Published online Jan 19, 2024. doi: 10.5498/wjp.v14.i1.15
Peer-review started: November 7, 2023
First decision: November 23, 2023
Revised: December 3, 2023
Accepted: December 26, 2023
Article in press: December 26, 2023
Published online: January 19, 2024
Processing time: 72 Days and 22.8 Hours
Numerous observational studies have documented a correlation between inflammatory bowel disease (IBD) and an increased risk of dementia. However, the causality of their associations remains elusive.
To assess the causal relationship between IBD and the occurrence of all-cause dementia using the two-sample Mendelian randomization (MR) method.
Genetic variants extracted from the large genome-wide association study (GWAS) for IBD (the International IBD Genetics Consortium, n = 34652) were used to identify the causal link between IBD and dementia (FinnGen, n = 306102). The results of the study were validated via another IBD GWAS (United Kingdom Biobank, n = 463372). Moreover, MR egger intercept, MR pleiotropy residual sum and outlier, and Cochran's Q test were employed to evaluate pleiotropy and heterogeneity. Finally, multiple MR methods were performed to estimate the effects of genetically predicted IBD on dementia, with the inverse variance wei-ghted approach adopted as the primary analysis.
The results of the pleiotropy and heterogeneity tests revealed an absence of significant pleiotropic effects or heterogeneity across all genetic variants in outcome GWAS. No evidence of a causal effect between IBD and the risk of dementia was identified in the inverse variance weighted [odds ratio (OR) = 0.980, 95%CI : 0.942-1.020, P value = 0.325], weighted median (OR = 0.964, 95%CI : 0.914-1.017, P value = 0.180), and MR-Egger (OR = 0.963, 95%CI : 0.867-1.070, P value = 0.492) approaches. Consistent results were observed in validation analyses. Reverse MR analysis also showed no effect of dementia on the development of IBD. Furthermore, MR analysis suggested that IBD and its subtypes did not causally affect all-cause dementia and its four subtypes, including dementia in Alzheimer's disease, vascular dementia, dementia in other diseases classified elsewhere, and unspecified dementia.
Taken together, our MR study signaled that IBD and its subentities were not genetically associated with all-cause dementia or its subtypes. Further large prospective studies are warranted to elucidate the impact of intestinal inflammation on the development of dementia.
Core Tip: Dementia is a major disease burden for public health and healthcare systems worldwide. This study used two-sample Mendelian randomization (MR) to assess the causal relationship between inflammatory bowel disease (IBD) and all-cause dementia. Multiple MR methods have failed to find that IBD increases the risk of developing all-cause dementia and its four subtypes. The present study suggests that genetically predicted IBD is not associated with risk of all-cause dementia and that dementia prevention interventions for patients with IBD can be similar to those for the healthy population.
- Citation: Liao OL, Xie SY, Ye J, Du Q, Lou GC. Association between inflammatory bowel disease and all-cause dementia: A two-sample Mendelian randomization study. World J Psychiatry 2024; 14(1): 15-25
- URL: https://www.wjgnet.com/2220-3206/full/v14/i1/15.htm
- DOI: https://dx.doi.org/10.5498/wjp.v14.i1.15
Dementia is a syndrome arising from brain disorders, usually of a chronic or progressive nature, characterized by acquired behavioral and cognitive deficits, including domains such as memory, communication and language abilities, concentration and attention span, reasoning and judgment, and visual perception[1]. It is a global health concern and has emerged as a pandemic in the aging population. Over 1315 million people are predicted to be affected by the mid-21st century[2]. Mounting evidence suggests that intestinal homeostasis is involved in psychiatric and neurologic disorders through the bidirectional microbiome-gut-brain axis. Besides conventional brain-gut disorders (e.g., functional gastrointestinal disorders), recent studies point toward a potential role of the interaction between the gut and central nervous system in depression, anxiety, Parkinson's disease, autism spectrum disorders, and other related disorders[3].
Inflammatory bowel disease (IBD), comprising ulcerative colitis (UC) and Crohn's disease (CD), refers to a chronic intestinal disorder featured by a relapsing and remitting course. Its cause remains to be elucidated, but its pathogenesis may involve environmental factors triggering an aberrant immune response between the gut microbiota and the intestinal immune system in genetically susceptible hosts, thereby eliciting intestinal mucosal inflammation[4]. The genome-wide association study (GWAS) identified gene loci associated with IBD susceptibility, influencing not only gut microbial recognition and clearance but also the maintenance of intestinal immune homeostasis[5].
Several studies have reported that IBD patients are at increased risk of neurodegenerative diseases such as Parkinson's disease, multiple sclerosis, and dementia[6-8]. A Taiwanese population-based cohort study determined a significantly increased risk of developing dementia among IBD patients, whereas a Danish study concluded that IBD patients had a marginally increased risk of all-cause dementia[9,10]. Contrastingly, a longitudinal cohort study found no association between IBD and dementia[8]. To date, the causal relationship between IBD and dementia remains underexplored. Furthermore, the aforementioned observational studies might be susceptible to various measurement errors, underlying biases, and confounding factors, which could have compromised the results or even reverse causality.
