©2013 Baishideng.
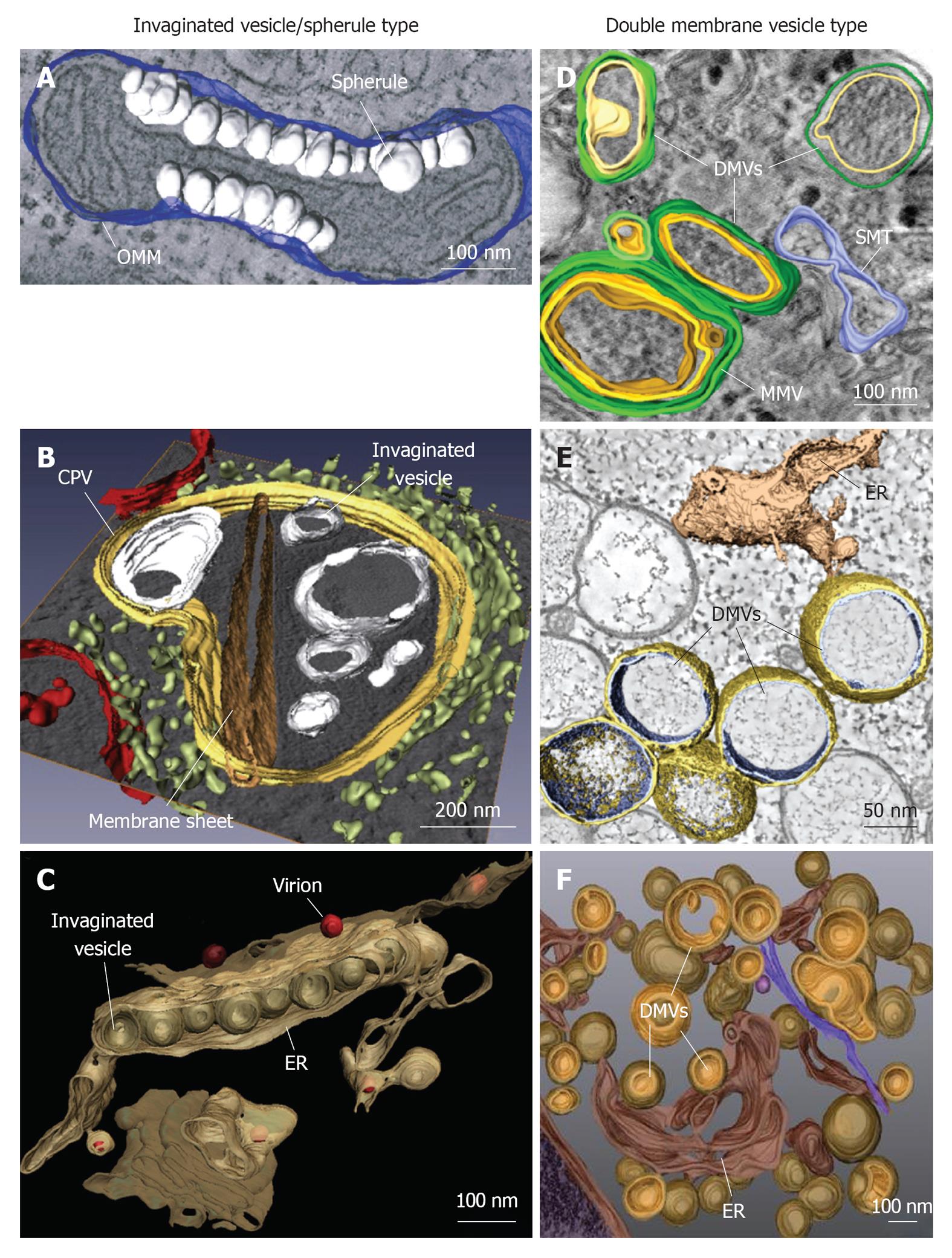
Figure 1 Classification and morphologies of plus-strand RNA virus-induced membrane alterations as revealed by electron tomography and three-dimensional reconstruction.
