Published online Aug 19, 2024. doi: 10.5498/wjp.v14.i8.1148
Revised: June 13, 2024
Accepted: July 9, 2024
Published online: August 19, 2024
Processing time: 129 Days and 20.7 Hours
Precision medicine is transforming psychiatric treatment by tailoring personalized healthcare interventions based on clinical, genetic, environmental, and lifestyle factors to optimize medication management. This study investigates how artificial intelligence (AI) and machine learning (ML) can address key challenges in integrating pharmacogenomics (PGx) into psychiatric care. In this integration, AI analyzes vast genomic datasets to identify genetic markers linked to psychiatric conditions. AI-driven models integrating genomic, clinical, and demographic data demonstrated high accuracy in predicting treatment outcomes for major depre
Core Tip: This paper explores the convergence of precision medicine and artificial intelligence (AI) in psychiatric care, focusing on tailoring treatments to individuals' genetic backgrounds. It underscores the complexity of psychiatric disorders, attributed to varied genetic, environmental, and lifestyle factors, and the role of AI in navigating these challenges by analyzing large genomic datasets. Despite obstacles such as data privacy, computational requirements, and model generalization, the study highlights the necessity for ethical guidelines and regulatory frameworks for AI use in psychiatric genetics. Furthermore, it stresses the importance of interdisciplinary collaboration to effectively address the AI-related implementation challenges in precision medicine.
- Citation: Okpete UE, Byeon H. Challenges and prospects in bridging precision medicine and artificial intelligence in genomic psychiatric treatment. World J Psychiatry 2024; 14(8): 1148-1164
- URL: https://www.wjgnet.com/2220-3206/full/v14/i8/1148.htm
- DOI: https://dx.doi.org/10.5498/wjp.v14.i8.1148
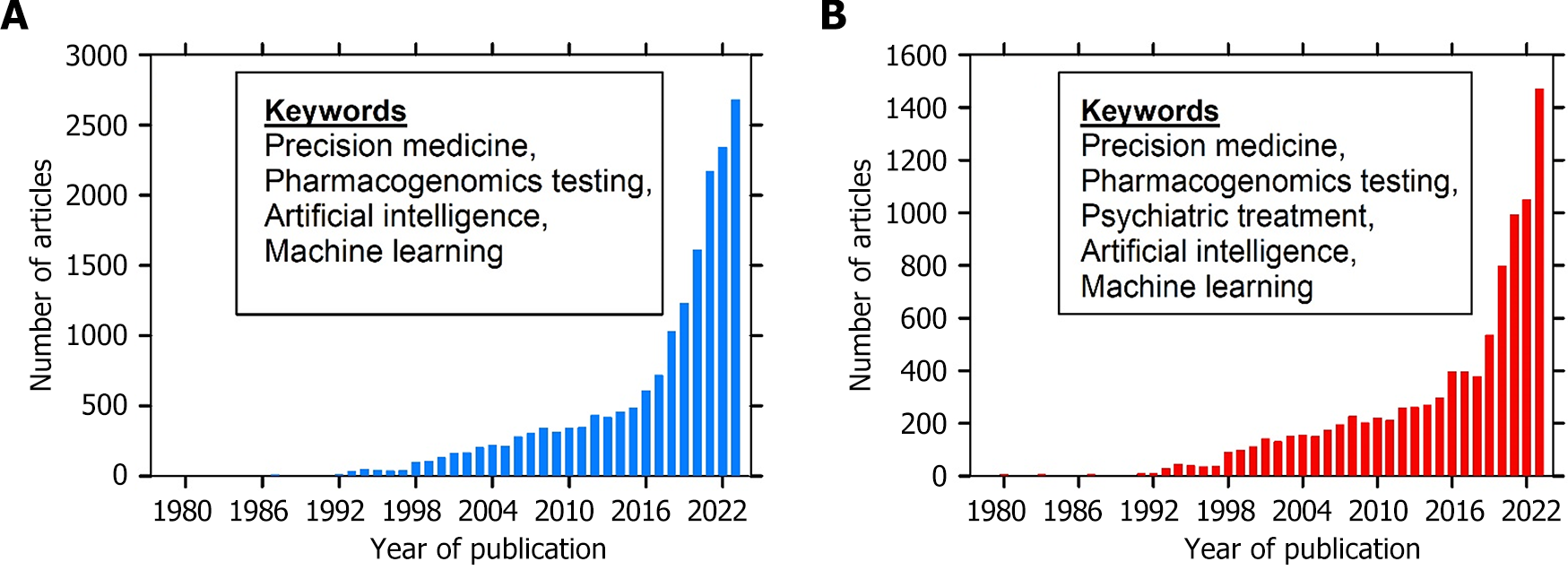
Precision medicine, characterized by the customization of healthcare interventions based on individual variability in genes, environment, and lifestyle, is revolutionizing various medical fields, including psychiatry, through improved informed clinical decision-making. Psychiatric disorders have complex etiology (both internal and external stressors) and heterogeneous clinical presentations that pose significant challenges to traditional treatment approaches[1,2]. Conventional therapies for refractory psychiatric disorders often fail to achieve remission in most cases, mainly due to the extended period required to observe any therapeutic benefits and their significant societal cost[3,4]. Hence, a more comprehensive understanding of the physiological mechanisms underlying mental illness is needed. The advent of the genomic revolution, driven by landmark projects such as the Human Genome Project, has provided unprecedented opportunities to unravel the genetic underpinnings of psychiatric disorders[5]. In recent years, there has been a paradigm shift toward leveraging genomic insights to enhance the diagnosis and treatment of psychiatric disorders. This is exemplified by the increasing number of published articles in this field (Figure 1). Advancements in genomic research have elucidated the genetic architecture of psychiatric illnesses and treatment response in patients with psychiatric disorders, particularly by identifying candidate genes, including transporter and receptor genes, implicated in disease pathogenesis and determining the efficacy of therapeutic intervention[6]. Clinicians and researchers in academia and biotech industries devote time to the development of interventions related to personalized treatment[7-9]. These discoveries have paved the way for pharmacogenomic (PGx) approaches aimed at predicting individual drug treatment outcomes, thus showing the potential of personalized therapy.
Genomic psychiatric treatment focuses on using an individual's genome to diagnose and treat mental health conditions. Using gene profiles from PGx testing can improve outcomes of psychiatric conditions and reduced cost of care for patients with a prior history of inadequate clinical response[10,11]. Guidelines concerning the utilization of PGx tests to inform dosing of commonly prescribed antidepressants and antipsychotics were published in several respected data-sharing consortia[12,13] and curated knowledge databases with comprehensive resources [e.g., Pharmacogenetics Knowledgebase (PharmGKB) and Sequence2Script] describing how genetic variations influence drug response[14,15]. Additionally, pharmacogenetics information approved by drug labels and testing agencies, such as the Food and Drug Administration (FDA) in the United States or the European Medicines Agency in Europe, is well documented in medication leaflets providing guidance for patients with a particular genotype/metabolizer phenotype taking related prescribed drugs. This information includes specific dosage recommendations, potential drug interactions, and guidance on the likelihood of therapeutic failure. Despite the considerable potential of genomic psychiatric treatment, its routine implementation into clinical practice remains largely unstandardized, inconsistent, and unregulated[16]. The key challenges associated with PGx testing for psychiatric medications include the identification of relevant genes and alleles, ensuring tests are accredited according to recognized standards, maximizing the clinical usefulness of PGx testing through careful interpretation and implementation of test results for medication selection, and ensuring that prescribing recommendations are supported by evidence-based guidelines [e.g., Clinical Pharmacogenetics Implementation Consortium (CPIC), Dutch Pharmacogenetics Working Group (DPWG), FDA, and Health Canada] to avoid potentially harmful outcomes associated with recommendations lacking sufficient clinical validity[17]. For example, of > 25 gene-based drug dosing guidelines developed by expert committees and published in guideline sources, only 7 genes (
| Drug category | Drug name | Gene | Genotype group | Pharmgkb top FDA label testing level | Available clinical guidelines | Major clinically relevant drug-gene interactions |
| Anti-dementia drugs | Donepezil | CYP2D6 | UM or PM | Actionable PGx | No data | May result in altered systemic concentrations |
| Anti-dementia drugs | Galantamine | CYP2D6 | PM | Informative PGx | No data | Results in higher drug exposure compared to NMs |
| Antidepressants | Amitriptyline | CYP2C19 | UM, IM or PM | No data | CPIC, DPWG | May result in altered conversion of tertiary amines to secondary amines |
| Antidepressants | Amitriptyline | CYP2D6 | UM, IM or PM | Actionable PGx | CPIC, DPWG | May result in altered systemic concentrations |
| Antidepressants | Bupropion | CYP2D6 | Testing required | Potential drug-drug interaction | ||
| Antidepressants | Citalopram | CYP2C19 | PM | Actionable PGx | CPIC, DPWG | Results in higher drug exposure and higher risk of adverse reaction (QT prolongation) compared to NMs |
| Antidepressants | Clomipramine | CYP2C19 | UM | Actionable PGx | CPIC, DPWG | Results in decreased drug exposure and increased risk of ineffectiveness compared to NMs |
| Antidepressants | Clomipramine | CYP2D6 | PM | Actionable PGx | CPIC, DPWG | May result in altered systemic concentrations |
| Antidepressants | Desvenlafaxine | CYP2D6 | PM | Informative PGx | CPIC | No difference in plasma concentration from NMs |
| Antidepressants | Doxepin | CYP2C19 | IM or PM | Actionable PGx | CPIC, DPWG | Results in higher drug exposure compared to NMs |
| Antidepressants | Doxepin | CYP2D6 | UM, IM or PM | Actionable PGx | CPIC, DPWG | May result in altered systemic concentrations |
| Antidepressants | Duloxetine | CYP2D6 | PM | Actionable PGx | CPIC, DPWG | Potential drug-drug Interaction. May result in higher drug exposure |
| Antidepressants | Escitalopram | CYP2C19 | UM, IM or PM | Actionable PGx | CPIC, DPWG | May result in altered systemic concentrations |
| Antidepressants | Fluvoxamine | CYP2D6 | PM | Actionable PGx | CPIC, DPWG | Results in higher drug exposure compared to NMs |
| Antidepressants | Imipramine | CYP2C19 | PM | No data | DPWG | Results in higher drug exposure and higher risk of adverse reaction compared to NMs. Avoid use in PMs |
| Antidepressants | Imipramine | CYP2D6 | UM, IM or PM | Actionable PGx | CPIC, DPWG | May result in altered systemic concentrations |
| Antidepressants | Nortriptyline | CYP2D6 | UM, IM or PM | Actionable PGx | CPIC, DPWG | May result in altered systemic concentrations |
| Antidepressants | Paroxetine | CYP2D6 | UM, IM or PM | Criteria Not Met | CPIC, DPWG | May result in altered systemic concentrations |
| Antidepressants | Sertraline | CYP2C19 | PM | No data | CPIC, DPWG | Results in higher drug exposure and higher risk of adverse reaction compared to NMs |
| Antidepressants | Venlafaxine | CYP2D6 | PM | Actionable PGx | CPIC, DPWG | Results in altered parent drug and metabolite concentrations |
| Antidepressants | Vortioxetine | CYP2D6 | PM | Actionable PGx | CPIC | Results in higher drug exposure compared to NMs |
| Antidepressants | Amoxapine | CYP2D6 | UM, IM or PM | Actionable PGx | No data | May result in altered systemic concentrations |
| Antidepressants | Desipramine | CYP2D6 | UM, IM or PM | Actionable PGx | CPIC | May result in altered systemic concentrations |
| Antidepressants | Trimipramine | CYP2D6 | UM, IM or PM | Actionable PGx | CPIC | May result in altered systemic concentrations |
| Antiepileptics | Carbamazepine | HLA-A | *31: 01 positive | Actionable PGx | CPIC, DPWG, CPNDS | Results in higher risk of adverse reaction risk (severe skin reactions) compared to NMs |
| Antiepileptics | Carbamazepine | HLA-B | *15: 02 positive | Testing required | CPIC, DPWG, CPNDS | Results in higher risk of adverse reaction risk (severe skin reactions) compared to NMs. Consider alternative therapies, or use only if potential benefits outweigh risks |