Mendelian randomization (MR) is conceptually similar to randomized controlled trials (RCTs), using genetic variants as instrumental variables (IVs) to infer causality between an exposure and an outcome based on the principle of random assortment of alleles during gamete formation and conception[11,12]. The former utilizes genetic data, such as single nucleotide polymorphisms (SNPs) associated with an exposure (in this study, IBD), as IVs to examine the causal effect of the exposure on the target outcome (in this study, dementia)[13]. The intrinsic nature of the random assortment of genetic variants at conception dictates that their effects on outcomes remain unaffected by postnatal environmental, behavioral, and economic confounders. Besides, they are not susceptible to reverse causality bias[14]. Given that these confounding factors are inherently balanced across subgroups at conception, MR closely mimics the randomization process in RCTs.
A large number of MR studies have been undertaken to investigate the causal effect of IBD on neurodegenerative diseases, encompassing Alzheimer's disease (AD), Parkinson's disease, and amyotrophic lateral sclerosis[15-17]. While Li and Wen[15] did not identify a correlation between IBD and AD, Guo et al[17] found that IBD exerted a genetically protective effect against AD. There are currently no MR studies or RCTs focusing on the effect of IBD on the risk of dementia subtypes other than AD. Based on the publicly available GWAS data from a large population, a two-sample MR analysis was adopted to identify the effect of IBD on all-cause dementia and its subtypes, which holds clinical implications for the formulation of interventions to delay cognitive decline and mitigate the burden of dementia in the IBD population. Our results may provide novel insights into the bidirectional interactions in the gut-brain axis.
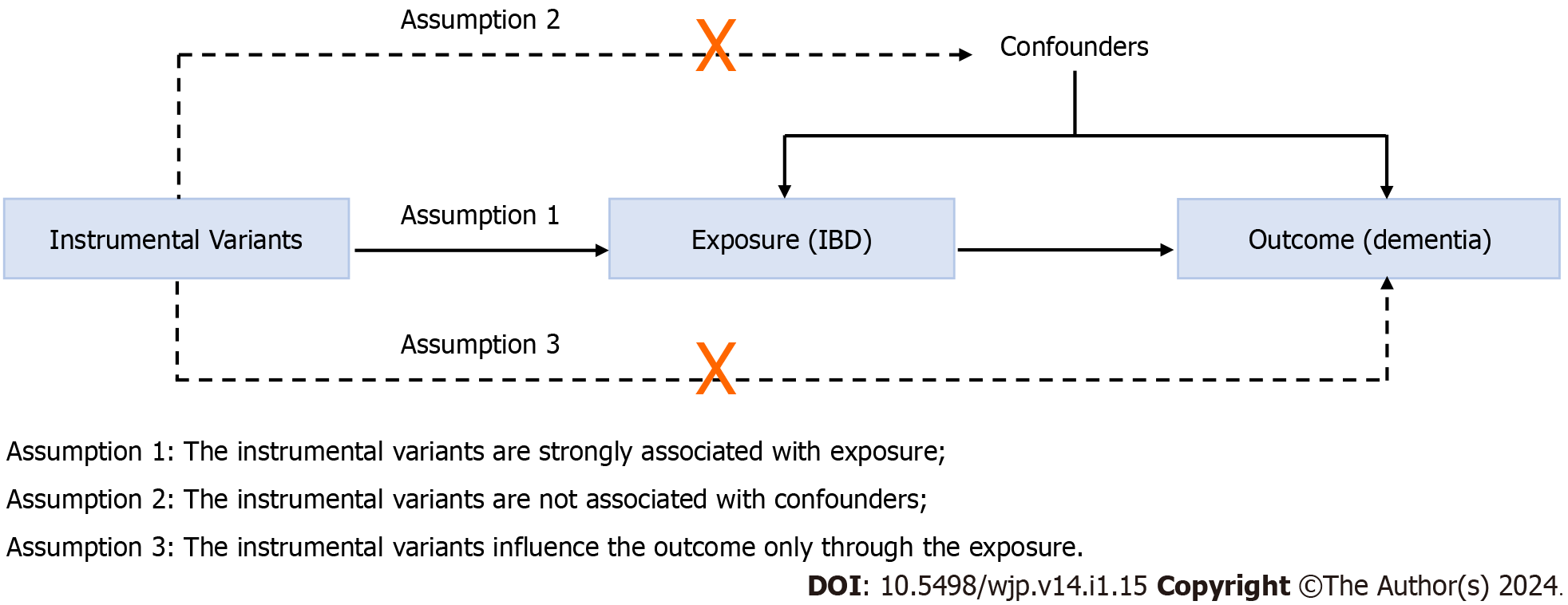
A two-sample MR method was conducted to evaluate the causal relationship between IBD and all-cause dementia. MR follows three key assumptions (Figure 1): (1) The IVs are strongly associated with the exposure (IBD); (2) the IVs are unrelated to confounder factors linked to the selected exposure and outcome, and (3) the IVs exclusively influence the outcome (dementia and its six subtypes) via IBD. Considering that this study used publicly available datasets from participant studies conducted in compliance with ethical standards, the requirement for ethical approval was waived.
