Published online May 25, 2022. doi: 10.5501/wjv.v11.i3.144
Peer-review started: December 16, 2021
First decision: February 21, 2022
Revised: February 21, 2022
Accepted: April 26, 2022
Article in press: April 26, 2022
Published online: May 25, 2022
Processing time: 154 Days and 11.4 Hours
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants are currently a new hazard. Since the first appearance of classical SARS-CoV-2 in late 2019, pathogen genetic alterations have continued to occur, and some new hazardous forms have already emerged. The underlying pathophysiological process leading to clinical issue is molecular change caused by genetic mutation.
To determine the change in the interaction between receptor binding domain of omicron variant SARS-CoV-2 and the angiotensin-converting enzyme 2 (ACE2).
The researchers investigated how alterations in the binding area of the SARS receptor CoV2 interacted electrostatically with the ACE2 receptor. In this report, three important coronavirus disease 2019 variants, beta, delta, and omicron, were investigated.
According to this study, there was a change of electrostatic interactions between the receptor binding domain of SARS-CoV-2 with the ACE2 receptor due to each studied variant. The most change was detected in omicron variant followed by delta variant and beta variant.
Our results may support the clinical finding that the omicron variant is more transmissible than the wild type and other variants.
Core Tip: Change of electrostatic interactions between receptor binding domain of severe acute respiratory syndrome coronavirus 2 with the angiotensin-converting enzyme 2 receptor can support the clinical observation that the omicron variant has increased transmissibility compared to the wild type and other variants.
- Citation: Mungmunpuntipantip R, Wiwanitkit V. Omicron variant and change of electrostatic interactions between receptor binding domain of severe acute respiratory syndrome coronavirus 2 with the angiotensin-converting enzyme 2 receptor. World J Virol 2022; 11(3): 144-149
- URL: https://www.wjgnet.com/2220-3249/full/v11/i3/144.htm
- DOI: https://dx.doi.org/10.5501/wjv.v11.i3.144
In late 2019, a novel coronavirus epidemic emerged in Asia and quickly spread throughout the world[1]. A pandemic occurred, resulting in millions of cases of coronavirus disease 2019 (COVID-19) all across the world. The disease has already infected over 200 million individuals worldwide, resulting in millions of deaths. Since the initial appearance of classical severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late 2019, scientists have been keeping a tight eye on the pathogen's genetic mutations all across the world[2]. Several pathogenic genetic mutations have been identified, and several variants have already proven to be troublesome novel variants[2,3].
The delta variant is one of the dangerous mutations that has spread globally[4,5]. Because transmission of the delta variation is higher than that of COVID-19, it can provide a concern in disease control. A newer form, the delta plus variant, has also been discovered, and it is now being considered in clinical practice[6,7]. The impact of novel variations on disease epidemiology and clinical characteristics is interesting. The newest troublesome variant of concern, the omicron variant, was discovered in Africa in November 2021[8]. There are various structural alterations in this new variant molecule. Omicron is spreading in a rapid manner, and many nations have already reported cases[9].
Clinically, the underlying pathophysiological mechanism that can result in a clinical disease is molecular change caused by genetic mutation. The impact of molecular changes is interesting, but it has received little research. The clinical impact of the omicron mutation is unknown. Pathogenesis may change as a result of molecular changes. A change in the interaction between receptor binding domain of SARS-CoV-2 with the ACE2 is an interesting issue. The authors conducted this study to see how mutations are associated with electrostatic interactions between the receptor binding domain of SARS-CoV-2 and the ACE2 receptor. In this report, three important COVID-19 variants, beta, delta, and omicron, are investigated.
The current research is in the field of medical molecular bioinformatics. It is part of a series of experiments aimed at determining the effects of molecular changes in mutants of SARS-CoV-2. The goal of this research is to see how electrostatic interactions between SARS-CoV-2 and ACE2 receptor change according to the emerging variants. For the investigation of change of electrostatic interactions between receptor binding domain of SARS-CoV-2 with the ACE2 receptor, the authors applied a conventional informatics technique, as described in a recent publication[10].
