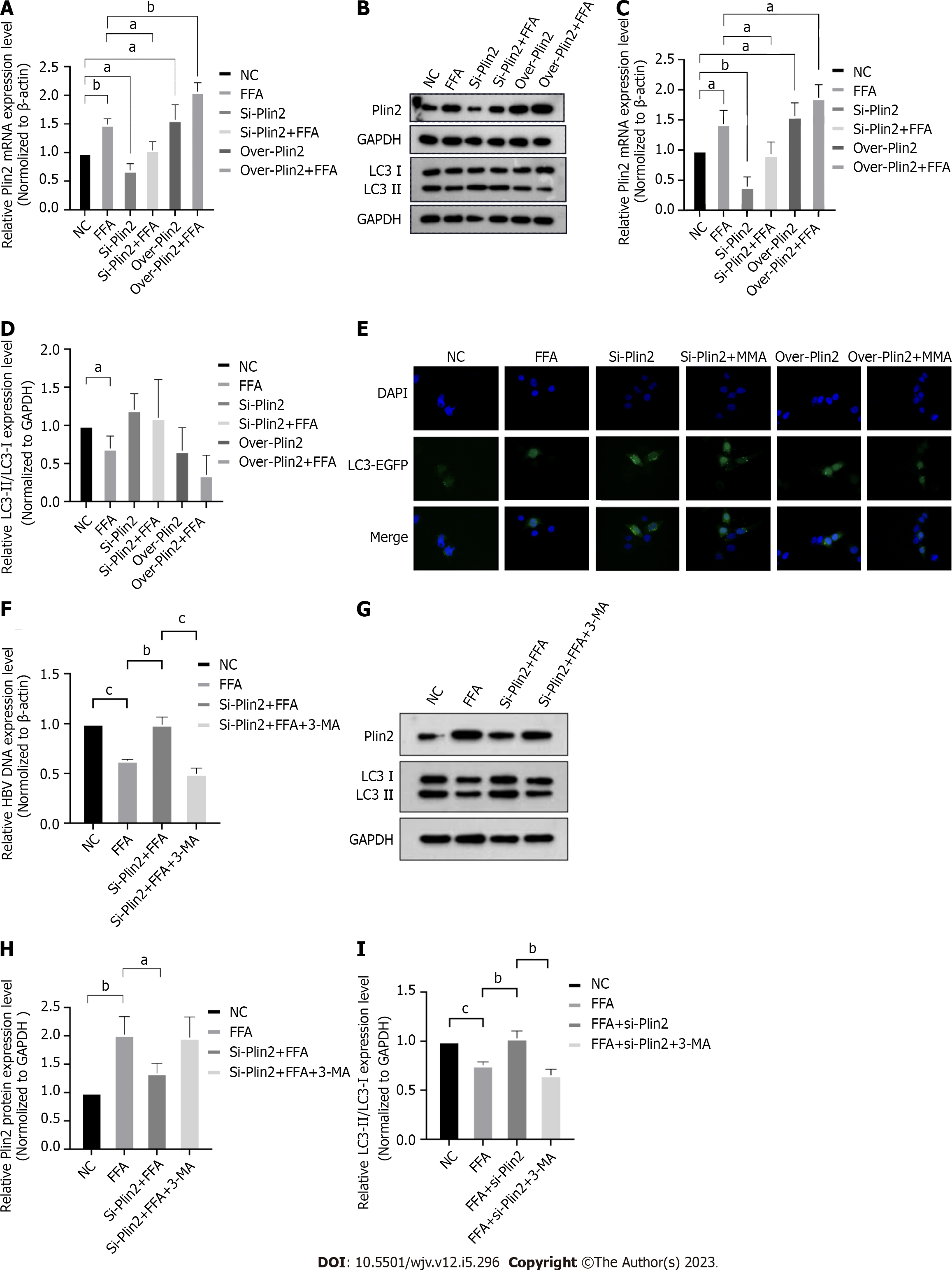
Western blotting was used to examine the expression of Plin2 and autophagy-related proteins LC3-II and LC3-I. The results showed that Plin2 expression was significantly upregulated under fatty acid stimulation conditions, and Plin2 knockdown under fatty acid stimulation conditions demonstrated a decreasing trend in upregulated Plin2 expression (Figure 3A-C). Autophagy-related proteins LC3-II/LC3-I were significantly decreased when HepG2.2.15 cells were stimulated with fatty acids, indicating autophagy inhibition. Autophagy expression increased when Plin2 expression was disturbed; after knockdown of Plin2 under high-fat conditions, autophagy was restored, whereas overexpression of Plin2 under both normal medium and high-fat stimulation significantly inhibited autophagy (Figure 3B and D). The number of autophagic vesicles significantly decreased when HepG2.2.15 cells were stimulated with fatty acids, indicating that autophagy was inhibited as seen through GFP-LC3 staining. The number of autophagic vesicles increased when Plin2 expression was interfered with and significantly decreased when Plin2 expression was overexpressed (Figure 3E).
