©The Author(s) 2015.
World J Virology. Aug 12, 2015; 4(3): 265-276
Published online Aug 12, 2015. doi: 10.5501/wjv.v4.i3.265
Published online Aug 12, 2015. doi: 10.5501/wjv.v4.i3.265
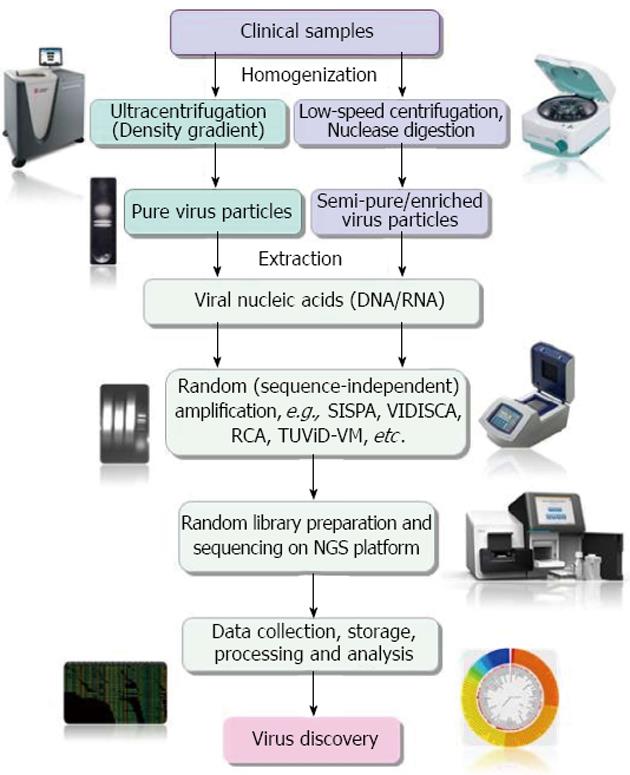
Figure 1 Diagrammatic representation of main steps of clinical virus discovery by next-generation sequencer based technologies.
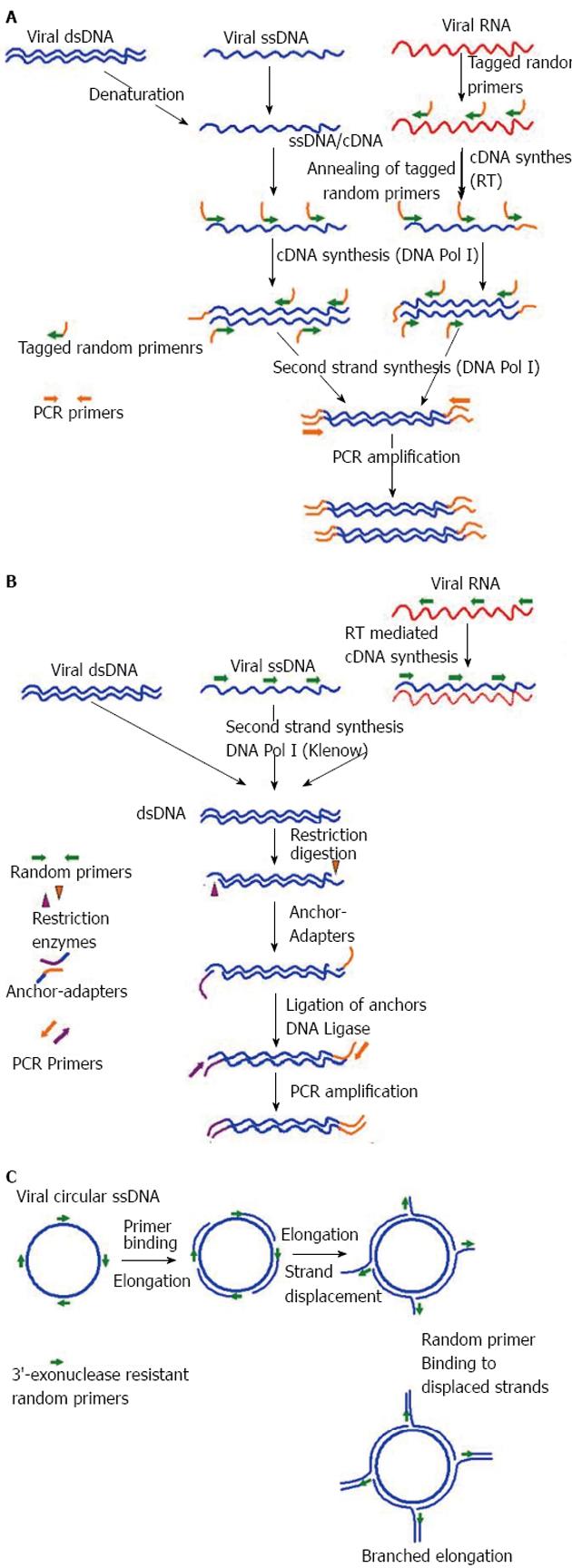
Figure 2 Different virus nucleic acid enrichment techniques.
A: Sequence-independent single-primer amplification. Initially viral RNA and ssDNA is transcribed into complementary DNA (cDNA) using reverse transcriptase (RT) and DNA Pol I respectively, with the help of tagged-primers having defined sequence at the 5’ end while random nucleotides at the 3’ end. Subsequently, second strand synthesis is performed using DNA Pol I (Klenow) to make the cDNA double stranded (dsDNA). Now all the nuceic acids present in the reaction are dsDNA fragments have tagged sequence at their ends. Finally, anchored dsDNA is amplified with primers annealing to the adapter specific sequences, PCR product are checked and ready for analysis through cloning-sequencing or direct sequencing through next-generation sequencers (NGS); B: Virus discovery based on cDNA-AFLP. Initially viral RNA is reverse transcribed into complementary DNA (cDNA) using RT and random primers. Subsequently, second strand synthesis is performed using DNA Pol I (Klenow) to make the cDNA double stranded (dsDNA). In this step, other viral single stranded DNA (ssDNA) viral is also converted to dsDNA. Now all the nuceic acids present in the reaction are dsDNA. In the next step dsDNA are digested with a set of frequent cutter restriction endonucleases, which produce asymmetric cuts. Now specially designed matching anchor-adapters are ligated ends of the restriction fragments using DNA Ligase. Finally, anchored dsDNA is amplified with primers annealing to the adapter specific sequences, PCR product are checked and ready for analysis through cloning-sequencing or direct sequencing through NGS; C: Rolling circle amplification. Amplification of multiply primed single stranded circular viral genomes. 3’-exonuclease resistant primers randomly bind the genome and are elongated by the Phi29 polymerase. The growing strand subsequently displaces the preceding strand of the DNA, making the strand available for binding of random primers and further elongation. This cyclic displacement and elongation leads to a highly branched structure of growing DNA, which is linear in topology. Rolling circle amplification has the capability to specifically enrich the circular ssDNA genomes in an environment of other genetic materials, and could then be characterized by NGS.
- Citation: Datta S, Budhauliya R, Das B, Chatterjee S, Vanlalhmuaka, Veer V. Next-generation sequencing in clinical virology: Discovery of new viruses. World J Virology 2015; 4(3): 265-276
- URL: https://www.wjgnet.com/2220-3249/full/v4/i3/265.htm
- DOI: https://dx.doi.org/10.5501/wjv.v4.i3.265