Various protein-protein interactions are known to be dominated by electrostatic interactions[11]. Analysis was performed according to the published protocol[10]. Briefly, we examined the impact of electrostatic interactions on binding energetics. At the molecular level, both molecular mechanics and Monte Carlo simulations were used to assess the interaction between the receptor binding domain of spike viral protein and ACE2. The protein structure was obtained from the protein data bank and used in all computations (PDB ID: 6m17). To begin, the crystal structure was optimized using the python-based open technique[12]. Then, using multiconformation continuum electrostatics[13], rotamers were created, with each rotatable bond rotated by 60 degrees to sample precisely the sidechain conformations. Finally, the Poisson Boltzmann equation was utilized to calculate electrostatic interactions using optimized protein structures with the most occupied conformers[10]. When DELPHI was used to calculate pairwise electrostatic interactions between conformers, it is referred to as DELPHI[10]. The Boltzmann distribution for all conformers was then estimated using Monte Carlo sampling for the WT and altered structures at pH 7 using multiconformation continuum electrostatics. For single and double mutant structures, as well as the wild type, the electrostatic and van der Waals contributions to the interaction energies of SARS-CoV-2/ACE2 were estimated[10].
The research type of SARS-CoV-2 included both wild type and mutation-free SARS-CoV-2. In silico mutation assignment was by PyMol (PyMol, version 2.4). The variants studied are: (1) Beta (K417N, E484K, and N501Y assigned mutations); (2) Delta (T478K, P681R, and L452R assigned mutations); and (3) Omicron (K417N, E484K, and N501Y assigned mutations) (A67V, T95I, G142D, L212I, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, and L981F assigned mutations).
The overall electrostatic interactions value for wild type was derived from the previously mentioned bioinformatic procedure. The already described molecular changes were used for simulation to get the overall electrostatic interactions value for each specific variant. We then calculated the effects of the aforementioned mutations and compared our findings to those of the wild type (native) protein. In brief, the effect of variant on electrostatic interactions was calculated based on a direct comparison to the baseline electrostatic interactions value in wild type. For calculation, the derived overall electrostatic interactions for wild type and each SARS-CoV-2 variant were used as basic parameters. For each type, the change of electrostatic interactions compared to wild type was calculated by the formula “change of electrostatic interactions comparing to wild type = 100 x (electrostatic interactions in that type/ electrostatic interactions of wild type)” and presented in percentage.
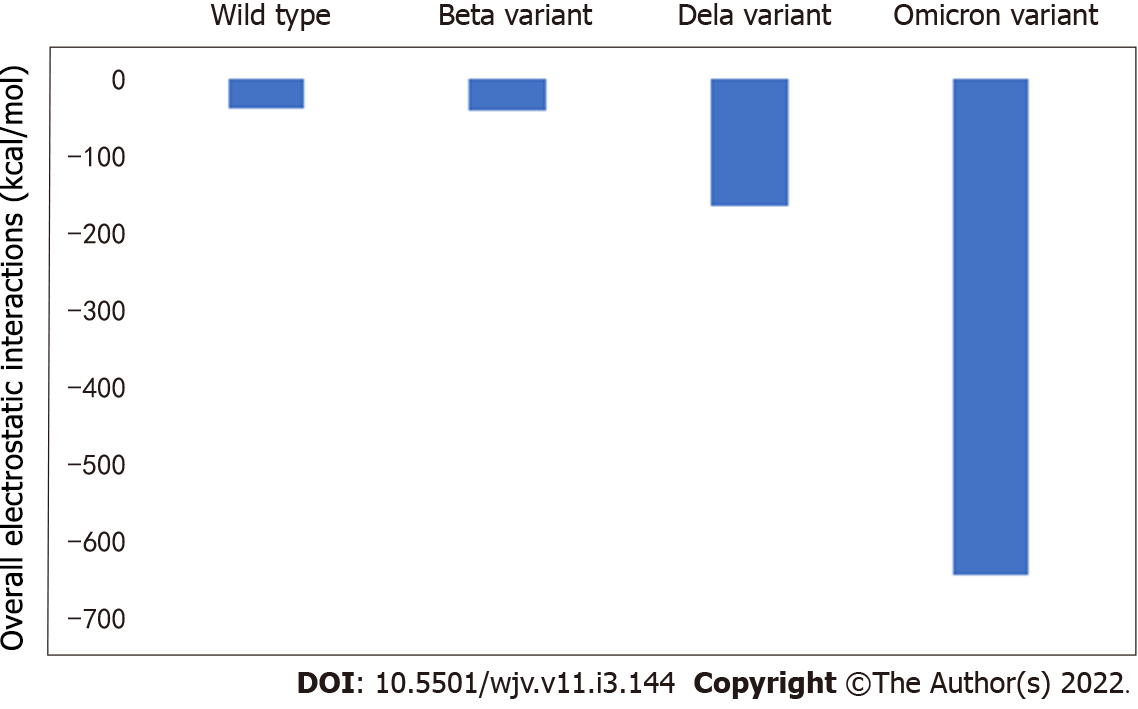
The electrostatic interactions between the receptor binding domain of SARS-CoV-2 with the ACE2 receptor for wild type, beta variant, delta variant, and omicron variant SARS-CoV-2 are presented in Figure 1. The values are equal to -39.38, -41.26, -163.82, and -643.71 kcal/mol, respectively.