Morphological similarities of membrane structures belonging to the invaginated vesicle (InV)/spherule type (A-C) or the DMV type (D-F). A: Flock-house virus (FHV); B: Rubella virus (RUBV); C: Dengue virus (DENV); D: Poliovirus (PV); E: Severe acute respiratory syndrome coronavirus (SARS-CoV); F: Hepatitis C virus (HCV). A: FHV-induced spherules (white) in the outer mitochondrial membrane (OMM; blue); B: InVs (white) and rigid membrane sheets (dark brown) in a modified endosome (the cytopathic vacuole, CPV) (yellow) in cells replicating RUBV. The rough endoplasmic reticulum (ER) is shown in light-green and mitochondria in red; C: Spherule-like invaginations (InVs) in the endoplasmic reticulum (ER) (brown) observed in DENV-infected cells. Newly formed progeny virions are shown in red; D: PV infection induces single membrane tubules (SMT) (blue), double membrane vesicles (DMVs) and multi membrane vesicles (MMVs). The DMV outer membrane is shown in green, the second membrane in yellow and a third membrane in case of MMVs in orange. These membranes are primarily derived from the Golgi; E: An interconnected network of ER-derived DMVs is found in SARS-CoV infected cells. The ER is colored in light-brown, interconnected outer membranes of DMVs in yellow and DMV inner membranes in blue; F: HCV-induced DMVs protruding from the ER (dark-brown). Outer membranes of DMVs are shown in light-brown and inner membranes in orange. Images are modified with permission from ©2007 Kopek et al[24] Plos Biol (A); ©2010 Elsevier Inc.[8] (B); ©2009 Elsevier Inc.[10] (C); ©2012 American Society for Microbiology[26] (D); ©2008 Knoops et al[11] Plos Biol (E); ©2012 Romero-Brey et al[28] Plos Pathog (F).
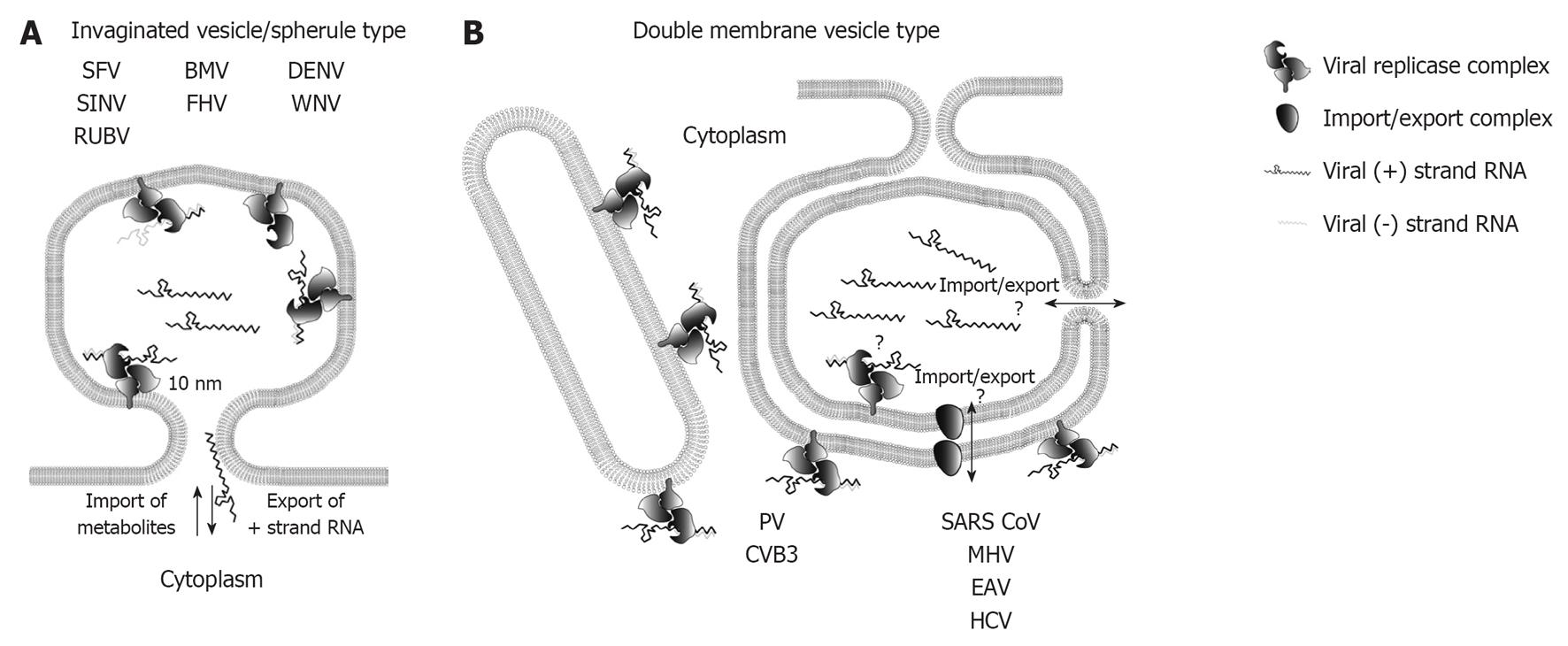
Figure 2 Topology of plus-strand RNA virus replication sites.