| Antiepileptics | Lamotrigine | HLA-B | *15: 02 positive | No data | DPWG | Results in higher risk of adverse reaction risk (lamotrigine-induced SJS/TEN). Avoid use in patients with *15: 02 positive allele |
| Antiepileptics | Oxcarbazepine | HLA-B | *15: 02 positive | Testing required | CPIC, DPWG | Results in increased risk of adverse reaction (severe skin reactions) |
| Antiepileptics | Phenytoin | CYP2C9 | IM or PM | Testing recommended | CPIC, DPWG | May result in higher drug exposure and higher risk of adverse reaction (CNS toxicity) compared to NMs |
| Antiepileptics | Phenytoin | HLA-B | *15: 02 positive | Testing recommended | CPIC, DPWG | May result in higher risk of adverse reaction (SJS/TEN) compared to NMs |
| Antiepileptics | Valproic acid | POLG | A467T and W748S mutations | Testing required | No data | Results in increased risk of adverse reaction (acute liver failure and resultant deaths). The use is contraindicated in patients with POLG mutations |
| Antimigraine preparations | Clonidine | CYP2D6 | UM, IM or PM | No data | DPWG | No significant effect (No recommendation). Possible alternative for atomoxetine in variant CYP2D6 metabolisers |
| Antipsychotics | Aripiprazole | CYP2D6 | PM | Actionable PGx | DPWG | Results in higher drug exposure compared to NMs and higher risk of adverse reaction |
| Antipsychotics | Clozapine | CYP2D6 | PM | Actionable PGx | DPWG | Results in higher drug exposure compared to NMs |
| Antipsychotics | Haloperidol | CYP2D6 | UM or PM | Actionable PGx | DPWG | Results in increased risk of adverse reaction In PMs and higher risk of reduced effectiveness In UMs |
| Antipsychotics | Olanzapine | CYP2D6 | PM | Informative PGx | DPWG | No significant effect (No recommendation) |
| Antipsychotics | Paliperidone | CYP2D6 | PM | Informative PGx | No data | No significant difference in exposure or clearance compared to NMs |
| Antipsychotics | Perphenazine | CYP2D6 | PM | Actionable PGx | No data | Results in higher drug exposure and higher risk of adverse reaction compared to NMs |
| Antipsychotics | Pimozide | CYP2D6 | PM | Testing required | DPWG | Results in higher drug exposure compared to NMs |
| Antipsychotics | Quetiapine | CYP3A4 | PM | No data | DPWG | Results in decreased conversion of systemic parent drug (quetiapine) to the active metabolite. Use alternative therapy |
| Antipsychotics | Risperidone | CYP2D6 | UM, IM or PM | Informative PGx | DPWG | Results in altered parent drug and metabolite concentrations |
| Anxiolytics | Clobazam | CYP2C19 | IM or PM | Actionable PGx | No data | Results in increased active metabolite concentrations and increased risk of adverse reaction as compared to NMs |
| Anxiolytics | Diazepam | CYP2C19 | PM | Actionable PGx | No data | May result in altered systemic concentrations |
| Psycholeptics and psychoanaleptics in combination | Fluoxetine | CYP2D6 | PM | Actionable PGx | CPIC, DPWG | Results in higher drug exposure compared to NMs |
| Psycholeptics and psychoanaleptics in combination | Fluoxetine | FKBP5 | PM | Actionable PGx | CPIC, DPWG | Results in higher drug exposure compared to NMs |
| Psychostimulants, agents used for adhd and nootropics | Atomoxetine | CYP2D6 | PM | Actionable PGx | CPIC, DPWG | Results in higher drug exposure and higher risk of adverse reaction compared to NMs |
| Psychostimulants, agents used for adhd and nootropics | Modafinil | CYP2D6 | PM | Actionable PGx | No data | May require dose modification when administered with medication metabolized by CYP2C19 |
Artificial intelligence (AI) and machine learning (ML) are two advanced computational techniques used extensively in analyzing large data volumes to identify patterns that can be used to make informed decisions about a patient's treatment[18]. ML uses data and algorithms to train a model to make predictions or decisions. Both AI and ML enable machines to learn and improve their performance on a specific task, playing a pivotal role in advancing genomic psychiatry treatment, particularly within the realm of PGx testing. AI and ML are two broad but similar concepts that are often used inter
Precision medicine is particularly important in psychiatry due to the complex and elusive nature of mental illnesses, which often defy traditional Diagnostic and Statistical Manual of Mental Disorders (DSM)-5 and ICD-10 categorical classification frameworks and exhibit extensive clinical heterogeneity. Unlike other fields where group-level analyses suffice, psychiatric disorders necessitate a personalized approach as conventional case-control designs often fail to account for individual-level brain abnormalities essential for accurate diagnosis and tailored treatment strategies[19,20]. Precision medicine aims to address psychiatric treatment selection by aligning individual disease characteristics with precise treatment approaches. Thus, clinicians can tailor treatments to specific patient groups by considering genetic, environmental, and lifestyle factors.
Several innovative approaches have been explored to improve treatment selection. Pharmacogenetics stands out as a crucial tool for clinicians, allowing for tailored pharmacological treatment based on DNA analysis of polymorphisms within key gene sequences[18,21]. Various factors, including issues related to compliance and genetic variations, contribute to the absence of a positive response to initial psychiatric medication in a significant proportion of patients[22]. Genetic modifications are estimated to account for a substantial portion of treatment response variability. Consequently, finding an effective medication through multiple trials can be time-consuming and frustrating for patients, leading to discontinuation of treatment and exacerbation of symptoms. Nine genes central to recent PGx investigations belong to three main categories: The cytochrome P450 family (CYP1A2, CYP2B6, CYP2C19, CYP2D6, CYP3A4, and CYP3A5), genes related to the serotonergic pathway (SLC6A4 and HTR2A), and those associated with the dopaminergic pathway (DRD2), of which 3 have shown inconclusive evidence (DRD2, SLC6A4, and HTR2A). The cytochrome P450 enzymes, responsible for drug metabolism, exhibit various phenotypes based on genetic polymorphisms, potentially influencing individual responses to psychotropic medications. Similarly, polymorphisms in serotonin transporter and receptor genes play a significant role in the pharmacodynamics of selective serotonin reuptake inhibitors (SSRIs). Additionally, variations in DRD2 can impact medication responses, particularly in the treatment of psychotic disorders. Computational models offer promising solutions to address the complexities of psychiatric treatment selection. For instance, the Antidepressant Response Prediction Network (ARPNet) utilizes a neural network incorporating various biomarkers and genetic factors to predict antidepressant response[23,24]. Genomic research also identified numerous genetic variants associated with antidepressant response and side effects. Understanding the influence of genetic variation on drug metabolism enzymes, such as cytochrome P450 enzymes (CYP2D6 and CYP2C19), offers opportunities for tailored treatment strategies based on individual genotypes. Moreover, polygenic risk-scoring frameworks integrating single nucleotide polymorphisms (SNPs) could revolutionize treatment selection by predicting outcomes ranging from treatment response to drug metabolism, potentially transforming psychiatric care. However, further validation and integration into clinical practice are necessary. Nevertheless, they represent significant strides toward personalized treatment considering individual genetic profiles, ultimately enhancing the efficacy and precision of psychiatric care.
Accurate identification and prioritization of genetic variants play a crucial role in PGx testing. Until the advent of precision medicine, previous methods heavily relied on in-silico prediction of significant pharmacogenes affecting response to or involved in the metabolism of one or multiple drugs, leading to limited sensitivity and challenges in result interpretation. Thus, annotating each variant and providing evidence for prioritization is crucial to address this issue. Several pharmacogenes with acceptable evidence of association with response to psychiatric medication are listed based on CPIC, PharmGKB, and DPWG[25]. For instance, CYP2D6 enzyme (responsible for the metabolism of most psychiatric medications, including amitriptyline, aripiprazole, atomoxetine, clomipramine, desipramine, doxepin, fluvoxamine, and haloperidol) has several genetic variants leading to individual variations in drug response. These genetic variants, including gene deletion, gene duplications, and hybrid alleles comprising portions of CYP2D6 and CYP2D7, are categorized into tiers by the American College of Medical Genetics and Genomics genome interpretation guidelines based on functional characterization, allele frequency, availability of reference materials, and technical feasibility for clinical detection. Tier classification facilitates the management of psychiatric medications by aiding in the identification of individuals with different metabolizing capacities, which can be assessed using explainable AI techniques[26]. For instance, Tier 1 alleles with known functional effects and significant prevalence are crucial for guiding medication dosing and selection. Conversely, Tier 2 alleles, including hybrid alleles, are important in treatment decision-making, especially when considering potential interactions and side effects, despite being technically challenging to characterize. Thus, clinicians can integrate genetic information and tier classifications to optimize medication regimens tailored to individual patient profiles by leveraging precision medicine (which relies heavily on AI or ML algorithms to analyze large volumes of diverse data), ultimately enhancing treatment efficacy and safety in psychiatric care.