There were differences in electrostatic interactions between the receptor binding domain of SARS-CoV-2 with the ACE2 receptor among the variants studied. The most change was detected in the omicron variant, followed by delta variant and beta variant (Table 1).
| Types | Mutations | Electrostatic interactions | |
| Overall, kcal/mol | Change compared to wild type, % | ||
| Wild type | No | -39.38 | 0 |
| Beta variant | T478K, P681R, and L452R | -41.26 | 104.8 |
| Dela variant | T478K, P681R, L452R, and K417N | -163.82 | 416.0 |
| Omicron variant | A67V, T95I, G142D, L212I, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, and L981F | -634.71 | 1611.8 |
In clinical genetics, a genetic change may occur, which may result in a new clinical condition. The clinical problem caused by the pathogen's genetic variation has already been noticed in COVID-19[4,5]. In clinical virology, a mutation in the SARS-CoV-2 virus could occur, and the new variety could be clinically significant. SARS-CoV-2 variations have been reported in a number of places. The changes occur at the receptor-binding region of the spike glycoprotein, which is critical for binding to the ACE2 receptor. The interaction between receptor and SARS-CoV-2 is a significant factor of sickness, according to pathophysiology.
Basically, several alterations have been discovered in the omicron variant's molecular structure. The mutations could lead to a shift in molecular pathogenesis. A key feature, electrostatic interaction with receptor, was evaluated in this study. The ability of SARS-CoV-2 to bind to a receptor is a critical factor in its transmission. There is no doubt that the new variant spreads quickly[7], which can be explained by the change in electrostatic interactions between receptor and SARS-CoV-2.
As a result, measuring changes in virus-receptor electrostatic interactions can help researchers better understand disease pathogenesis. According to this study, there has been a significant change in electrostatic interactions. The change of electrostatic interaction has been well described in the delta variant[10], and a change was also observed in the omicron variant. In delta variant, a replacement due to mutation resulted in electrostatic interaction change, and the increased magnitude of electrostatic interactions corresponded to the increased transmissibility of the virus[14].
According to this study, there is a different change of electrostatic interactions between receptor binding domain of SARS-CoV-2 and the ACE2 receptor due to different SARS-CoV-2 variants. The most change was detected in omicron variant, followed by delta variant and beta variant. According to Table 1, the greatest percentage of change compared to wild type was detected in omicron variant. The greatest degree of change indicates the most changes in electrostatic interactions, which can also indicate major changes in clinical features. When compared to wild type, the omicron variant poses around 16 times more electrostatic interactions, implying a significantly stronger connection between the virus and its receptor.
This finding can support the clinical observation that the omicron variant has an increased transmissibility compared to the wild type and other variants. The data from this preliminary study are useful for explaining the pathogenesis of the omicron variant. Further studies on the detailed flexibility of molecular binding, molecular mass change, and immunological epitope change will add to our under-standing of the virological properties of the variant.
Each studied variant affects the electrostatic interactions between the SARS-CoV-2 receptor binding domain and the ACE2 receptor, according to this study. The omicron form demonstrated the greatest change, followed by the delta and beta variants. These results could support the clinical finding that the omicron variant is more contagious than the wild type and other SARS-CoV-2 variants.
According to this study, each investigated variant altered the electrostatic interactions between the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) receptor binding domain and the angiotensin-converting enzyme 2 (ACE2) receptor. The omicron variant showed the biggest alteration, followed by the delta and beta variants. This finding could back up the clinical observation that the omicron variant is more transmissible than the wild type and other SARS-CoV-2 variants.
Each studied variant affected the electrostatic interactions between the SARS-CoV-2 receptor binding domain and the ACE2 receptor. The omicron form, followed by the delta and beta variants, displays the most change. This could support the clinical finding that the omicron variant is more contagious than the wild type and other SARS-CoV-2 variants.