A: RNA replication of plus-strand RNA viruses inducing invaginated vesicles/spherules is thought to occur in the spherule lumen. Examples of viruses inducing this type of remodeled membranes are denoted. The neck like opening to the cytoplasm allows export of progeny RNA destined for translation or packaging, as well as import of metabolites required for RNA replication; B: Possible topologies of RNA replication site for double membrane vesicle (DMV)-type factories. Enterovirus replication is thought to take place on the outer membrane of single membrane tubules and DMVs, whereas the exact replication site of nidovirales and hepatitis C virus (HCV) is still unclear as indicated by the question marks. Replication in the DMV lumen would require import and export across two membranes, which could be mediated via a proteinacous transport complex, or via an opening to the cytosol of not yet fully closed DMVs (see main text for details). SFV: Semliki Forrest virus; SINV: Sindbis virus; RUBV: Rubella virus; BMV: Brome mosaic virus; FHV: Flock-house virus; DENV: Dengue virus; WNV: West Nile virus; PV: Poliovirus; CVB3: Coxsackievirus B3; SARS-CoV: Severe acute respiratory syndrome coronavirus; MHV: Mouse hepatitis virus; EAV: Equine arterivirus.
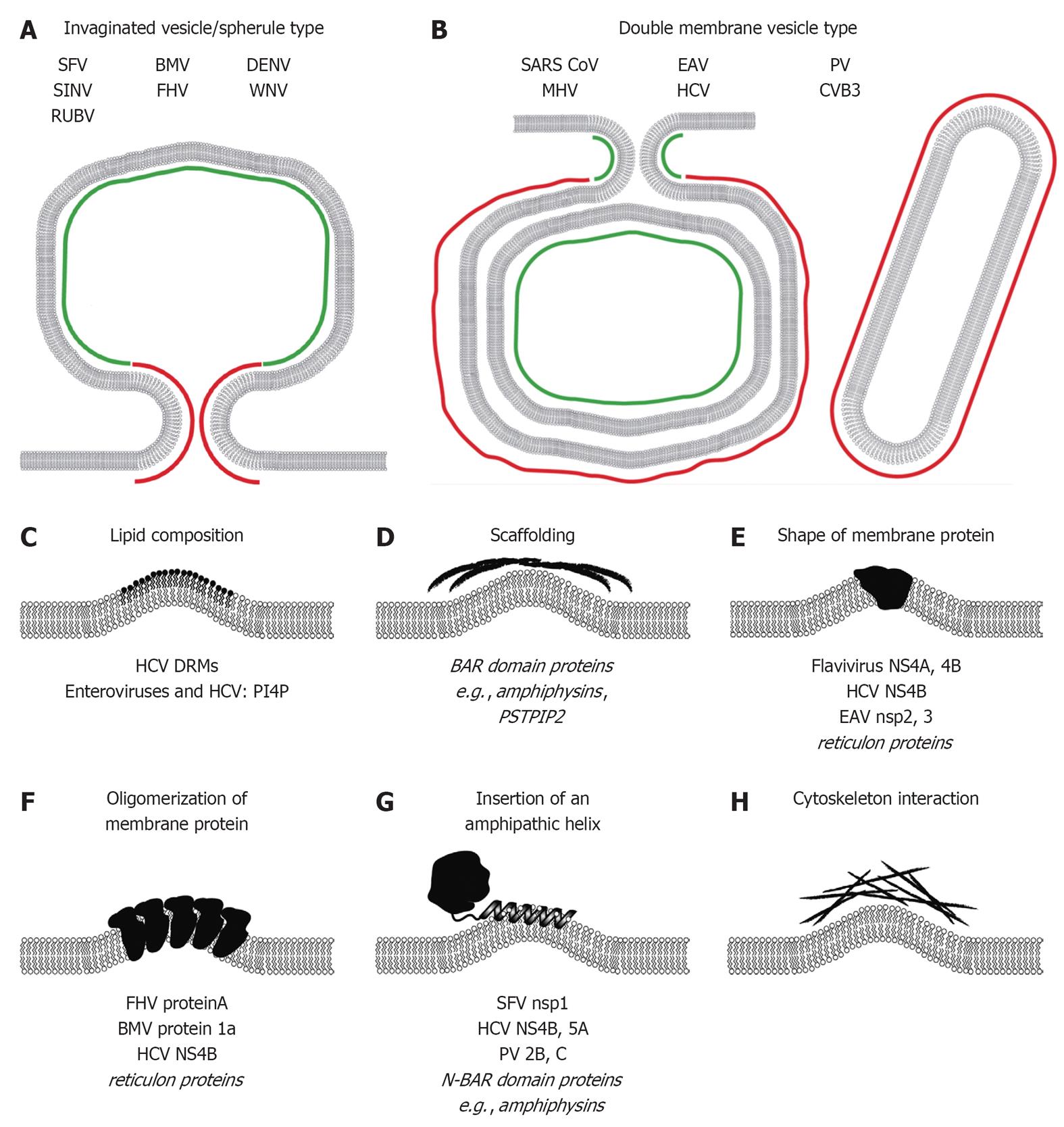
Figure 3 Mechanisms responsible for induction of membrane curvature likely contributing to formation of plus-strand RNA virus replication factories.
Positive and negative membrane curvature for invaginated vesicle/spherule type (A) and double membrane vesicle type (B) replication factories is indicated in red and green, respectively. Mechanisms of membrane modifications/bending are schematically depicted in (C-H). Examples of viral and cellular (in italics) proteins are given at the bottom of each panel (see main text for details). SFV: Semliki Forrest virus; SINV: Sindbis virus; RUBV: Rubella virus; BMV: Brome mosaic virus; FHV: Flock-house virus; DENV: Dengue virus; WNV: West Nile virus; SARS-CoV: Severe acute respiratory syndrome coronavirus; MHV: Mouse hepatitis virus; EAV: Equine arterivirus; HCV: Hepatitis C virus; PV: Poliovirus; CVB3: Coxsackievirus B3; DRM: Detergent-resistant-membrane; BAR: Bin-Amphiphysin-Rvs; PSTPIP2: Proline-serine-threonine phosphatase interacting protein 2; NS4A: Non-structural protein 4A; NS4B: Non-structural protein 4B; N-BAR: N-terminal amphipathic helix containing BAR; PI4P: Phosphatidylinositol-4-phosphate.