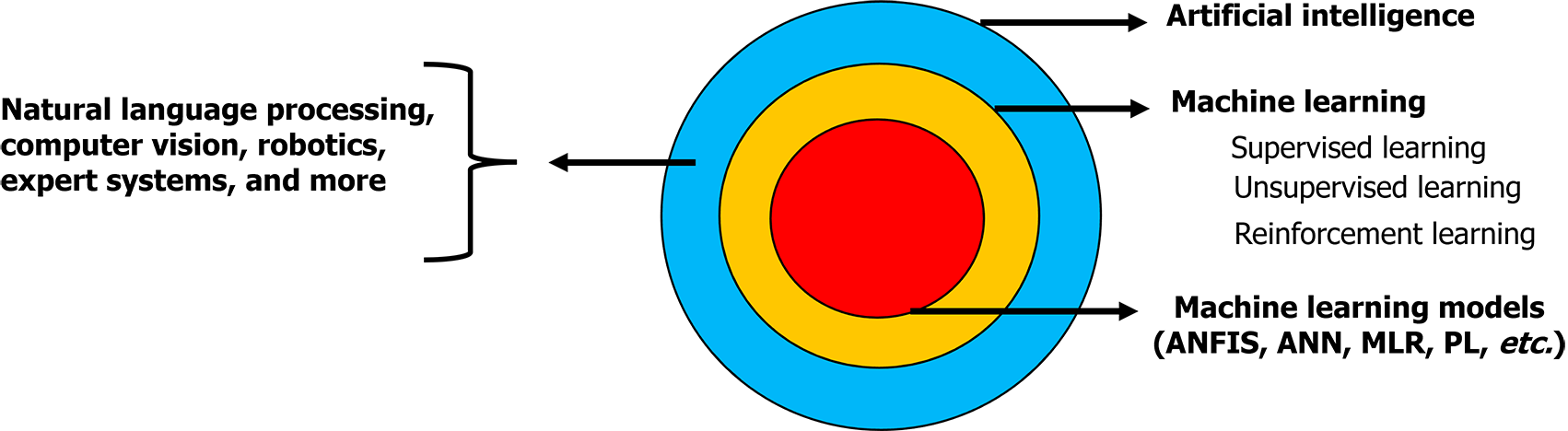
Although AI and ML are closely related concepts, the key difference between them is that AI encompasses other techniques, such as natural language processing, computer vision, robotics, and expert systems, in addition to ML (Figure 2).
ML techniques encompass various methods, such as random forest (RF), support vector machine (SVM), gradient boosting, and extreme gradient boosting. These methods can be employed to detect and evaluate genetic risk factors in individuals, particularly those susceptible to diseases. ML algorithms can be categorized into supervised, unsupervised, and reinforcement learning (Table 2), each serving different learning scenarios and tasks[27-29]. In supervised learning, the algorithm learns from labeled examples to train a model to predict future outcomes with high accuracy. Supervised learning algorithms, such as RF, SVM, and artificial neural networks, can assess databases of patients with psychiatric disorders to predict treatment responses and hospital readmission[27,30]. These algorithms aid clinicians in selecting personalized therapies by analyzing gene expression data and genetic variants associated with altered responses to psychiatric medication. Unsupervised learning techniques, unlike supervised learning, aim to unveil underlying patterns, relationships, or groupings inherent within the data. This approach is commonly utilized for exploratory data analysis, dimensionality reduction, and clustering tasks. It serves as a powerful tool for deriving insights from unlabeled datasets, enabling the extraction of meaningful information without prior guidance. Various algorithms are utilized in unsupervised learning, including k-means clustering, hierarchical clustering, principal component analysis, and auto-encoders, assisting in identifying patterns and relationships within patient data and contributing to treatment planning and prognostic analyses[28,31]. Reinforcement learning algorithms, such as Q-learning and policy gradients, optimize treatment selection by maximizing cumulative rewards over time, particularly in diseases such as bipolar disorder in which predicting lithium treatment response is critical. A decision tree rule using ML methods, such as RF, decision trees, and elastic nets, can also be applied to forecast antidepressant treatment responses, featuring the multifaceted approach to individualized psychiatric care.
| Machine learning category | Description | Machine learning techniques used | Algorithm | Uses | Ref. |
| Supervised learning | Supervised learning algorithm learns from labelled examples to train a model to predict future outcomes with high accuracy | Random forests, support vector machines, artificial neural networks | Classification, regression, sequence labelling | Predict treatment responses based on genomic profiles, aid in therapy selection | Nasteski et al[27] |
| Unsupervised learning | Unsupervised machine learning discerns patterns in unlabelled datasets to predict relationships and meaningful patterns | K-means clustering, principal component analysis | Clustering, dimensionality reduction | Identify patterns and relationships within patient data for treatment planning and prognostic analyses | Ghahramani[28] |
| Reinforcement learning | Reinforcement learning integrates user feedback to refine decision-making, enhancing the model's performance | Q-learning, Policy gradients | Sequential decision making | Optimize treatment selection by maximizing cumulative rewards over time | Sutton et al[29] |
The main idea of utilizing AI and ML in the PGx approach to psychiatric treatment is to assist clinicians in selecting medications that offer optimal efficacy with minimal side effects. Past research demonstrated the utility of ML algorithms across a spectrum of psychiatric disorders, including Alzheimer's disease, autism, major depressive disorder (MDD), and schizophrenia. A critical analysis of AI applications revealed diverse methodologies employed for disease prediction, biomarker identification, genetic feature prioritization, and selection of psychiatric medication[10,11,32-35]. Studies utilized raw genomic data, including RNA sequencing (RNA-seq) and whole-genome sequencing (WGS/WES), in conjunction with ML algorithms to identify genetic variants associated with treatment response. For instance, a predictive model based on differential gene expression analysis was developed to classify male and female lithium responders using gene expression data from the Lithium Treatment-Moderate dose Use Study. The Linear Models for Microarray and RNA-Seq (limma) package in R was utilized to explore how specific genes linked to lithium response variability relate to psychiatric symptomatology in bipolar disorders. Results showed that genes RBPMS2 and LILRA5 effectively classified male lithium responders, while ABRACL, FHL3, and NBPF14 genes classified female lithium responders. The predictive models showed high sensitivity (96% for males and 92% for females) in identifying lithium responders based on pre-treatment gene expression signatures[36].
Moreover, AI techniques for predicting drug responses in psychiatric treatments are still in their early stages due to limited human studies investigating predictive models for treatment evaluation. Both conventional AI methods and deep learning approaches have been employed to predict antidepressant responders using genetic analysis datasets and demographic and clinical information. In a comprehensive literature review focusing on ML and deep learning in PGx studies related to antidepressant treatment in MDD, researchers identified predictive genomic variants and biomarkers associated with antidepressant treatment outcomes. They introduced the concept of intermediate endophenotypes to quantify behavioral phenotypes, establishing potential connections between genes and behavior[16]. These endophenotypes represent quantitative biomarkers of brain activity acquired through neuroimaging, offering insights into neurobiological changes related to psychiatric disorders and treatment.
Research combining PGx and neuroimaging, especially for antidepressant outcomes in MDD, remains scarce. Studies such as the ARPNet employed traditional ML techniques (linear regression model) to identify key predictive features from patient data, achieving an 84% accuracy in recommending effective antidepressants[37]. The ARPNet model helped doctors prescribe the most suitable antidepressant for patients by identifying common features among patients and recognizing comparable treatment outcomes from previous patient records. Additionally, Pei et al[38] demonstrated that a neuroimaging PGx approach, combining functional magnetic resonance imaging (MRI) data with multi-omics data, successfully predicted an early response to antidepressant treatment within the first two weeks. They initially identified key predictive variables using the SVM-recursive feature elimination model, including three SNPs (DRD5 rs1967550, HTR2C rs1801412, and TOR1A rs3842225) and six brain regions of interest in functional MRI data. Subsequently, an SVM algorithm was applied to achieve an 86% accuracy in predicting early-stage antidepressant treatment response[38].
Furthermore, deep learning architecture was applied in MDD to estimate individual-specific antidepressant response prediction. Lin et al[39] utilized a deep learning framework integrating diverse data types, including genetic (SNPs), demographic (marital status, age, and sex), and clinical datasets (baseline Hamilton Rating Scale for Depression, depressive episodes, and suicide attempt status), to capture complex interactions between biomarkers and antidepressant response using multi-layer feedforward neural networks (MFNNs). MFNN predictive models exhibited high accuracy, with the area under the receiver operating characteristic curve (AUC) of 0.82 for response and 0.81 for remission, respectively[39].
Despite these advancements, challenges persist, including limited human studies and the need for robust validation of predictive models in clinical settings. Furthermore, a comprehensive understanding of past research dynamics will inform future development trends in AI-driven genomic psychiatric treatment. Table 3, as well as the following cited resources, provide further insights into the utilization and performance of ML algorithms in diagnostic, prognostic, and treatment prediction contexts[40-46].