Based on these findings, it can be concluded that fatty acid stimulation alters the autophagic trend by affecting the expression of Plin2, thereby affecting HBV DNA replication.
Plin2 knockdown under fatty acid stimulation was observed to restore the inhibited HBV replication, whereas the restored DNA expression load was again inhibited after adding 3-MA to inhibit autophagy (Figure 3F). Plin2 protein expression remained unchanged upon the addition of autophagy inhibitor, as observed through Western blot analysis (Figure 3G and H). In contrast, the values of autophagy-related protein LC3-II/LC3-I were significantly downregulated (Figure 3G and I), indicating that autophagy was clearly inhibited. These results suggest that fatty acid stimulation inhibits autophagy and HBV DNA replication via Plin2.
HBV, a DNA virus that causes immune-mediated liver disease, can be transmitted through blood and body fluids. Although immunomodulatory drugs such as interferon and antiviral drugs have a good safety profile and are also effective in controlling HBV replication in patients with chronic hepatitis B, they rarely eliminate HBV completely and do not completely eliminate liver cancer risk[14]. Therefore, there is an urgent requirement to explore the deeper regulatory mechanisms of HBV in order to seek the development of new drugs for HBV clearance.
Hepatic steatosis is characterized by an excessive accumulation of triglycerides in hepatocytes[15]. In previous studies, hepatic steatosis has been found in patients with chronic HBV infection, causing a reduction in their response to antiviral therapy[16]. However, there is mixed evidence on the association between hepatic steatosis and HBV replication. Several studies have shown that hepatic steatosis induces HBsAg clearance and reduces HBV replication[17,18]. Chia-Ming Chu's study showed that in patients with increased body mass index, hepatic steatosis accelerated HBsAg serological clearance by approximately 5 years[19]. Conversely, Lesmana et al[20] found no difference in HBV replication between HBV patients with and without hepatic steatosis. Our results indicated that serum triglycerides, HDL levels, and the rate of hepatic steatosis were significantly lower in the HBV-DNA high load (815 cases) group compared to the HBV-DNA low load (788 cases) group. The findings of logistic regression demonstrated that serum TG levels (OR 0.83 95%CI 0.70–0.98, P = 0.027), apoA levels (OR 0.47, 95%CI 0.26–0.83, P = 0.009), LDL-C levels (OR 0.59, 95%CI 0.45–0.77, P < 0.001) and hepatic steatosis (OR 0.15, 95%CI 0.10–0.23, P < 0.001) showed significant negative correlation with HBV-DNA load. As a result, we conducted a stratified analysis according to HBeAg serostatus. The results of logistic regression showed that hepatic steatosis serum triglyceride load was negatively correlated with blood HBV-DNA load in both HBeAg positive or negative groups (P < 0.001). These findings are consistent with a study reported by Jarcuska et al[21], which stated a significantly lower HBV-DNA load in patients with hypertriglyceridemia. These findings indicate that increased lipid metabolism in the body can inhibit HBV replication.
Previous studies have demonstrated that autophagy is closely associated with HBV DNA replication, and various factors in HBV infection, such as interferon Alpha and endoplasmic reticulum stress, can affect HBV replication by inducing autophagy[22]. Yongjun Tian et al[23] found that after liver-specific Atg5 knockdown in the HBV Tg05 mouse, the serum levels of HBeAg and HBsAg were decreased by about 50% and 60%, respectively. However, this autophagy inhibition decreased HBV DNA levels by more than 90%. A previous study showed that IFNα-2a treatment promoted autophagy initiation and blocked autophagy degradation, leading to a slight enhancement of HBV replication[24]. Autophagy disorders often result in metabolic abnormalities and play an essential role in the pathogenesis of numerous metabolic liver diseases, such as alcoholic liver disease and NAFLD[25]. Singh et al[26] coined the term "lipophagy" after identifying autophagy-mediated lipolytic functions in the LIPA pathway; TSAI T H showed that specific knockdown of Plin2 decreased triglyceride levels in mice by approximately 60%[27]. In another study of liver-specific Plin2 knockout mice, a significant increase in LC3 and p62-positive spots were detected in the livers of Plin2-deficient mice fed WTD[28]. In the study by Tsai et al[27], it was found that the down-regulation of Plin2 stimulated TG catabolism by upregulating autophagy expression through direct knockdown of Plin2. Therefore, we speculated that the high-fat environment regulates the Plin2-autophagy pathway and thus inhibits HBV DNA replication. Our experiments first examined the relationship between Plin2 and autophagy. Autophagy expression increased both under normal medium and under high-fat stimulation when Plin2 protein expression was down-regulated, and overexpression of Plin2 protein resulted in significant inhibition of autophagy. Fluorescence microscopy showed an increase in autophagy vesicles when Plin2 protein expression was down-regulated. Then the association between Plin2 and autophagy under fatty acid stimulation was further explored. Plin2 expression was significantly upregulated in HepG2.2.15 cells, and cellular autophagy was significantly inhibited. Compared to the control group, the expression of supernatant HBV DNA load and serological markers in HepG2.2.15 cells were significantly lower in the Plin2 knockdown group, whereas the overexpression of Plin2 was reversed. Finally, we inhibited the growth of autophagy with 3-MA in parallel with the knockdown of Plin2. At that time, we found that the growth of autophagy-related proteins was significantly reduced. In addition, the increased HBV DNA replication was also reduced, and HBsAg and HBeAg were similarly altered. All these results indicate that the Plin2-autophagy pathway is involved in the regulation of high-fat inhibition of HBV replication.