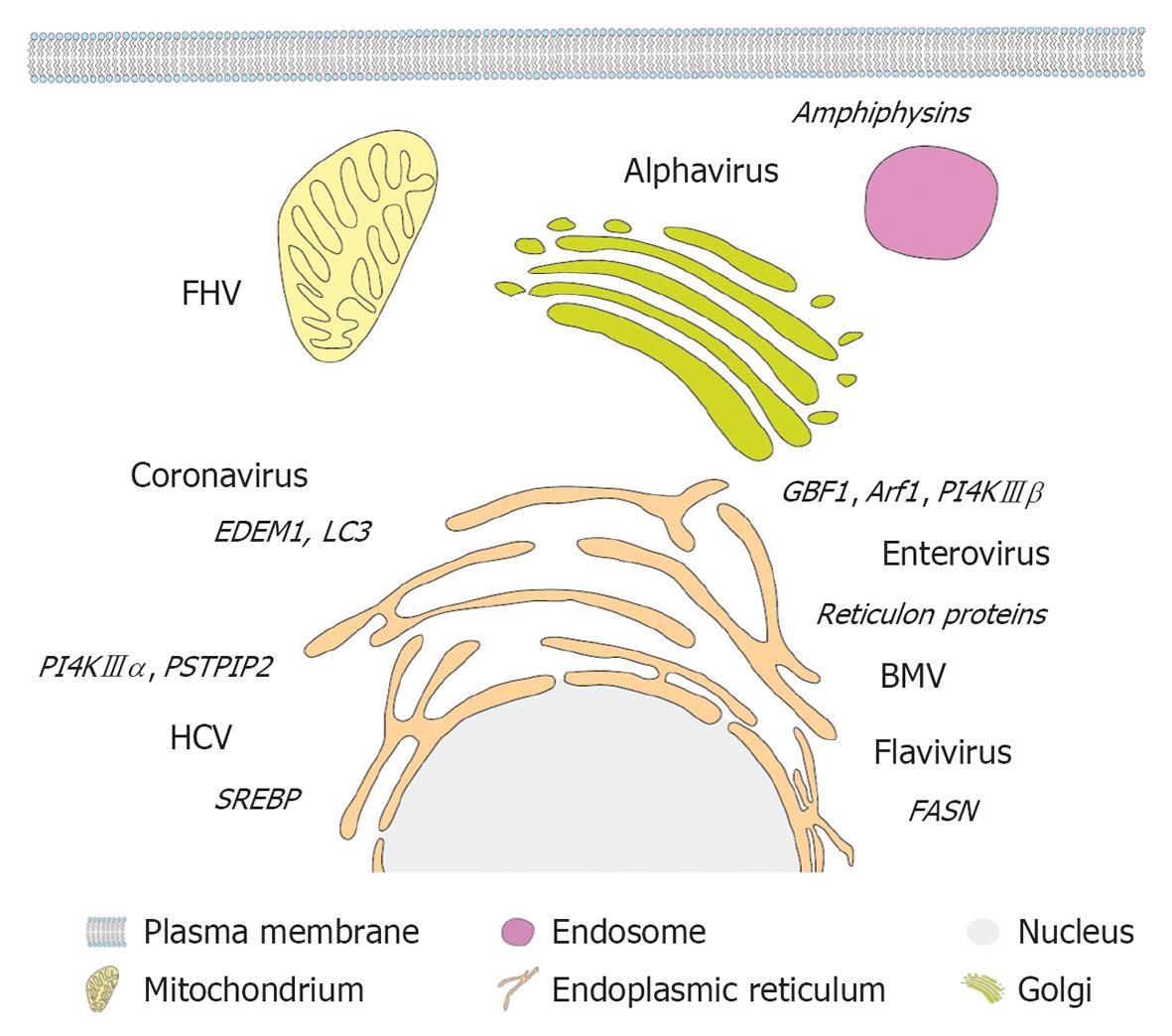
Figure 4 Primary membrane source and host cell factors subverted by different plus-strand RNA viruses.
A eukaryotic cell and (endo)membrane organelles are depicted schematically as indicated on the bottom. Viruses are displayed next to their primary membrane source organelle and recruited host cell factors involved in lipogenesis and membrane remodeling are denoted in italics adjacent to each virus name. FHV: Flock-house virus; PSTPIP2: Proline-serine-threonine phosphatase interacting protein 2; HCV: Hepatitis C virus; SREBP: Sterol regulatory element-binding protein; PI4K: Phosphatidylinositol-4-kinase; GBF1: Golgi-specific BFA-resistance guanine nucleotide-exchange factor 1; BMV: Brome mosaic virus; FASN: Fatty acid synthase.
- Citation: Paul D, Bartenschlager R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol 2013; 2(2): 32-48
- URL: https://www.wjgnet.com/2220-3249/full/v2/i2/32.htm
- DOI: https://dx.doi.org/10.5501/wjv.v2.i2.32