| Psychiatric disorder | Machine learning method | Datatypes | Dataset features | Findings | Ref. |
| Bipolar disorder | Decision tree, random forest | Gene expression | RBPMS2, LILRA5 (male responders); ABRACL, FHL3, NBPF14 (female responders) | Predicted lithium responders in bipolar patients with AUC = 0.92 | Eugene et al[36] |
| Major depressive disorder | ARPNet model-linear regression | SNPs, DNA methylation, demographic | Neuroimaging biomarkers, Genetic variants, DNA methylation, demographic information | Predicted the most effective antidepressant with 84% accuracy | Chang et al[37] |
| Major depressive disorder | Deep learning-MFNNs | SNPs, demographic, clinical | Genome-wide associations, marital status, age, sex, suicide attempt status, baseline hamilton rating scale for depression score, depressive episodes | Conducted GWAS to identify SNP associations with antidepressant treatment response and remission. MFNN models achieved high accuracy (AUC = 0.82 for response, AUC = 0.81 for remission). | Lin et al[39] |
| Major depressive disorder | Tree-based ensemble structure | Clinical, demographic | Clinical variables (patients with depression from STAR*D) | Predicted clinical antidepressant remission with 59% accuracy | Chekroud et al[40] |
| Major depressive disorder | Elastic net | Clinical, demographic | Clinical variables: Patients with major depressive disorder (GENDEN participants) | Forecasted antidepressant response with AUC = 0.72 | Iniesta et al[41] |
| Treatment-resistant depression | Random forest | SNPs, clinical | SNP (rs6265 (BDNF gene), rs6313 (HTR2A gene), rs7430 (PPP3CC gene), Clinical variable - Melancholia | Predicted antidepressant treatment outcome with 25% accuracy | Kautzky et al[42] |
| Major depressive disorder | SVM, decision trees | SNPs | rs2036270 SNP (RARB gene), rs7037011 SNP (LOC105375971 gene) | Estimated antidepressant treatment response with 52% accuracy | Maciukiewicz et al[43] |
| Bipolar disorder | Random forest | Clinical | Clinical variables (patients with bipolar disorder treated primarily with lithium) | Predicted responders for lithium treatment outcome with AUC = 0.8 | Nunes et al[44] |
| Late-life depression | Alternating decision tree | Clinical, demographic | Mini-mental status examination scores, age, structural imaging | Predicted antidepressant treatment response with 89% accuracy | Patel et al[45] |
| Major depressive disorder | Random forest | SNPs | SNPs (rs5743467, rs2741130, rs2702877, rs696692, rs17137566, rs10516436) | Predicted antidepressant therapy response with AUC > 0.7 and accuracy > 69% | Athreya et al[46] |
Another important application of AI techniques focused on assessing the significance of multiple pharmacokinetic genes in predicting blood levels of commonly used SSRIs while considering demographic factors such as age, sex, body weight, and genetic variations known to influence drug metabolism. Studies examined the effectiveness of a combinatorial PGx algorithm in predicting citalopram, escitalopram, and sertraline blood levels among patients in the Genomics Used to Improve DEpression Decisions (GUIDED) trial[10,11]. The combinatorial PGx test using a weighted assessment of three genes (CYP2C19, CYP2D6, and CYP3A4) outperformed single-gene approaches in predicting medication response by correlating pharmacokinetic assessments with medication blood levels. The combinatorial PGx algorithm predicted sertraline blood levels better than individual genes (CYP2C19 and CYP2B6) across multivariate analyses. Specifically, the observed F-statistic value of 33.3 indicated that the combinatorial PGx algorithm (approximately 2-7 times larger than for individual genes) significantly contributes to explaining the variability in sertraline blood levels, suggesting its potential to optimize medication management in patients with MDD[10]. Similar findings were reported in a previous GUIDED trial with 191 MDD patients treated with citalopram/escitalopram. An evaluation of the predictive capacity of individual pharmacokinetic genes (CYP2C19, CYP2D6, and CYP3A4) and a comprehensive PGx test incorporating all three genes showed that the F-statistic for the combinatorial PGx test (when adjusted for age and smoking status) also exceeded that of individual genes by 1.7-2.9 times. When assessing both individual genes and the combinatorial test together, only the combinatorial PGx test retained significance, indicating its superiority in predicting citalopram/escitalopram blood levels compared to individual genes[11].
Patient stratification entails the intricate integration of diverse biomedical, demographic, and sociometric data to classify patients into subpopulations for clinical trial design and practice. Data mining of electronic health records (EHRs) was proposed as an efficient method to identify eligible patients for clinical trials based on relevance[47]. Studies involving EHR-linked DNA repositories demonstrated the utility of integrating PGx and sociometric data for predictive modeling to optimize dosage and reduce dosing errors. Healthcare providers and researchers can identify better treatment options by leveraging clinically available information for each psychiatric patient, such as age, gender, and education. In 2013, the FDA issued guidelines recommending the incorporation of PGx testing into early-phase clinical trials to identify specific populations, cohorts, and individuals who may require adjusted drug doses or titration intervals based on genetic factors influencing drug exposure, response, and adverse reactions[48].
The integration of AI methods for clinical data–based patient stratification and the development of proprietary ML algorithms to classify patients using both structured and unstructured data from EHRs is gaining traction in academic research, clinical practice, and pharmaceutical trials. Linking EHRs to genomic data in biobanks can address the critical need for large sample sizes in genetic research. Studies demonstrated that diagnostic algorithms with high positive predictive value can be derived from EHRs, especially when structured data are combined with text mining[49]. These algorithms facilitate semi-automated phenotyping for large-scale case-control studies. Additionally, the vast scale of EHR databases enables the identification of phenotypic subgroups and the development of algorithms for longitudinal risk prediction. EHR-derived genomic data are ideal for rapid replication of putative risk genes, studies of pleiotropy, investigations of genetic networks, and PGx research. Ethnicity stratification can be performed at various levels by leveraging genomic data along with information from resources such as HapMap and the 1000 Genomes Project, enhancing patient categorization. However, EHR-based genomic characterization or phenotyping remains underutilized in psychiatric genomic research. Moreover, addressing the underlying issues is crucial for advancing precision psychiatry.
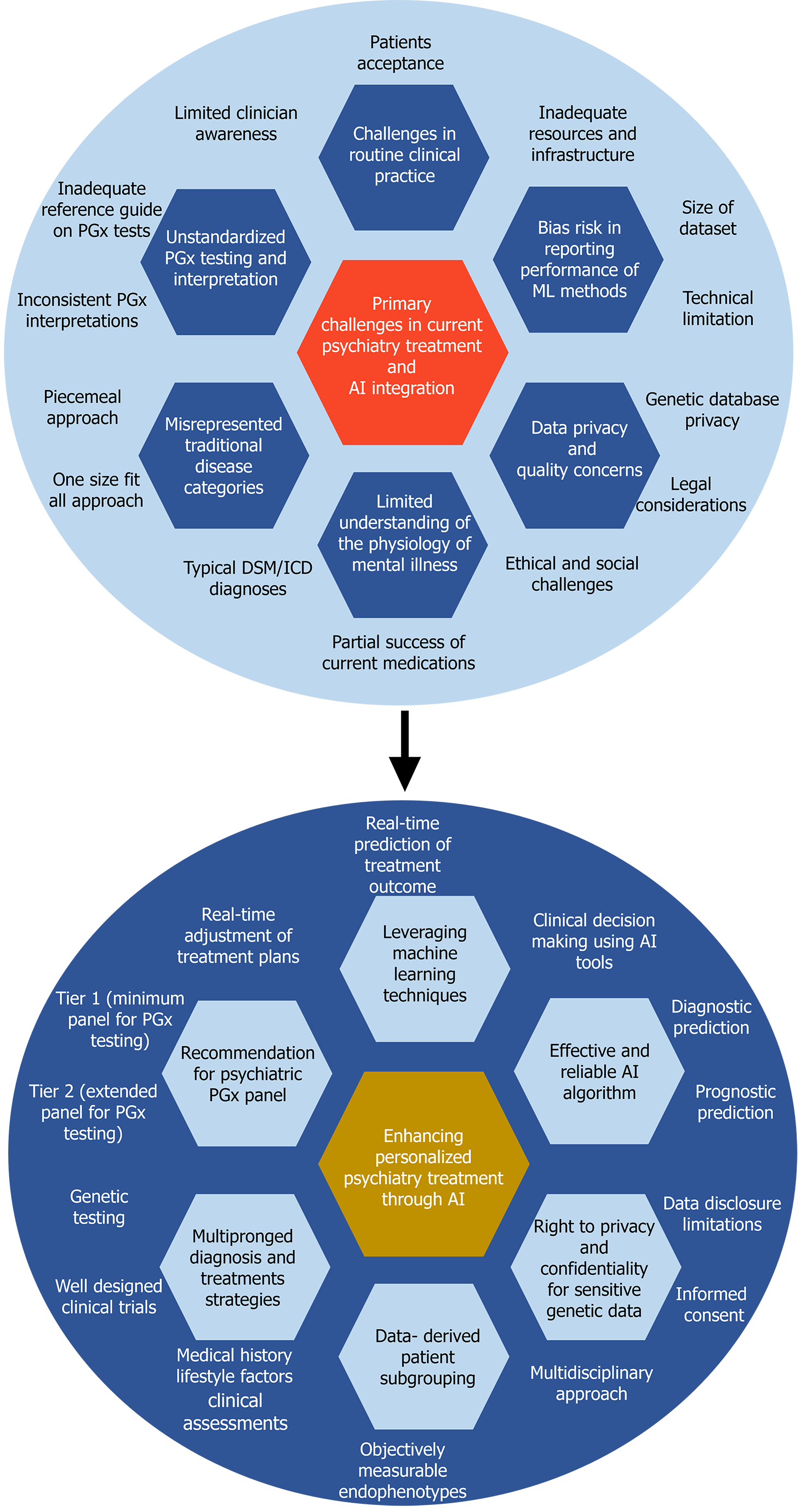
Despite recognizable advancements, the integration of ML and AI into genomic psychiatric treatment presents several challenges, which are categorized in this study into conceptual and practical challenges. Conceptual challenges pertain to understanding the underlying mechanisms of mental illness, diagnostic ambiguity, treatment personalization, interpretability, clinical integration, lack of uniform guidance, and clinicians’ limited knowledge of genomics. Practical challenges relate to data quality, privacy, and algorithmic performance, as well as integration with precision medicine. Addressing these challenges requires interdisciplinary collaboration, methodological innovation, and a nuanced understanding of the complexities of mental health and genomic data (Figure 3).
Limited understanding of the physiology of mental illness and diagnostic ambiguity: A fundamental challenge in precision psychiatry is the need for a more comprehensive understanding of the physiological mechanisms underlying mental illness. Research shows that a particular drug or psychotherapy might work well for one subgroup of patients but not for another subgroup with the same diagnosis[16,50]. Without a thorough grasp of these underlying mechanisms, the development of accurate predictive models and personalized treatment strategies remains elusive. The current standard for diagnosing and treating mental disorders, the DSM developed by the American Psychiatric Association, heavily relies on clinicians' observations, behavioral symptoms, and patient reports, all of which can vary greatly[51,52]. Therefore, developing objective neurobiological markers for mental disorders is crucial, considering their diversity and co-occurrence with other conditions that have gradual, long-term effects over 2-4 weeks. For instance, depression assessments based on the DSM inquire about physiological activity. While serotonin regulation may directly impact symptoms such as psychomotor retardation, it may have minimal effects on feelings of guilt. Many other mental disorders present as spectrums with overlapping symptoms. Variability in symptoms across demographics and genetic factors complicates standardized approaches to diagnosis and treatment. Identifying biomarkers or genetic variants associated with specific disorders can be difficult due to the heterogeneity of symptoms and genetic factors. This discrepancy highlights the need to identify subgroups with predictable treatment responses, requiring innovative statistical and scientific methods.