The authors conducted a study to see how mutations are associated with alterations of electrostatic interactions between receptor binding domain of SARS-CoV-2 with the ACE2 receptor.
The researchers investigated how mutations affect electrostatic interactions between the SARS-CoV-2 receptor binding domain and the ACE2 receptor. In this report, three important coronavirus disease 2019 variants, beta, delta, and omicron, were investigated.
There was a change of electrostatic interactions between the receptor binding domain of SARS-CoV-2 with the ACE2 receptor due to each studied variant compared to wild type. The most change was detected for the omicron variant, followed by delta variant and beta variant.
Our findings can support the clinical observation that the omicron variant has an increased transmissibility comparable to the wild type and other variants.
Our findings are consistent with the clinical observation that the omicron variation is more transmissible than the wild type and other variants.
| 1. | Hsia W. Emerging new coronavirus infection in Wuhan, China: situation in early 2020. Case Study Case Rep. 2020;10:8-9. |
| 2. | Callaway E. Fast-spreading COVID variant can elude immune responses. Nature. 2021;589:500-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 83] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 3. | Lauring AS, Hodcroft EB. Genetic Variants of SARS-CoV-2-What Do They Mean? JAMA. 2021;325:529-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 422] [Article Influence: 84.4] [Reference Citation Analysis (0)] |
| 4. | Hendaus MA, Jomha FA. Delta variant of COVID-19: A simple explanation. Qatar Med J. 2021;2021:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 5. | Torjesen I. Covid-19: Delta variant is now UK's most dominant strain and spreading through schools. BMJ. 2021;373:n1445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 82] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 6. | Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021;600:21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 342] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 7. | Torjesen I. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ. 2021;375:n2943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 205] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 8. | Rahman FI, Ether SA, Islam MR. The "Delta Plus" COVID-19 variant has evolved to become the next potential variant of concern: mutation history and measures of prevention. J Basic Clin Physiol Pharmacol. 2021;33:109-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 40] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 9. | Kannan SR, Spratt AN, Cohen AR, Naqvi SH, Chand HS, Quinn TP, Lorson CL, Byrareddy SN, Singh K. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J Autoimmun. 2021;124:102715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 188] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 10. | Goher SS, Ali F, Amin M. The Delta Variant Mutations in the Receptor Binding Domain of SARS-CoV-2 Show Enhanced Electrostatic Interactions with the ACE2. Med Drug Discov. 2021;100114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 11. | Li B, Deng A, Li K, Hu Y, Li Z, Shi Y, Xiong Q, Liu Z, Guo Q, Zou L, Zhang H, Zhang M, Ouyang F, Su J, Su W, Xu J, Lin H, Sun J, Peng J, Jiang H, Zhou P, Hu T, Luo M, Zhang Y, Zheng H, Xiao J, Liu T, Tan M, Che R, Zeng H, Zheng Z, Huang Y, Yu J, Yi L, Wu J, Chen J, Zhong H, Deng X, Kang M, Pybus OG, Hall M, Lythgoe KA, Li Y, Yuan J, He J, Lu J. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. Nat Commun. 2022;13:460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 226] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 12. | Eastman P, Swails J, Chodera JD, McGibbon RT, Zhao Y, Beauchamp KA, Wang LP, Simmonett AC, Harrigan MP, Stern CD, Wiewiora RP, Brooks BR, Pande VS. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput Biol. 2017;13:e1005659. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 867] [Cited by in RCA: 1863] [Article Influence: 207.0] [Reference Citation Analysis (0)] |
| 13. | Song Y, Mao J, Gunner MR. MCCE2: improving protein pKa calculations with extensive side chain rotamer sampling. J Comput Chem. 2009;30:2231-2247. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 134] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 14. | Pascarella S, Ciccozzi M, Zella D, Bianchi M, Benedetti F, Benvenuto D, Broccolo F, Cauda R, Caruso A, Angeletti S, Giovanetti M, Cassone A. SARS-CoV-2 B.1.617 Indian variants: Are electrostatic potential changes responsible for a higher transmission rate? J Med Virol. 2021;93:6551-6556. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 77] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Infectious diseases
Country/Territory of origin: Thailand
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Casaca W, Brazil; Nazari N, Iran; Ren S, China S-Editor: Ma YJ L-Editor: Filipodia P-Editor: Ma YJ