The GWAS summary data for IBD were extracted from the International IBD Genetics Consortium (IIBDGC)[18] that is composed of 15 cohorts of European ancestry (enrolled cohorts are listed in Supplementary Table 1) and contained data on IBD as a whole (12882 cases; 21770 controls) and also on CD (5956 cases; 14927 controls) and UC (6968 cases; 20464 controls). All cases were confirmed using standard clinical, endoscopic, and histopathological criteria. The validation analysis incorporated summary statistics acquired from a United Kingdom Biobank GWAS involving European participants (7045 self-reported cases and 456327 controls) wherein IBD cases were identified using the International Classification of Diseases 10th Revision (ICD-10) codes F50 and F51[19]. Information about the GWAS of exposure is summarized in Table 1.
| Phenotype | Consortium | Year | Ncase | Ncontrol | Population |
| IBD | IIBDGC | 2015 | 12882 | 21770 | European |
| UC | IIBDGC | 2015 | 6968 | 20464 | European |
| CD | IIBDGC | 2015 | 5956 | 14927 | European |
| IBD (val) | United Kingdom Biobank | 2021 | 7045 | 449282 | United Kingdom |
| All-cause dementia | FinnGen | 2022 | 11602 | 294500 | Finnish |
| Dementia in AD | FinnGen | 2022 | 3540 | 294500 | Finnish |
| VaD | FinnGen | 2022 | 1602 | 297552 | Finnish |
| Dementia in other diseases classified elsewhere | FinnGen | 2022 | 882 | 294500 | Finnish |
| Unspecified dementia | FinnGen | 2022 | 2729 | 294500 | Finnish |
Independent genetic IVs were extracted from the respective exposure based on several criteria. To begin, SNPs strongly associated with the exposure were selected (P-value < 5 × 10-8). Following this, to ensure the inclusion of IVs without linkage disequilibrium (LD), the clumping procedure (R2 < 0.001, window size = 10000 kb) was executed utilizing European samples to calculate the LD. Thirdly, SNPs associated with other potential risk factors at genome-wide significance, which may act as confounders and interfere with the effect of IBD (including UC and CD) on dementia, were excluded using PhenoScanner (http://www.phenoscanner.medschl.cam.ac.uk/). Known risk factors for dementia include diabetes, hypertension, atrial fibrillation or flutter, obesity, chronic obstructive pulmonary disease, cerebrovascular disease, smoking, and hypothyroidism[8-10]. SNPs associated with dementia due to other diseases identified by the ICD-10 code F02 were also excluded in this analysis. The strength of the relationship between IVs and exposure was estimated using the F statistic (F statistics < 10 indicating a weak IV bias)[20]. For each IV, the F statistic was calculated using the following formula: F= [R2 × (N-2)/(1 -R2), where N denotes the sample size, R2 represents the variance of exposure explained by the IVs (R2 = 2 × beta2 × eaf × (1-eaf)/[2 × beta2 × eaf × (1-eaf) + 2 × se × N × eaf × (1-eaf)], eaf stands for the effect allele frequency, se represents standard error, and beta is the estimated effect of the SNP)[21-23].
All-cause dementia was defined as ICD-10 codes F00 (dementia in AD), F01 [vascular dementia [VaD)], F02 (dementia in other diseases classified elsewhere) and F03 (unspecified dementia)[24]. The GWAS summary statistics for dementia were utilized as a whole and its subtypes from the FinnGen study[25]. The sample of all-cause dementia consisted of 11602 cases (ICD-10 F00-F03, ICD-9 290|3310|4378A, ICD-8 290) and 294500 controls; the sample of dementia in AD consisted of 3540 cases (ICD-10 F00, ICD-8 29010) and 294500 controls; the sample of VaD consisted of 1602 cases (ICD-10 F01, ICD-9 4378) and 297552 controls; the sample of dementia in other diseases classified elsewhere consisted of 882 cases (ICD-10 F02) and 294500 controls; and the sample of unspecified dementia consisted of 2729 cases (ICD-10 F03, ICD 290|2941, ICD-8 2900|29019) and 294500 controls. Details on the outcome of GWAS are listed in Table 1.
IVs were sequentially extracted from the outcome GWASs as described above, while outcome-related SNPs were eliminated. Subsequently, ambiguous SNPs with incompatible alleles (e.g., A/G vs. A/C) and palindromic SNPs (e.g., A/T or G/C) were excluded when harmonizing exposure and outcome datasets[26]. SNPs absent in the outcome data were substituted by proxy SNPs obtained from the online platform LDlink (https://Ldlink.nih.gov/) based on high LD from European data. Proxies were required to have a minimum R2 value of 0.8, and palindromic SNP strands were aligned using a minor allele frequency of up to 0.3[27]. The summary characteristics of all genetic IVs are illustrated in Supplementary Tables 2-4. The correlations between IBD (including UC and CD) genetic IVs and the GWAS datasets for dementia and its subtypes are displayed in Supplementary Tables 5-7. Additionally, a comprehensive summary of IVs associated with the validation analysis is presented in Supplementary Tables 8 and 9.
MR egger intercept and MR pleiotropy residual sum and outlier (MR-PRESSO) tests are typically used to assess horizontal pleiotropy[28]. If the selected IVs are not pleiotropic, the MR Egger intercept term tends to approach zero with an increase in sample size[29]. MR-PRESSO can correct horizontal pleiotropy by eliminating underlying outliers prior to each MR analysis[28]. The harmonized SNPs underwent the MR-PRESSO test (NbDistribution=10000). A P > 0.05 in the MR-PRESSO global test indicates no significant pleiotropy of all IBD-associated IVs in the dementia GWAS dataset. In the MR-PRESSO outlier test, outliers with a P-value less than 0.05 should be removed. The Cochran's Q statistic was employed to evaluate heterogeneity, which is extensively employed in MR Egger and inverse variance weighted (IVW) analyses[30,31]. A P > 0.05 indicates the absence of significant heterogeneity. The summarized results of the pleiotropy and heterogeneity tests are shown in Supplementary Table 10.