Unstandardized PGx testing and interpretation with lack of uniform guidance: One major challenge in psychiatric genomics is the complexity of genotyping, particularly in pharmacogenetic testing involving genes such as CYP2D6 and CYP2C19, which are linked with the highest number of drugs (Table 1) for which PGx guidelines suggest modifications to medical treatment[53]. Identifying clinically significant variants poses challenges due to differences in variant analysis among laboratories, requiring clear reporting of investigated SNPs[54-56]. Additionally, the conversion of SNPs into variant alleles and assigning activity scores for predicting metabolic phenotypes presents difficulties in clinical interpretation, hindering clinicians' ability to translate genetic test results into practical decisions[57]. Next-generation sequencing offers advantages in detecting rare variants, but its implementation in clinical settings is challenging in assigning clinical relevance to newly identified variants[13,57]. Technical complexities, such as gene deletions, multiplications, and hybrid alleles, further complicate accurate genotyping, necessitating harmonization efforts. Inconsistent guidelines and standards for pharmacogenetic testing hampers the development of effective ML models and contribute to uncertainty in treatment decisions. Gaps in understanding and clinician awareness of genetic complexities, such as those associated with CYP2D6, further exacerbate challenges in analysis and interpretation. PGx test results can be difficult to interpret, particularly when multiple genes influence drug therapy or when unusual genetic variations are present. Ensuring that clinicians can accurately interpret and apply these results in the context of individual patient care is crucial but challenging, especially given the time constraints and intellectual demands of clinical practice.
Treatment personalization: The multifactorial nature of mental disorders and the described variability in treatment outcomes pose a major challenge to identifying biomarkers or genetic predictors of treatment response. Genomic psychiatry, centered on personalized treatment, seeks to move away from the "one size fits all" (implementing uniform strategies regardless of individual or community differences) or piecemeal approach (addressing specific issues with tailored interventions, potentially overlooking broader systemic factors) and instead tailors therapies to individual patients[58]. However, it is important to recognize that individuality in healthcare is constantly evolving and shaped by advances in biomedicine and our understanding of genetics. Tailoring treatment solely based on patients' genetic and clinical profiles may prove inefficient. A critical challenge in psychiatric treatment lies in considering factors such as race and lifestyle during treatment planning. This approach not only focuses on treating existing conditions but also emphasizes disease prevention by identifying and addressing risk factors early. Furthermore, previous research primarily focused on the collective effects of neurobiology, overlooking individual brain abnormalities crucial for personalized medicine development. Additionally, many psychiatric disorders can present across multiple dimensions, with high rates of co-occurrence indicating different symptom patterns resulting from shared risk factors and potentially similar underlying disease mechanisms[19,20]. This high level of comorbidity adds complexity to efforts aimed at understanding and effectively treating psychiatric disorders.
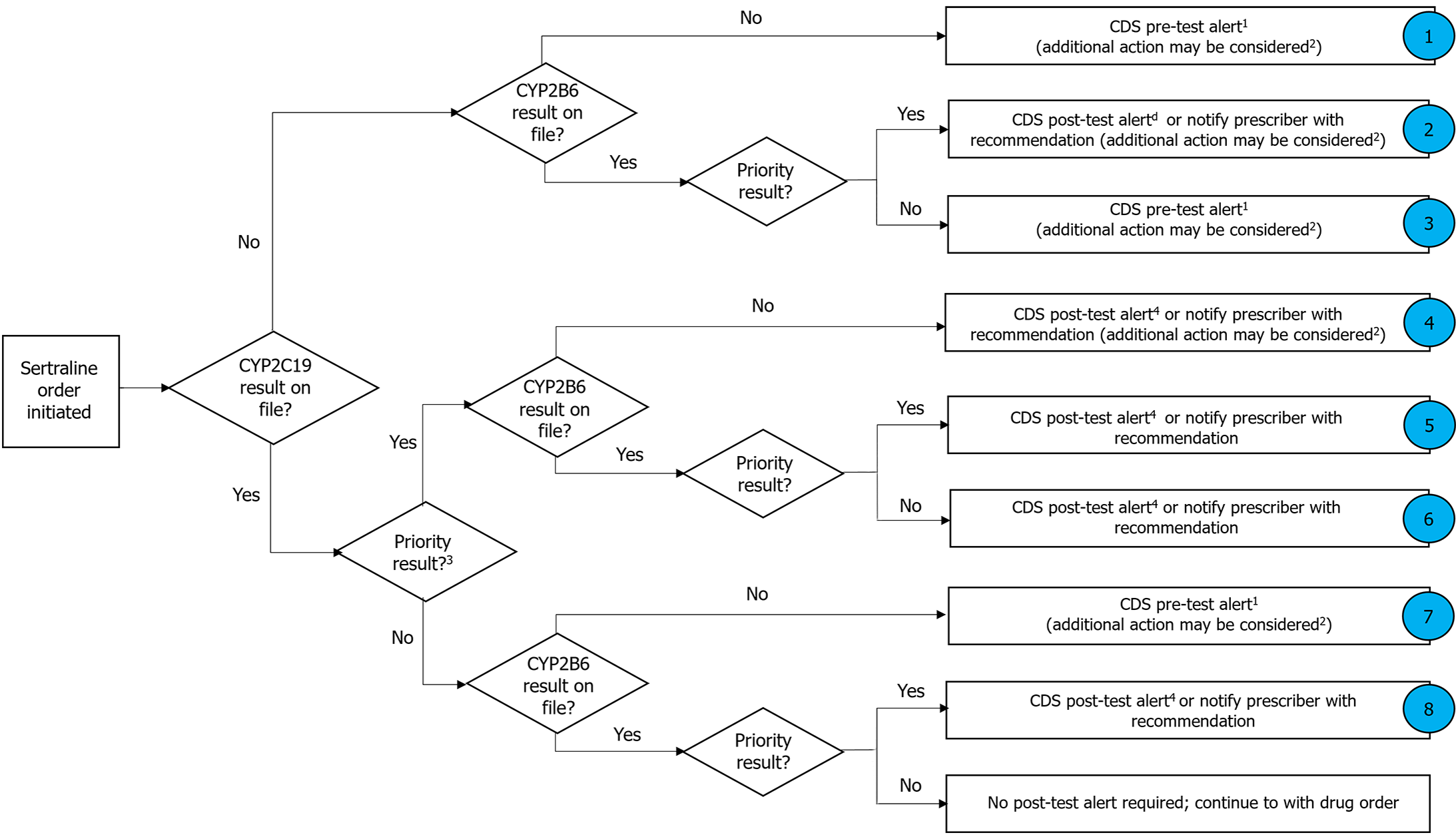
EHR customization and implementation efforts: As research progresses, new gene-drug interactions and PGx relationships continue to emerge with the expanding volume of PGx clinical knowledge. Keeping up with this rapidly evolving knowledge base and incorporating it into EHR systems in a timely manner poses a significant challenge[59,60]. Integrating PGx clinical decision support (CDS) into EHRs often requires customization and significant implementation efforts at the institutional level. Institutions must organize relevant medication and genomic knowledge, define clinical workflows, and develop alert texts tailored to their specific needs and contexts. This customization can be resource-intensive and require collaboration across multiple departments and stakeholders within the healthcare organization. Test results from years ago might still impact drug selection and dosing decisions in the future. Managing and recalling these historical results within the context of ongoing patient care workflows is challenging, especially considering the potential for changes in clinical guidelines and therapeutic options for psychiatric medication over time[61,62]. CPIC, established in 2009, offers clinical practice guidelines facilitating the translation of genetic laboratory test outcomes into practical drug prescription decisions. These guidelines adhere closely to the IOM’s Standards for Developing Trustworthy Clinical Practice. CPIC members possess diverse expertise in pharmacogenetics, with many actively involved in clinical PGx implementation. CPIC emphasizes the importance of a curated and machine-readable pharmacogenetics database suitable for integration into EHRs with CDS, recognizing the necessity of adopting pharmacogenetics into routine clinical care. In 2013, CPIC formed the CPIC Informatics Working Group to support guideline adoption in clinical electronic environments. Starting with HLA-B genotype and abacavir use, the CPIC systematically incorporates implementation resources into all guidelines, intending to make them applicable across various EHR systems. Currently, there are over 24 psychiatric medications with clinical guideline recommendations of which 14 drugs (amitriptyline and CYP2D6, CYP2C19; atomoxetine and CYP2D6; carbamazepine and HLA-A, HLA-B; citalopram/escitalopram and CYP2C19; fosphenytoin/phenytoin and CYP2C9, HLA-B; nortriptyline and CYP2D6; oxcarbazepine and HLA-B; paroxetine and CYP2D6; sertraline and CYP2B6; CYP2C19, vortioxetine and CYP2D6; fluvoxamine and CYP2D6; venlafaxine and CYP2D6) have pre- and post-CDS alert text and CDS flow chart indicating the need for a PGx test for a specific genotype before drug administration[13,62-65]. CPIC also provides guidelines for managing drugs with multiple gene interactions (Figure 4). ML-powered CDS systems integrated into EHRs can provide real-time guidance to clinicians at the point of care. These systems can alert providers to potential drug-gene or drug-drug interactions, recommend alternative medications or dosages based on a patient's genetic profile, and facilitate informed decision-making regarding pharmacotherapy. Additionally, AI algorithms can generate clinically applicable precision medicine tools at the bedside by leveraging big data from biobanks and EHRs, optimizing diagnosis, therapeutic intervention, and prognosis[66].
Ethical considerations and data privacy concerns: Applications of ML in psychiatry face ethical challenges similar to those in other areas of medicine and computer science. In this study, the challenges of genomic psychiatry treatment are categorized into two main issues regarding privacy in PGx testing and ML applications in psychiatric treatment. PGx testing requires the establishment of extensive genomic databases to facilitate the development of predictive models for drug response. These databases, containing sensitive genetic information possibly linkable to genetic diseases or predispositions, present unique privacy challenges compared to conventional medical data. Concerns arise regarding data security, unauthorized access, and a potential breach of confidentiality due to the aggregation of vast genomic data[67]. The European legal framework emphasizes the right to privacy and the duty of confidentiality for sensitive genetic data. Strict legal protection, including consent requirements and limitations on data disclosure, is essential to safeguard patient privacy and prevent unauthorized use of genetic information[68]. Despite efforts to technically enhance data confidentiality, genetic databases may remain vulnerable. The potential for individual identification from medically relevant genomic sequences stored in databases also raises concerns. Rigorous adherence to strict guidelines and legal protections is imperative to mitigate confidentiality risks and ensure ethical conduct in PGx studies[69].
Another paramount ethical and social challenge of PGx testing is ensuring adequate protection against potential discrimination based on predictive medical information. Ethical considerations, such as informed consent, data protection, and safeguarding patient autonomy, demand careful attention. Additionally, effectively translating PGx evidence into clinical practice is needed to ensure equitable access across diverse populations[70].