In summary, our data suggest that fatty acid stimulation inhibits HBV replication by upregulating Plin2 expression, inhibiting hepatocyte autophagy. This process associates with lipid metabolism, autophagy pathway, and HBV replication. Further study of lipid metabolism-Plin2-autophagy is important to understand HBV host interactions and pathogenesis better and suggests a possible route for treating patients with chronic HBV infection combined with NAFLD.
The relationship between lipid metabolism and hepatitis B virus (HBV) deoxyribonucleic acid (DNA) replication and its underlying mechanisms are not well understood.
To investigate the relationship between lipid metabolism and HBV DNA replication and its underlying mechanisms.
We speculated that the Plin2-autophagy pathway might be involved in regulating lipid deposition and HBV replication in hepatocytes.
We first explored the relationship between patients' lipid levels and HBV DNA load. Also, we constructed an HBV infection combined with a hepatic steatosis cell model in vitro.
Stratified analysis by HBeAg showed significant negative correlations between HBV-DNA load and hepatic steatosis ratio in both HBeAg-positive group and in HBeAg-negative group. The results of in vitro experiments suggested that fatty acid treatment increased the lipid droplets deposition and decreased the cell supernatant HBsAg, HBeAg expression and HBV DNA load.
Fatty acid stimulation inhibits HBV replication by upregulating Plin2 expression, inhibiting hepatocyte autophagy.
A possible route for treating patients with chronic HBV infection combined with nonalcoholic fatty liver disease.
Thank you, Professor Song Guirong, for your support in the statistical design of our experiment.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Cell biology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Spera AM, Italy; Kelleni MT, Egypt S-Editor: Liu JH L-Editor: A P-Editor: Zhang XD
| 1. | Zhang WL, Ji ZH, Fu T, Zhang L, Su HX, Yan YP. [Meta analysis on HBsAg-positive rate among general populations aged 1-59 years, 2007-2016, China]. Zhonghua Liu Xing Bing Xue Za Zhi. 2017;38:1278-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 2. | Iannacone M, Guidotti LG. Immunobiology and pathogenesis of hepatitis B virus infection. Nat Rev Immunol. 2022;22:19-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 328] [Article Influence: 82.0] [Reference Citation Analysis (0)] |
| 3. | European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67:370-398. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3745] [Cited by in RCA: 3990] [Article Influence: 443.3] [Reference Citation Analysis (1)] |
| 4. | Wong VW, Wong GL, Chu WC, Chim AM, Ong A, Yeung DK, Yiu KK, Chu SH, Chan HY, Woo J, Chan FK, Chan HL. Hepatitis B virus infection and fatty liver in the general population. J Hepatol. 2012;56:533-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 197] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 5. | Itabe H, Yamaguchi T, Nimura S, Sasabe N. Perilipins: a diversity of intracellular lipid droplet proteins. Lipids Health Dis. 2017;16:83. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 266] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 6. | Chiramel AI, Best SM. Role of autophagy in Zika virus infection and pathogenesis. Virus Res. 2018;254:34-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 96] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 7. | Xie M, Yang Z, Liu Y, Zheng M. The role of HBV-induced autophagy in HBV replication and HBV related-HCC. Life Sci. 2018;205:107-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 60] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 8. | Takahashi Y, Shinoda A, Kamada H, Shimizu M, Inoue J, Sato R. Perilipin2 plays a positive role in adipocytes during lipolysis by escaping proteasomal degradation. Sci Rep. 2016;6:20975. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 86] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 9. | Zhang X, Wang Y, Liu P. Omic studies reveal the pathogenic lipid droplet proteins in non-alcoholic fatty liver disease. Protein Cell. 2017;8:4-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 10. | Najt CP, Senthivinayagam S, Aljazi MB, Fader KA, Olenic SD, Brock JR, Lydic TA, Jones AD, Atshaves BP. Liver-specific loss of Perilipin 2 alleviates diet-induced hepatic steatosis, inflammation, and fibrosis. Am J Physiol Gastrointest Liver Physiol. 2016;310:G726-G738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 114] [Article Influence: 11.4] [Reference Citation Analysis (1)] |