Three different MR methods (IVW, weighted median, and MR Egger) were performed to estimate the effect of the exposure on outcome susceptibility. IVW was selected as the primary method, with the remaining MR methods assessing the sensitivity of our findings with robust estimates. IVW represents the weighted average of Wald ratio estimates of the causal impact for each variant and provides the most accurate estimate when all IVs are valid[32]. The weighted median yields consistent estimates even if up to 50% of selected SNPs are not valid[28,29]. MR Egger accounts for pleiotropy among all IVs but requires that the associations between genetic variants and exposure remain independent of the effects of genetic variants on the outcome[33]. Additionally, a "leave one out" analysis was carried out to systematically exclude each SNP individually to examine the influence of SNPs on the MR estimate[34].
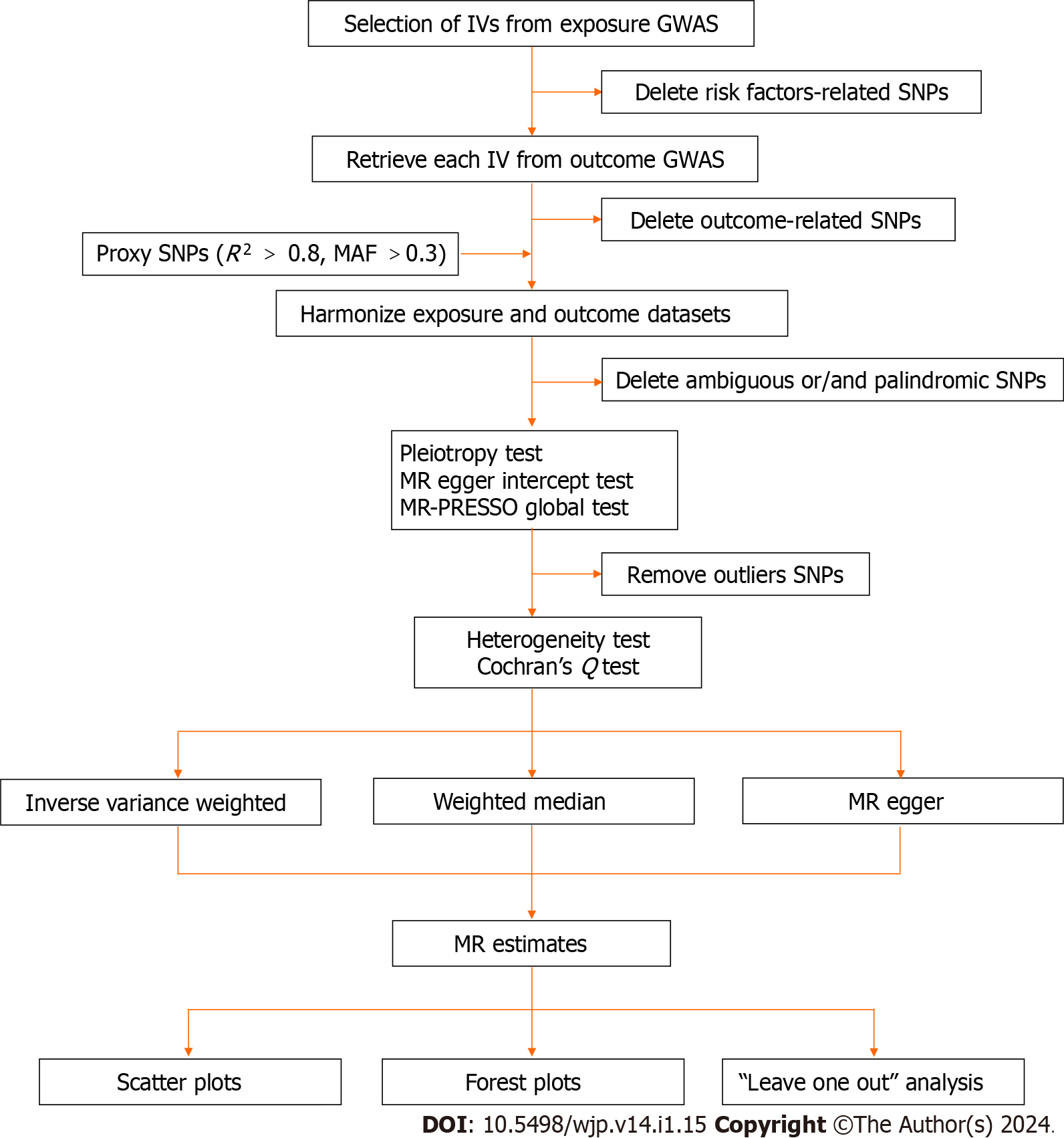
All analyses in this MR study were performed using the Package "TwoSampleMR version 0.5.6" in R version 4.2.2. The significance threshold was set at P < 0.05/X/Y = 0.05/3/5 = 0.003, corrected by the Bonferroni method (X: the number of exposures, Y: the number of outcomes). An overview of our study's process is presented in Figure 2.