The integration of ML into genomic psychiatry presents several challenges with ethical implications, including issues such as responsibility in decision-making, avoiding the dehumanization of patients, respecting clinicians' judgment, and ensuring transparency and fairness in algorithmic decision-making. As ML programs become integrated into clinical practice, physicians and ML tools are envisioned as collaborative "teammates" in treatment selection. However, determining who holds authority and ethical responsibility over the decisions made poses a significant question[71]. A competent human agent (especially a clinician) is required to review and ultimately assume final responsibility for the suggestions made by ML, as only humans possess consciousness and empathy, a comprehensive understanding of the contextual environment. Second, a risk of dehumanization exists as ML may overlook the patient's subjective experience[72]. While ML can incorporate various psychological, environmental, and social variables, allowing patients the opportunity to fully express their concerns is essential, ensuring accurate diagnosis, improving health outcomes, and maintaining humane care. Third, decision-making complexities can arise, with non-experts possibly relying too heavily on protocols and overlooking tacit knowledge possessed by experienced clinicians[73]. Respecting clinicians' judgment and not disempowering them are crucial. Additionally, clinicians might become overly reliant on ML outcomes, particularly in complex cases, risking the loss of clinical judgment. However, training clinicians in ML applications can mitigate this risk. Transparency in ML algorithms is vital to ensure understanding and trust among human teammates, reducing resistance and empowering patients. Furthermore, there is a concern about bias in training datasets, potentially leading to erroneous predictions for underrepresented groups. Few retrospective studies conducted for heart failure and type I and II diabetes incorporated adequate external validation procedures[74-76]. However, prospective studies assessing the clinical feasibility and effectiveness of predictive models are scarce[77,78]. Some studies in mental disorders, including bipolar disorder, obtained predictive models above chance but without validation in independent samples, limiting their clinical applicability[17]. Stricter guidelines and legal protections are necessary to address these privacy concerns and ensure ethical conduct. Overall, addressing these challenges requires a comprehensive approach involving ethical, legal, and technical considerations to maintain patient confidentiality and autonomy in genomic psychiatry.
Challenges of sample size used in ML: The effectiveness of ML models hinges significantly on the size and quality of the datasets used for training. While large datasets offer numerous benefits, including enhanced model training, improved generalization, and better performance, working with small datasets poses several challenges that can hinder the model's effectiveness and generalization ability[79,80]. One significant challenge associated with small datasets is limited sample size restricting the number of available training samples. With fewer examples to learn from, the model may not capture complex patterns and relationships present in the data, leading to suboptimal performance. In psychiatric treatment, small sample sizes in pharmacogenetic studies resulted in contradictory findings, hindering implementation due to the lack of prescriber confidence and interpretive skills[81,82]. Moreover, small datasets often exhibit higher variability that can result in overfitting; hence, the model learns to fit the training data too closely, capturing noise or idiosyncrasies specific to the limited samples[83]. Furthermore, small datasets may lack sufficient representation of all possible variations or classes present in the real-world scenario, leading to biased or incomplete learning. This limitation can restrict the model's ability to accurately generalize in unseen instances. Additionally, small datasets may not provide enough instances of events or classes in scenarios where rare events or imbalanced class distributions exist, making it challenging for the model to effectively learn their characteristics and make accurate predictions[84]. Enhanced prediction accuracy is also determined by the amount and quality of data. Mere dataset size does not assure superior outcomes; rather, the caliber and relevance of data, encompassing issues of noise, bias, and diversity, considerably influence overall model performance. Thus, meticulous attention to data quality, alongside quantity, is essential[85,86].
Bias risk in reporting performance of ML methods and performance variability: Bias in ML, also referred to as algorithmic bias or AI bias, arises when an algorithm generates outcomes exhibiting systematic prejudice because of flawed assumptions within the ML process. Biases inherent in data and algorithms can lead to unfair or inaccurate predictions, particularly in sensitive domains, such as healthcare. ML methods are employed to make predictions in psychiatry from genotypes. The performance of these ML methods is highly varied, with different ranges of AUC, which utilizes probability to assess how effectively a model distinguishes between classes[87]. In a study in which 63 complete texts were evaluated from a pool of 652 abstracts, information was gathered for 77 models across 13 studies. The performance of ML techniques varied considerably (AUC: 0.48-0.95) and showed discrepancies among schizophrenia (AUC: 0.54-0.95), bipolar disorder (AUC: 0.48-0.65), autism (AUC: 0.52-0.81), and anorexia (AUC: 0.62-0.69). This variability is likely attributed to the substantial risk of bias identified in study designs and analysis of the reported outcomes. Factors such as predictor selection, hyperparameter exploration, validation methodology, and exposure to the test set during training were common contributors to the elevated risk of bias in the analysis. Variability in the performance of ML models across different studies and datasets hinders their reliability and generalizability to diverse populations.
Global optimism toward the integration of AI and ML into genomic psychiatry is driven by several key factors. First, the expanding knowledge base of PGx and personalized medicine, combined with the growing need for data preprocessing from diverse sources, necessitates the need for psychiatrists to incorporate AI into patient assessments for guiding diagnoses and treatment strategies. Second, understanding the neural underpinnings of human cognition and behavior requires effective analysis of both inter- and intra-individual variability. The emergence of advanced Explainable AI tools shows promise in deciphering complex neural behaviors, providing unbiased risk diagnoses, and offering personalized medicine recommendations. Modern ML capabilities extend beyond predicting treatment outcomes. They now include explaining predictions and addressing questions about why specific outcomes are chosen, their reliability, potential failures, and reasons for incorrect predictions. Third, the concept of genomic data exchange, allowing selective sharing of sensitive genomic or phenotypic data, necessitates secure handling of vast amounts of sensitive data. Conventional data management systems often cannot securely handle large quantities of sensitive data[17]. In response to this challenge, innovative systems managing information on patient outcomes, enhancing real-time communication on the significance of test results between patients and doctors, and providing AI assistance on personalized medication must be developed. To guide future research, desirable recommendations include the development of enhanced AI-driven predictive models leveraging multi-omics data and advanced ML techniques. Privacy-preserving techniques for genomic data exchange should be enhanced to enable selective sharing of sensitive genomic and phenotypic data. Two prominent techniques in this domain, differential privacy (DP) and federated learning (FL), offer unique approaches to safeguarding sensitive information while enabling effective data analysis[88]. DP, a cornerstone of privacy-preserving ML, operates by injecting noise into datasets, thereby obscuring individual identities while retaining overall data utility. On the other hand, FL revolutionizes the traditional paradigm of centralized data processing by allowing multiple parties to collaboratively train ML models without sharing raw data. This decentralized approach mitigates privacy concerns associated with data aggregation and facilitates seamless collaboration among institutions.
Designing innovative informatics systems is required to manage patient outcomes, facilitate real-time patient-doctor communication, and offer AI assistance for personalized medication recommendations. Implementing PGx CDS within EHRs is crucial for integrating PGx into routine psychiatric care. This integration enables the curation and dissemination of patient-specific PGx data at the point of care, facilitating optimal drug therapy based on genetic profiles. Efficient integration of PGx CDS into EHRs addresses challenges related to expanding PGx knowledge and complex interpretation of results. Furthermore, introducing risk-scoring systems based on comprehensive data integration (clinical, genomic, demographic, and drug interactions) can enhance medication safety and personalize prescriptions. Clinicians can make informed decisions to minimize adverse reactions and optimize patient outcomes by leveraging AI and ML for risk assessment and clinical decision-making, refined through iterative analysis.
In summary, the integration of PGx into routine psychiatric care through EHRs is pivotal for advancing precision medicine. Utilizing informatics, especially CDS within EHRs, is critical for this integration. Efficient implementation of such technologies can address challenges related to the expanding volume of PGx knowledge and the complexity of result interpretation, ultimately enhancing patient care and outcomes.