| 11. | Fader Kaiser CM, Romano PS, Vanrell MC, Pocognoni CA, Jacob J, Caruso B, Delgui LR. Biogenesis and Breakdown of Lipid Droplets in Pathological Conditions. Front Cell Dev Biol. 2021;9:826248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 12. | Lin Y, Deng W, Pang J, Kemper T, Hu J, Yin J, Zhang J, Lu M. The microRNA-99 family modulates hepatitis B virus replication by promoting IGF-1R/PI3K/Akt/mTOR/ULK1 signaling-induced autophagy. Cell Microbiol. 2017;19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 78] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 13. | Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, Roggendorf M, Lu M. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology. 2011;53:1476-1485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 14. | Udompap P, Kim WR. Development of Hepatocellular Carcinoma in Patients With Suppressed Viral Replication: Changes in Risk Over Time. Clin Liver Dis (Hoboken). 2020;15:85-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 15. | Idilman IS, Ozdeniz I, Karcaaltincaba M. Hepatic Steatosis: Etiology, Patterns, and Quantification. Semin Ultrasound CT MR. 2016;37:501-510. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 93] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 16. | Jin X, Chen YP, Yang YD, Li YM, Zheng L, Xu CQ. Association between hepatic steatosis and entecavir treatment failure in Chinese patients with chronic hepatitis B. PLoS One. 2012;7:e34198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 17. | Hui RWH, Seto WK, Cheung KS, Mak LY, Liu KSH, Fung J, Wong DK, Lai CL, Yuen MF. Inverse relationship between hepatic steatosis and hepatitis B viremia: Results of a large case-control study. J Viral Hepat. 2018;25:97-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 91] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 18. | Machado MV, Oliveira AG, Cortez-Pinto H. Hepatic steatosis in hepatitis B virus infected patients: meta-analysis of risk factors and comparison with hepatitis C infected patients. J Gastroenterol Hepatol. 2011;26:1361-1367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 19. | Chu CM, Lin DY, Liaw YF. Clinical and virological characteristics post HBsAg seroclearance in hepatitis B virus carriers with hepatic steatosis versus those without. Dig Dis Sci. 2013;58:275-281. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 20. | Lesmana LA, Lesmana CR, Pakasi LS, Krisnuhoni E. Prevalence of hepatic steatosis in chronic hepatitis B patients and its association with disease severity. Acta Med Indones. 2012;44:35-39. [PubMed] |
| 21. | Jarčuška P, Janičko M, Kružliak P, Novák M, Veselíny E, Fedačko J, Senajová G, Dražilová S, Madarasová-Gecková A, Mareková M, Pella D, Siegfried L, Kristián P, Kolesárová E; HepaMeta Study Group. Hepatitis B virus infection in patients with metabolic syndrome: a complicated relationship. Results of a population based study. Eur J Intern Med. 2014;25:286-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 22. | Chen Q, Fang W, Cui K, Chen Q, Xiang X, Zhang J, Zhang Y, Mai K, Ai Q. Endoplasmic reticulum stress induces hepatic steatosis by transcriptional upregulating lipid droplet protein perilipin2. FASEB J. 2021;35:e21900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 17] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 23. | Tian Y, Sir D, Kuo CF, Ann DK, Ou JH. Autophagy required for hepatitis B virus replication in transgenic mice. J Virol. 2011;85:13453-13456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 118] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 24. | Li J, Kemper T, Broering R, Chen J, Yuan Z, Wang X, Lu M. Interferon Alpha Induces Cellular Autophagy and Modulates Hepatitis B Virus Replication. Front Cell Infect Microbiol. 2022;12:804011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 25. | Byrnes K, Blessinger S, Bailey NT, Scaife R, Liu G, Khambu B. Therapeutic regulation of autophagy in hepatic metabolism. Acta Pharm Sin B. 2022;12:33-49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 106] [Article Influence: 26.5] [Reference Citation Analysis (0)] |
| 26. | Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131-1135. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3239] [Cited by in RCA: 3213] [Article Influence: 189.0] [Reference Citation Analysis (0)] |
| 27. | Tsai TH, Chen E, Li L, Saha P, Lee HJ, Huang LS, Shelness GS, Chan L, Chang BH. The constitutive lipid droplet protein PLIN2 regulates autophagy in liver. Autophagy. 2017;13:1130-1144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 189] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 28. | Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci USA. 2010;107:4383-4388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 257] [Article Influence: 16.1] [Reference Citation Analysis (0)] |