Genetic IVs were selected from the dementia GWAS summary data based on several criteria: (1) P-value (genome-wide significance threshold < 5 × 10-8); (2) an LD R2 of < 0.001, and < 10000 kb from the index variant; and (3) no effects on potential risk factors, including inflammation, immune response, and gut microbiota. Then, IVs were extracted from the IBD GWAS. The IBD GWAS and dementia GWAS were sourced from the IIBDGC and FinnGen, respectively (Table 1). The summary characteristics of dementia IVs and their association with IBD GWAS are depicted in Supplemen
The MR Egger intercept and PRESSO methods were applied to determine the pleiotropy of dementia-associated IVs in IBD GWAS, whilst MR Egger and IVW in Cochran's Q statistic were employed to determine the heterogeneity of dementia-associated IVs in IBD GWAS (Supplementary Table 10). The methods of reverse MR analysis were consistent with those described above.
In the primary analysis investigating the causal impact of IBD on dementia, 65 SNPs were screened as potential genetic IVs, of which 19 SNPs related to other potential risk factors were excluded, nine SNPs could not be extracted from the outcome GWASs, and five SNPs were ambiguous or/and palindromic (Supplementary Table 2). In the analysis concerning the impact of UC on dementia, 39 SNPs were screened as potential genetic IVs, of which 13 SNPs related to other potential risk factors were excluded, four SNPs could not be extracted from the outcome GWASs, and three SNPs were ambiguous or/and palindromic (Supplementary Table 3). In the analysis of the impact of CD on dementia, 53 SNPs were initially identified as potential genetic IVs, among which 19 SNPs related to other potential risk factors were excluded, two SNPs could not be extracted from the outcome GWASs, and five SNPs were ambiguous or/and palindromic (Supplementary Table 4). Besides, 19 SNPs were identified as IVs in the validation analyses (Supplementary 8). The selected IVs could explain 6.75%, 4.36%, and 9.06% variance of IBD, UC, and CD, respectively. Additionally, the accounted variance by IVs was 0.19% in the validation analyses. The F-statistic of all selected IVs was > 10, demonstrating a marginal possibility of a weak instrument bias (Supplementary Table 2-4 and 8).
MR egger intercept and MR-PRESSO global tests both exposed the absence of significant pleiotropy (Supple
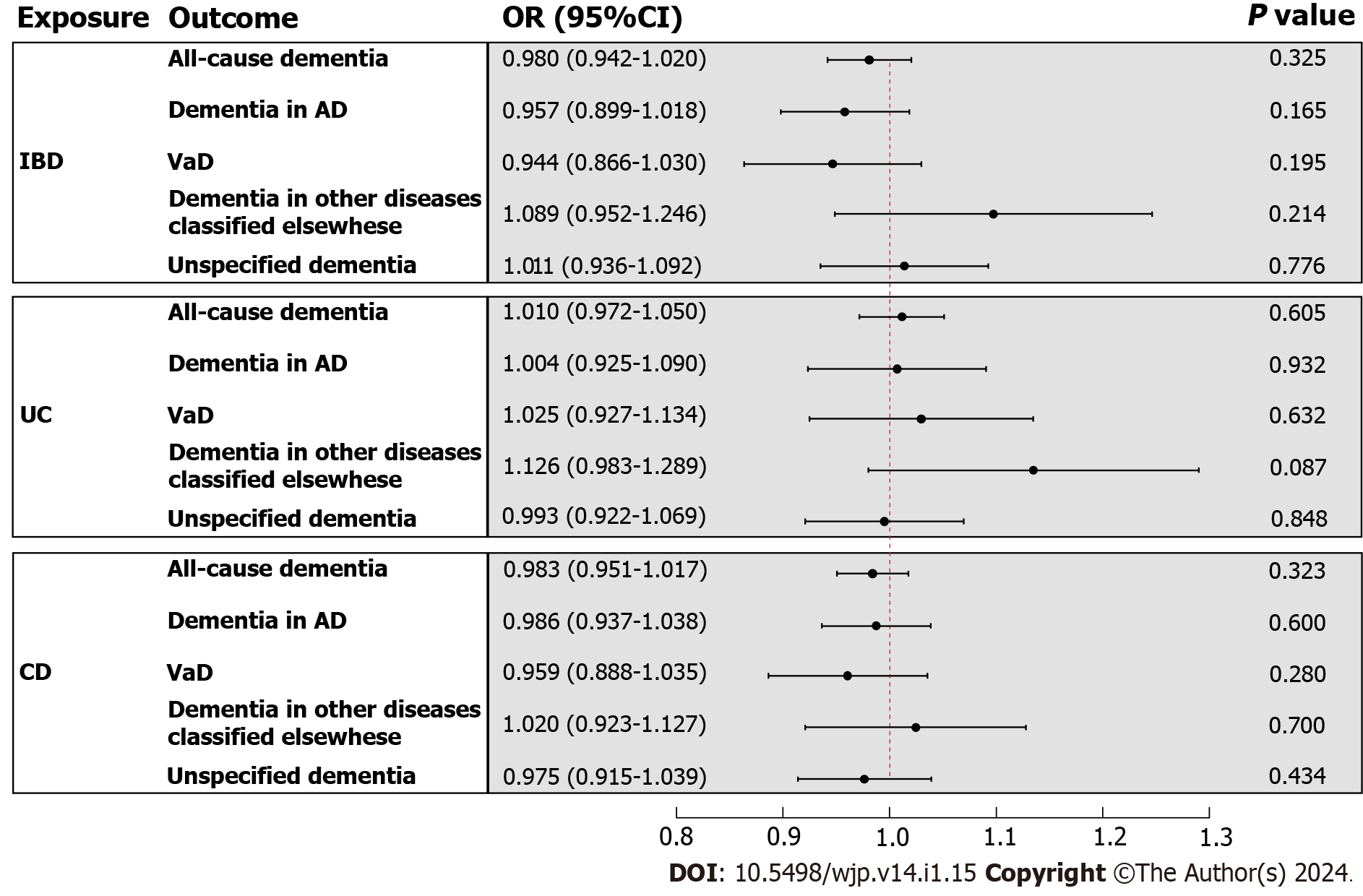
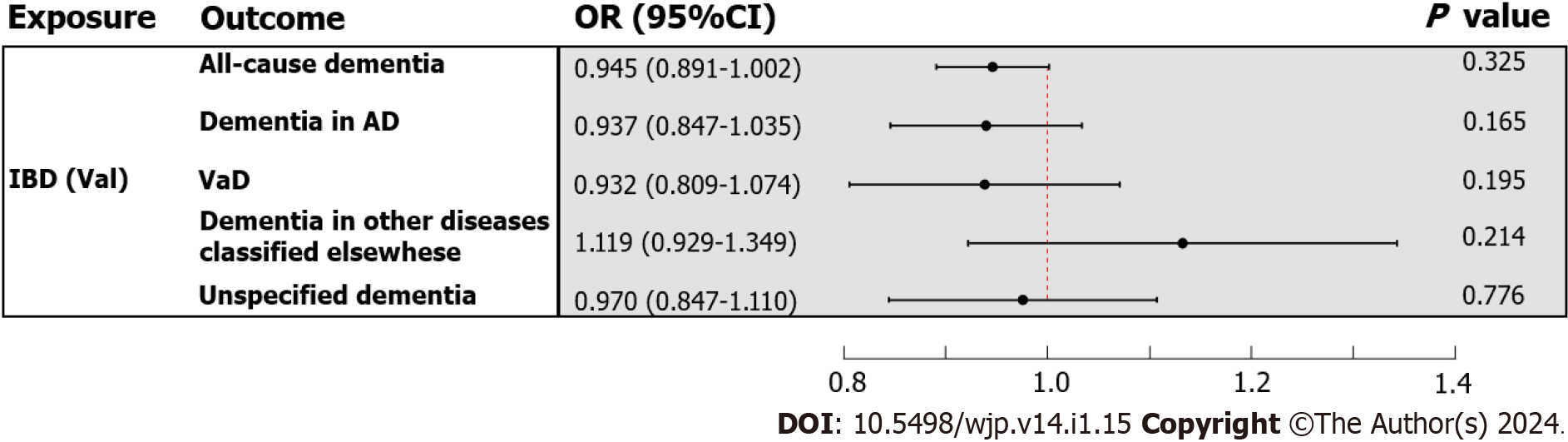
In the primary analysis, the IVW method determined that IBD was not causally related to all-cause dementia [odds ratio (OR) = 0.980, 95%CI : 0.942-1.020, P value = 0.325] (Figure 3). Subsequently, the causal relationship between IBD and the four subtypes of dementia was examined. Subgroup analyses did not support a significant association between IBD and dementia in AD (OR = 0.957, 95%CI : 0.899-1.018, P value = 0.165), VaD (OR = 0.944, 95%CI : 0.866-1.030, P value = 0.195), dementia in other diseases classified elsewhere (OR = 1.089, 95%CI : 0.952-1.246, P value = 0.214), and unspecified dementia (OR = 1.011, 95%CI : 0.936-1.092, P value = 0.776) (Figure 3). Similarly, the weighted median and MR Egger methods provided no evidence of a genetic causal relationship between IBD and all-cause dementia and its subtypes (Supplementary Table 13). As anticipated, these results were corroborated by the validation sample (Figure 4; Supplementary Table 13). The scatter plots and forest plots of the single SNP effect and combined effects are displayed in Supplementary Figure 1-4. The "leave one out" sensitivity analysis indicated that no individual SNP influenced the MR estimates (Supplementary Figures 5 and 6).