| 1. | Bogdan R, Salmeron BJ, Carey CE, Agrawal A, Calhoun VD, Garavan H, Hariri AR, Heinz A, Hill MN, Holmes A, Kalin NH, Goldman D. Imaging Genetics and Genomics in Psychiatry: A Critical Review of Progress and Potential. Biol Psychiatry. 2017;82:165-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 99] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 2. | Kwon S, Cheon SY. Influence of the inflammasome complex on psychiatric disorders: clinical and preclinical studies. Expert Opin Ther Targets. 2021;25:897-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 3. | Al-Harbi KS. Treatment-resistant depression: therapeutic trends, challenges, and future directions. Patient Prefer Adherence. 2012;6:369-388. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 368] [Cited by in RCA: 527] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 4. | Myers AJ, Nemeroff CB. New Vistas in the Management of Treatment-Refractory Psychiatric Disorders: Genomics and Personalized Medicine. FOC. 2010;8:525-535. [RCA] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 5. | Green ED, Watson JD, Collins FS. Human Genome Project: Twenty-five years of big biology. Nature. 2015;526:29-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 161] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 6. | Ptacek R, Kuzelova H, Stefano GB. Genetics in Psychiatry - up-to-date review 2011. Neuro Endocrinol Lett. 2011;32:389-399. [PubMed] |
| 7. | Mroz P, Michel S, Allen JD, Meyer T, McGonagle EJ, Carpentier R, Vecchia A, Schlichte A, Bishop JR, Dunnenberger HM, Yohe S, Thyagarajan B, Jacobson PA, Johnson SG. Development and Implementation of In-House Pharmacogenomic Testing Program at a Major Academic Health System. Front Genet. 2021;12:712602. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 8. | Ramsey LB, Namerow LB, Bishop JR, Hicks JK, Bousman C, Croarkin PE, Mathews CA, Van Driest SL, Strawn JR. Thoughtful Clinical Use of Pharmacogenetics in Child and Adolescent Psychopharmacology. J Am Acad Child Adolesc Psychiatry. 2021;60:660-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 24] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 9. | van Schaik RHN, Müller DJ, Serretti A, Ingelman-Sundberg M. Pharmacogenetics in Psychiatry: An Update on Clinical Usability. Front Pharmacol. 2020;11:575540. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 56] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 10. | Parikh SV, Law RA, Hain DT, Rothschild AJ, Thase ME, Dunlop BW, DeBattista C, Forester BP, Shelton RC, Macaluso M, Cogan ES, Brown K, Lewis DJ, Jablonski MR, Greden JF. Combinatorial pharmacogenomic algorithm is predictive of sertraline metabolism in patients with major depressive disorder. Psychiatry Res. 2022;308:114354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 11. | Rothschild AJ, Parikh SV, Hain D, Law R, Thase ME, Dunlop BW, DeBattista C, Conway CR, Forester BP, Shelton RC, Macaluso M, Brown K, Lewis D, Gutin A, Jablonski MR, Greden JF. Clinical validation of combinatorial pharmacogenomic testing and single-gene guidelines in predicting psychotropic medication blood levels and clinical outcomes in patients with depression. Psychiatry Res. 2021;296:113649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 12. | Beunk L, Nijenhuis M, Soree B, de Boer-Veger NJ, Buunk AM, Guchelaar HJ, Houwink EJF, Risselada A, Rongen GAPJM, van Schaik RHN, Swen JJ, Touw D, van Westrhenen R, Deneer VHM, van der Weide J. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction between CYP2D6, CYP3A4 and CYP1A2 and antipsychotics. Eur J Hum Genet. 2024;32:278-285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 91] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 13. | Bousman CA, Stevenson JM, Ramsey LB, Sangkuhl K, Hicks JK, Strawn JR, Singh AB, Ruaño G, Mueller DJ, Tsermpini EE, Brown JT, Bell GC, Leeder JS, Gaedigk A, Scott SA, Klein TE, Caudle KE, Bishop JR. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6, CYP2C19, CYP2B6, SLC6A4, and HTR2A Genotypes and Serotonin Reuptake Inhibitor Antidepressants. Clin Pharmacol Ther. 2023;114:51-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 252] [Article Influence: 84.0] [Reference Citation Analysis (0)] |
| 14. | Bousman CA, Wu P, Aitchison KJ, Cheng T. Sequence2Script: A Web-Based Tool for Translation of Pharmacogenetic Data Into Evidence-Based Prescribing Recommendations. Front Pharmacol. 2021;12:636650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 15. | Thorn CF, Klein TE, Altman RB. PharmGKB: the Pharmacogenomics Knowledge Base. Methods Mol Biol. 2013;1015:311-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 230] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 16. | Bzdok D, Meyer-Lindenberg A. Machine Learning for Precision Psychiatry: Opportunities and Challenges. Biol Psychiatry Cogn Neurosci Neuroimaging. 2018;3:223-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 276] [Article Influence: 30.7] [Reference Citation Analysis (4)] |
| 17. | Amr A, Hinderer M, Griebel L, Deuber D, Egger C, Sedaghat-Hamedani F, Kayvanpour E, Huhn D, Haas J, Frese K, Schweig M, Marnau N, Krämer A, Durand C, Battke F, Prokosch HU, Backes M, Keller A, Schröder D, Katus HA, Frey N, Meder B. Controlling my genome with my smartphone: first clinical experiences of the PROMISE system. Clin Res Cardiol. 2022;111:638-650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 18. | Johnson KB, Wei WQ, Weeraratne D, Frisse ME, Misulis K, Rhee K, Zhao J, Snowdon JL. Precision Medicine, AI, and the Future of Personalized Health Care. Clin Transl Sci. 2021;14:86-93. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 691] [Article Influence: 115.2] [Reference Citation Analysis (0)] |
| 19. | Feczko E, Miranda-Dominguez O, Marr M, Graham AM, Nigg JT, Fair DA. The Heterogeneity Problem: Approaches to Identify Psychiatric Subtypes. Trends Cogn Sci. 2019;23:584-601. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 289] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 20. | Satterthwaite TD, Feczko E, Kaczkurkin AN, Fair DA. Parsing Psychiatric Heterogeneity Through Common and Unique Circuit-Level Deficits. Biol Psychiatry. 2020;88:4-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 21. | Hare H, Sneed KB, Pathak Y. Applications of Precision Medicine in the Treatment of Psychiatric Disorders: A Literature Review. Chem Pharm Res. 2021;3. [DOI] [Full Text] |
| 22. | Jarvis JP, Peter AP, Shaman JA. Consequences of CYP2D6 Copy-Number Variation for Pharmacogenomics in Psychiatry. Front Psychiatry. 2019;10:432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 23. | Morel D, Yu KC, Liu-Ferrara A, Caceres-Suriel AJ, Kurtz SG, Tabak YP. Predicting hospital readmission in patients with mental or substance use disorders: A machine learning approach. Int J Med Inform. 2020;139:104136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 32] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 24. | An Q, Rahman S, Zhou J, Kang JJ. A Comprehensive Review on Machine Learning in Healthcare Industry: Classification, Restrictions, Opportunities and Challenges. Sensors (Basel). 2023;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 101] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 25. | Alshabeeb MA, Alyabsi M, Aziz MA, Abohelaika S. Pharmacogenes that demonstrate high association evidence according to CPIC, DPWG, and PharmGKB. Front Med (Lausanne). 2022;9:1001876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Kim HH, Kim DW, Woo J, Lee K. Explicable prioritization of genetic variants by integration of rule-based and machine learning algorithms for diagnosis of rare Mendelian disorders. Hum Genomics. 2024;18:28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 27. | Nasteski V. An overview of the supervised machine learning methods. Horizons. 2017;4:51-62. [DOI] [Full Text] |
| 28. | Ghahramani Z. Unsupervised Learning. Adv Lect Mach Learn. 2004;. [RCA] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 51] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 29. | Sutton RS, Barto AG. Reinforcement learning: An introduction. MIT press. 2018. Available from: https://www.andrew.cmu.edu/course/10-703/textbook/BartoSutton.pdf. |
| 30. | Bonaccorso G. Machine learning algorithms: A reference guide to popular algorithms for data science and machine learning. Packt. 2017. Available from: https://balasahebtarle.wordpress.com/wp-content/uploads/2020/01/machine-learning-algorithms_text-book.pdf. |
| 31. | Jain A, Patel H, Nagalapatti L, Gupta N, Mehta S, Guttula S, Mujumdar S, Afzal S, Sharma Mittal R, Munigala V. Overview and Importance of Data Quality for Machine Learning Tasks. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining (KDD '20). Association for Computing Machinery, New York, NY, USA; 2020: 3561–3562. [DOI] [Full Text] |
| 32. | Lin PI, Moni MA, Gau SS, Eapen V. Identifying Subgroups of Patients With Autism by Gene Expression Profiles Using Machine Learning Algorithms. Front Psychiatry. 2021;12:637022. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 12] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | Sardaar S, Qi B, Dionne-Laporte A, Rouleau GA, Rabbany R, Trakadis YJ. Machine learning analysis of exome trios to contrast the genomic architecture of autism and schizophrenia. BMC Psychiatry. 2020;20:92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 34. | Starke G, De Clercq E, Borgwardt S, Elger BS. Computing schizophrenia: ethical challenges for machine learning in psychiatry. Psychol Med. 2021;51:2515-2521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 31] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 35. | Trakadis YJ, Sardaar S, Chen A, Fulginiti V, Krishnan A. Machine learning in schizophrenia genomics, a case-control study using 5,090 exomes. Am J Med Genet B Neuropsychiatr Genet. 2019;180:103-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 34] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 36. | Eugene AR, Masiak J, Eugene B. Predicting lithium treatment response in bipolar patients using gender-specific gene expression biomarkers and machine learning. F1000Res. 2018;7:474. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 16] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 37. | Chang B, Choi Y, Jeon M, Lee J, Han KM, Kim A, Ham BJ, Kang J. ARPNet: Antidepressant Response Prediction Network for Major Depressive Disorder. Genes (Basel). 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 38. | Pei C, Sun Y, Zhu J, Wang X, Zhang Y, Zhang S, Yao Z, Lu Q. Ensemble Learning for Early-Response Prediction of Antidepressant Treatment in Major Depressive Disorder. J Magn Reson Imaging. 2020;52:161-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 32] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 39. | Lin E, Kuo PH, Liu YL, Yu YW, Yang AC, Tsai SJ. A Deep Learning Approach for Predicting Antidepressant Response in Major Depression Using Clinical and Genetic Biomarkers. Front Psychiatry. 2018;9:290. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 40. | Chekroud AM, Zotti RJ, Shehzad Z, Gueorguieva R, Johnson MK, Trivedi MH, Cannon TD, Krystal JH, Corlett PR. Cross-trial prediction of treatment outcome in depression: a machine learning approach. Lancet Psychiatry. 2016;3:243-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 447] [Article Influence: 44.7] [Reference Citation Analysis (0)] |
| 41. | Iniesta R, Malki K, Maier W, Rietschel M, Mors O, Hauser J, Henigsberg N, Dernovsek MZ, Souery D, Stahl D, Dobson R, Aitchison KJ, Farmer A, Lewis CM, McGuffin P, Uher R. Combining clinical variables to optimize prediction of antidepressant treatment outcomes. J Psychiatr Res. 2016;78:94-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 115] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 42. | Kautzky A, Baldinger P, Souery D, Montgomery S, Mendlewicz J, Zohar J, Serretti A, Lanzenberger R, Kasper S. The combined effect of genetic polymorphisms and clinical parameters on treatment outcome in treatment-resistant depression. Eur Neuropsychopharmacol. 2015;25:441-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 43. | Maciukiewicz M, Marshe VS, Hauschild AC, Foster JA, Rotzinger S, Kennedy JL, Kennedy SH, Müller DJ, Geraci J. GWAS-based machine learning approach to predict duloxetine response in major depressive disorder. J Psychiatr Res. 2018;99:62-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 44. | Nunes A, Ardau R, Berghöfer A, Bocchetta A, Chillotti C, Deiana V, Garnham J, Grof E, Hajek T, Manchia M, Müller-Oerlinghausen B, Pinna M, Pisanu C, O'Donovan C, Severino G, Slaney C, Suwalska A, Zvolsky P, Cervantes P, Del Zompo M, Grof P, Rybakowski J, Tondo L, Trappenberg T, Alda M. Prediction of lithium response using clinical data. Acta Psychiatr Scand. 2020;141:131-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 45. | Patel MJ, Andreescu C, Price JC, Edelman KL, Reynolds CF 3rd, Aizenstein HJ. Machine learning approaches for integrating clinical and imaging features in late-life depression classification and response prediction. Int J Geriatr Psychiatry. 2015;30:1056-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 107] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 46. | Athreya AP, Neavin D, Carrillo-Roa T, Skime M, Biernacka J, Frye MA, Rush AJ, Wang L, Binder EB, Iyer RK, Weinshilboum RM, Bobo WV. Pharmacogenomics-Driven Prediction of Antidepressant Treatment Outcomes: A Machine-Learning Approach With Multi-trial Replication. Clin Pharmacol Ther. 2019;106:855-865. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 82] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 47. | Shickel B, Tighe PJ, Bihorac A, Rashidi P. Deep EHR: A Survey of Recent Advances in Deep Learning Techniques for Electronic Health Record (EHR) Analysis. IEEE J Biomed Health Inform. 2018;22:1589-1604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 714] [Cited by in RCA: 563] [Article Influence: 70.4] [Reference Citation Analysis (0)] |