Furthermore, the causal effects of UC and CD on all-cause dementia and its four subtypes were assessed in a similar approach. The IVW method revealed that UC and CD were not causally related to all-cause dementia and its four subtypes, including dementia in AD, VaD, dementia in other diseases classified elsewhere, and unspecified dementia (Figure 3). The results of the weighted median and MR Egger are presented in Supplementary Table 13. All scatter plots, forest plots, and "leave one out" analysis plots for MR analyses of UC and CD on dementia are shown in Supple
To explore reverse causality, 12 SNPs were selected from dementia GWAS summary statistics as potential IVs, of which three SNPs associated with potential risk factors were excluded, whilst another SNP was excluded due to palindrome after harmonization of dementia GWAS and IBD GWAS (Supplementary Tables 11 and 12). The intercept term from the MR Egger regression and MR-PRESSO global test demonstrated no significant pleiotropy among the eight independent dementia-associated IVs in IBD GWAS. Importantly, Cochran's Q test did not identify significant heterogeneity among the effects of dementia-associated SNPs on IBD (Supplementary Table 10). Therefore, all eight dementia-associated SNPs could be regarded as valid genetic IVs for the ensuing MR analysis. In the reverse MR analysis, the results of IVW, weighted median, and MR Egger uncovered no genetically causal effect of dementia on IBD (Supplementary Table 14). The scatter plot, forest plot, and "leave one out" analysis plot for reverse MR analysis are shown in Supplemen
Herein, a two-sample MR approach was employed to comprehensively evaluate the causal relationship between genetically predicted IBD (including UC and CD) and the risk of all-cause dementia and its subtypes, namely, AD, VaD, dementia in other diseases classified elsewhere and unspecified dementia. The results of several methods of MR analyses did not indicate that IBD played a genetic role in the development of dementia (Supplementary Table 13). The findings were further confirmed by conducting a validation analysis in another summary statistics of IBD GWAS (Supple
IBD is etiologically related to gut microbiota dysbiosis, which induces proinflammatory activity in the gut that is transmitted to the nervous system via the microbiome-gut-brain axis, eventually resulting in neuroinflammation[35,36]. Recently, compelling evidence from population-based observational studies has insinuated an association between IBD and an increased risk of dementia. For instance, a longitudinal cohort study including 1742 patients with IBD and 17420 controls from the Taiwanese population demonstrated an increased risk of all-cause dementia following the diagnosis of IBD [hazard ratio (HR) = 2.54, 95%CI : 1.91-3.37], especially at younger ages, compared to controls[9]. Two other retrospective cohort studies from Germany and Denmark reported significant but less pronounced effects of IBD on the risk of dementia[10,37]. A recent systematic review and meta-analysis based on six studies including 2334472 subjects suggested an increased risk for developing dementia in IBD patients (HR = 1.27, 95%CI : 1.10-1.47)[38]. Notably, this result was in line with the findings of other systematic reviews and meta-analyses[39,40]. Furthermore, a large case-control study established systemic inflammation as a potential risk factor for AD, while the latest meta-analysis concluded that chronic elevation in the level of the inflammatory biomarker C-reactive protein was directly correlated with the lifetime risk of developing dementia[41,42]. Interestingly, drugs for the treatment of IBD, such as tumor necrosis factor blocking agents, might be associated with a lower risk of developing AD[41,43]. In a mouse model of IBD induced by sodium dextran sulfate, Kaneko et al[44] observed that neutrophils infiltrated the brain parenchyma of AD mice and accelerated amyloid plaque accumulation during acute colitis. Meanwhile, He et al[45] found that intestinal inflammation disrupted glymphatic clearance and triggered neuroinflammation, resulting in increased amyloid-β deposition and, ultimately, cognitive impairment.
However, the results of observational studies are largely inconsistent. A longitudinal cohort study of 497775 participants recruited from 2006 to 2010 in the UK Biobank highlighted an HR of 1.14 for incident dementia among IBD patients, but the differences were not significant (95%CI : 0.94-1.39, P value = 0.182). Besides, there was no statistically significant difference in the anatomical and tissue-specific volumes of their brains on magnetic resonance images[8]. Furthermore, a recent meta-analysis including seven observational studies (six cohort studies and one case-control study) and 20174 cases did not identify a significant association between UC [relative risks (RR) = 1.16, 95%CI : 0.96-1.41) or CD (RR = 1.17, 95%CI : 0.84-1.62] and the risk of AD[43]. Another meta-analysis encompassing nine studies, including seven cohort studies, one cross-sectional study, and one case-control study, described that a previous diagnosis of IBD did not influence the risk of subsequent all-cause dementia (RR = 1.32, 95%CI : 0.98-1.77) and AD (RR = 1.62, 95%CI : 0.96-2.76)[46]. Of note, subgroup analysis based on the study of the above meta-analysis implied that IBD increased the risk of all-cause dementia but not AD in the cohort study, UC increased the risk of subsequent all-cause dementia and AD, and CD only increased the risk of all-cause dementia[46]. So far, the causal relationship between IBD and dementia has not been established.
In the present MR study, no causal relationship was discovered between genetically predicted IBD and subentities and all-cause dementia and its four subtypes, which contradicts the results of the above-mentioned studies implicating an association between IBD and dementia (Supplementary Table 13). What's more, our finding is not in agreement with that of Guo et al[17]. It is worthwhile emphasizing that their MR study had some limitations, including sample selection bias in the selected AD dataset that included older clinically diagnosed patients but excluded patients with shortened life expectancy due to IBD-related comorbidities, thus reducing or even reversing the MR estimated effect. In addition, Guo et al[17] used univariable MR to estimate the causal roles of UC and CD in AD, which might have led to horizontal pleiotropy due to IV overlapping. Excitingly, a recent study published in Neurology with a large sample size carried out an observational analysis combined with MR analysis corroborated our findings. The observational analysis using data from the United Kingdom Clinical Practice Research Datalink described that the overall incidence of AD was higher in patients with IBD (HR = 1.17, 95%CI : 1.15-1.19, P value = 2.1 × 10-4). Nonetheless, their MR analysis yielded no association between IBD and AD, suggesting that confounding factors may compromised the observed association[47]. Observational studies are susceptible to inherent methodological shortcomings, such as bias and confounding variables. For instance, the recruitment of the majority of participants from Medicare databases or inpatient registries could have increased the risk of selection bias. Surveillance bias also may increase the likelihood of a positive correlation. The gut microbiota, obesity, and other factors have been established as risk factors for both IBD and dementia in previous studies[5,48-50]. On the other side, the use of medications such as proton pump inhibitors and tumor necrosis factor blocking agents might interfere with the results when assessing the association between IBD and dementia[51,52]. At the same time, shared genetic components, such as PPARG and NOS2, could also increase genetic susceptibility to both diseases[53]. Furthermore, meta-analyses typically exhibit statistical heterogeneity arising from differences in study populations, study designs, and inclusion criteria. Finally, two large-scale GWAS comprehensively evaluated the genetic overlap between cognitive traits or AD and gastrointestinal disorders, with neither detecting significant genetic overlap and correlation with IBD[54,55].