| 48. | Federal Register. Guidance for Industry on Clinical Pharmacogenomics: Premarket Evaluation in Early-Phase Clinical Studies and Recommendations for Labeling; Availability. Available from: https://www.federalregister.gov/documents/2013/01/28/2013-01638/guidance-for-industry-on-clinical-pharmacogenomics-premarket-evaluation-in-early-phase-clinical. |
| 49. | Smoller JW. The use of electronic health records for psychiatric phenotyping and genomics. Am J Med Genet B Neuropsychiatr Genet. 2018;177:601-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 50. | Gabrieli JDE, Ghosh SS, Whitfield-Gabrieli S. Prediction as a humanitarian and pragmatic contribution from human cognitive neuroscience. Neuron. 2015;85:11-26. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 402] [Article Influence: 36.5] [Reference Citation Analysis (0)] |
| 51. | Insel TR, Cuthbert BN. Medicine. Brain disorders? Precisely. Science. 2015;348:499-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 461] [Article Influence: 41.9] [Reference Citation Analysis (0)] |
| 52. | Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:617-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7240] [Cited by in RCA: 7370] [Article Influence: 351.0] [Reference Citation Analysis (0)] |
| 53. | Hippman C, Nislow C. Pharmacogenomic Testing: Clinical Evidence and Implementation Challenges. J Pers Med. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 73] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 54. | Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. 2015;526:343-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 593] [Article Influence: 59.3] [Reference Citation Analysis (0)] |
| 55. | Shuldiner AR, Relling MV, Peterson JF, Hicks JK, Freimuth RR, Sadee W, Pereira NL, Roden DM, Johnson JA, Klein TE; Pharmacogenomics Research Network Translational Pharmacogenetics Program Group, Shuldiner AR, Vesely M, Robinson SW, Ambulos N Jr, Stass SA, Kelemen MD, Brown LA, Pollin TI, Beitelshees AL, Zhao RY, Pakyz RE, Palmer K, Alestock T, O'Neill C, Maloney K, Branham A, Sewell D, Relling MV, Crews K, Hoffman J, Cross S, Haidar C, Baker D, Hicks JK, Bell G, Greeson F, Gaur A, Reiss U, Huettel A, Cheng C, Gajjar A, Pappo A, Howard S, Hudson M, Pui CH, Jeha S, Evans WE, Broeckel U, Altman RB, Gong L, Whirl-Carrillo M, Klein TE, Sadee W, Manickam K, Sweet KM, Embi PJ, Roden D, Peterson J, Denny J, Schildcrout J, Bowton E, Pulley J, Beller M, Mitchell J, Danciu I, Price L, Pereira NL, Weinshilboum R, Wang L, Johnson JA, Nelson D, Clare-Salzler M, Elsey A, Burkley B, Langaee T, Liu F, Nessl D, Dong HJ, Lesko L, Freimuth RR, Chute CG. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: overcoming challenges of real-world implementation. Clin Pharmacol Ther. 2013;94:207-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 144] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 56. | Weinshilboum RM, Wang L. Pharmacogenomics: Precision Medicine and Drug Response. Mayo Clin Proc. 2017;92:1711-1722. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 151] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 57. | Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med. 2017;19:69-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 274] [Cited by in RCA: 387] [Article Influence: 38.7] [Reference Citation Analysis (0)] |
| 58. | Quazi S. Artificial intelligence and machine learning in precision and genomic medicine. Med Oncol. 2022;39:120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 263] [Cited by in RCA: 153] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 59. | Troiano D, Jones MA, Smith AH, Chan RC, Laegeler AP, Le T, Flynn A, Chaffee BW; American Society of Health-System Pharmacists. ASHP Guidelines on the Design of Database-Driven Clinical Decision Support: Strategic Directions for Drug Database and Electronic Health Records Vendors. Am J Health Syst Pharm. 2015;72:1499-1505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 60. | Hicks JK, Dunnenberger HM, Gumpper KF, Haidar CE, Hoffman JM. Integrating pharmacogenomics into electronic health records with clinical decision support. Am J Health Syst Pharm. 2016;73:1967-1976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 122] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 61. | Hicks JK, Swen JJ, Thorn CF, Sangkuhl K, Kharasch ED, Ellingrod VL, Skaar TC, Müller DJ, Gaedigk A, Stingl JC; Clinical Pharmacogenetics Implementation Consortium. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther. 2013;93:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 318] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 62. | Hicks JK, Sangkuhl K, Swen JJ, Ellingrod VL, Müller DJ, Shimoda K, Bishop JR, Kharasch ED, Skaar TC, Gaedigk A, Dunnenberger HM, Klein TE, Caudle KE, Stingl JC. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102:37-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 476] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 63. | Brown JT, Bishop JR, Sangkuhl K, Nurmi EL, Mueller DJ, Dinh JC, Gaedigk A, Klein TE, Caudle KE, McCracken JT, de Leon J, Leeder JS. Clinical Pharmacogenetics Implementation Consortium Guideline for Cytochrome P450 (CYP)2D6 Genotype and Atomoxetine Therapy. Clin Pharmacol Ther. 2019;106:94-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 141] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 64. | Karnes JH, Rettie AE, Somogyi AA, Huddart R, Fohner AE, Formea CM, Ta Michael Lee M, Llerena A, Whirl-Carrillo M, Klein TE, Phillips EJ, Mintzer S, Gaedigk A, Caudle KE, Callaghan JT. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C9 and HLA-B Genotypes and Phenytoin Dosing: 2020 Update. Clin Pharmacol Ther. 2021;109:302-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 113] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 65. | Phillips EJ, Sukasem C, Whirl-Carrillo M, Müller DJ, Dunnenberger HM, Chantratita W, Goldspiel B, Chen YT, Carleton BC, George AL Jr, Mushiroda T, Klein T, Gammal RS, Pirmohamed M. Clinical Pharmacogenetics Implementation Consortium Guideline for HLA Genotype and Use of Carbamazepine and Oxcarbazepine: 2017 Update. Clin Pharmacol Ther. 2018;103:574-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 201] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 66. | Uddin M, Wang Y, Woodbury-Smith M. Artificial intelligence for precision medicine in neurodevelopmental disorders. NPJ Digit Med. 2019;2:112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 102] [Cited by in RCA: 120] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 67. | Vaszar LT, Cho MK, Raffin TA. Privacy issues in personalized medicine. Pharmacogenomics. 2003;4:107-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 68. | Romeo-Malanda S, Nicol D. Pharmacogenetic testing: legal considerations for consent, privacy and disclosure. Per Med. 2008;5:155-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 69. | Howard K. Lack of data privacy could hamper pharmacogenomics studies. Nat Rev Drug Discov. 2004;3:725. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 70. | Vijverberg SJ, Pieters T, Cornel MC. Ethical and social issues in pharmacogenomics testing. Curr Pharm Des. 2010;16:245-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 71. | Trujillo AC, Gregory IM, Ackerman KA. Evolving Relationship between Humans and Machines. IFAC-Papers On Line. 2019;51:366-371. [DOI] [Full Text] |
| 72. | Haque OS, Waytz A. Dehumanization in Medicine: Causes, Solutions, and Functions. Perspect Psychol Sci. 2012;7:176-186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 202] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 73. | Borrell-Carrió F, Suchman AL, Epstein RM. The biopsychosocial model 25 years later: principles, practice, and scientific inquiry. Ann Fam Med. 2004;2:576-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 777] [Cited by in RCA: 685] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 74. | Desai RJ, Wang SV, Vaduganathan M, Evers T, Schneeweiss S. Comparison of Machine Learning Methods With Traditional Models for Use of Administrative Claims With Electronic Medical Records to Predict Heart Failure Outcomes. JAMA Netw Open. 2020;3:e1918962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 186] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 75. | Lynam AL, Dennis JM, Owen KR, Oram RA, Jones AG, Shields BM, Ferrat LA. Logistic regression has similar performance to optimised machine learning algorithms in a clinical setting: application to the discrimination between type 1 and type 2 diabetes in young adults. Diagn Progn Res. 2020;4:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 75] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 76. | Mahboob Alam T, Iqbal MA, Ali Y, Wahab A, Ijaz S, Imtiaz Baig T, Hussain A, Malik MA, Raza MM, Ibrar S, Abbas Z. A model for early prediction of diabetes. Inform Med Unlocked. 2019;16:100204. [RCA] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 77. | Chekroud AM, Bondar J, Delgadillo J, Doherty G, Wasil A, Fokkema M, Cohen Z, Belgrave D, DeRubeis R, Iniesta R, Dwyer D, Choi K. The promise of machine learning in predicting treatment outcomes in psychiatry. World Psychiatry. 2021;20:154-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 280] [Article Influence: 56.0] [Reference Citation Analysis (0)] |
| 78. | Lin E, Lin CH, Lane HY. Precision Psychiatry Applications with Pharmacogenomics: Artificial Intelligence and Machine Learning Approaches. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 88] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 80. | Lipovetsky S. Statistical and Machine-Learning Data Mining: Techniques for Better Predictive Modeling and Analysis of Big Data. Technometrics. 2022;64:145-148. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 81. | Shad MU. Genetic Testing for Antipsychotic Pharmacotherapy: Bench to Bedside. Behav Sci (Basel). 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 82. | Alchakee A, Ahmed M, Eldohaji L, Alhaj H, Saber-Ayad M. Pharmacogenomics in Psychiatry Practice: The Value and the Challenges. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
| 83. | Vemuri VK. The Hundred-Page Machine Learning Book. J Inf Technol Case Appl Res. 2020;22:136-138. [DOI] [Full Text] |
| 84. | Krawczyk B. Learning from imbalanced data: open challenges and future directions. Prog Artif Intell. 2016;5:221-232. [RCA] [DOI] [Full Text] [Cited by in Crossref: 912] [Cited by in RCA: 581] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 85. | Saha B, Srivastava D. Data quality: The other face of Big Data. IEEE Xplore. 2014;. [DOI] [Full Text] |
| 86. | Cai L, Zhu Y. The Challenges of Data Quality and Data Quality Assessment in the Big Data Era. CODATA. 2015;14:2. [RCA] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 137] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 87. | Bracher-Smith M, Crawford K, Escott-Price V. Machine learning for genetic prediction of psychiatric disorders: a systematic review. Mol Psychiatry. 2021;26:70-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 88. | Khalid N, Qayyum A, Bilal M, Al-Fuqaha A, Qadir J. Privacy-preserving artificial intelligence in healthcare: Techniques and applications. Comput Biol Med. 2023;158:106848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 145] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: Https://creativecommons.org/Licenses/by-nc/4.0/