A major strength of our study is that the causal effects between IBD and the risk of all-cause dementia and its six subtypes were assessed by utilizing a two-sample MR design, which mitigates limitations inherent noted in observational studies, including measurement error, residual confounding, and reverse causation bias. This MR study incorporated independent and robust genetic variants as IVs, not only to limit the effect of LD and weak instrument bias but also to circumvent the time-consuming and labor-intensive challenges generally encountered in observational studies (Supplementary Tables 2-9). Furthermore, our methodology utilized an iterative approach that is conservative and resilient against the influence of outliers (Supplementary Table 10). A series of pleiotropy and heterogeneity tests were also conducted to ensure the consistency of causal estimates and to confirm the robustness of the present findings (Supplementary Table 10). Finally, our findings were validated through a second, largely independent GWAS that yielded concordant results (Supplementary Table 13). Nevertheless, our study has several limitations. Despite the strength of all selected IVs, they collectively accounted for only 6.75% of the variance in the IBD sample (Supple
Herein, no association was identified between the risk of all-cause dementia and genetically predicted IBD. While there is no clear genetic evidence to support IBD as a risk factor for dementia, the possibility of a potential association between the two diseases cannot be ruled out. Further research is necessitated to identify factors that exert a causal effect on the development of dementia.
Evidence from observational studies has not been able to establish a causal link between inflammatory bowel disease (IBD) and dementia.
Gut homeostasis is implicated in many psychiatric and neurological disorders through the bidirectional microbiome-gut-brain axis.
The aim was to find out whether IBD was causally related to all-cause dementia.
Based on the publicly available genome-wide association study data from large population, multiple methods of Mendelian randomization (MR) were performed to estimate the effects of genetically predicted IBD on dementia, and inverse variance weighted was considered as the primary analysis. MR egger intercept, MR pleiotropy residual sum and outlier, and Cochran's Q test were used to test pleiotropy and heterogeneity.
No evidence for a causal effect of IBD on dementia risk was found in three MR methods of MR, which was consistent with validation analyses. Furthermore, MR analysis suggested that IBD and subentities did not causally affect all-cause dementia and its four subtypes.
Our MR study found no association between the risk of all-cause dementia and genetically predicted IBD.
Genetically predicted IBD is not associated with all-cause dementia risk, and dementia prevention interventions for patients with IBD can be similar to those in healthy populations.
We want to acknowledge the participants and investigators of the IIBDGC and FinnGen study. We also thank the United Kingdom Biobank for providing summary statistics for these analyses.
| 1. | Maloney B, Lahiri DK. Epigenetics of dementia: understanding the disease as a transformation rather than a state. Lancet Neurol. 2016;15:760-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 118] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 2. | Shah H, Albanese E, Duggan C, Rudan I, Langa KM, Carrillo MC, Chan KY, Joanette Y, Prince M, Rossor M, Saxena S, Snyder HM, Sperling R, Varghese M, Wang H, Wortmann M, Dua T. Research priorities to reduce the global burden of dementia by 2025. Lancet Neurol. 2016;15:1285-1294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 280] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 3. | Martin CR, Osadchiy V, Kalani A, Mayer EA. The Brain-Gut-Microbiome Axis. Cell Mol Gastroenterol Hepatol. 2018;6:133-148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 492] [Cited by in RCA: 872] [Article Influence: 109.0] [Reference Citation Analysis (0)] |
| 4. | Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin Immunopathol. 2015;37:47-55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 602] [Article Influence: 50.2] [Reference Citation Analysis (1)] |
| 5. | Lee M, Chang EB. Inflammatory Bowel Diseases (IBD) and the Microbiome-Searching the Crime Scene for Clues. Gastroenterology. 2021;160:524-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 453] [Article Influence: 90.6] [Reference Citation Analysis (0)] |
| 6. | Brudek T. Inflammatory Bowel Diseases and Parkinson's Disease. J Parkinsons Dis. 2019;9:S331-S344. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 100] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 7. | Wang X, Wan J, Wang M, Zhang Y, Wu K, Yang F. Multiple sclerosis and inflammatory bowel disease: A systematic review and meta-analysis. Ann Clin Transl Neurol. 2022;9:132-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 8. | Sun Y, Geng J, Chen X, Chen H, Wang X, Chen J, Li X, Hesketh T. Association Between Inflammatory Bowel Disease and Dementia: A Longitudinal Cohort Study. Inflamm Bowel Dis. 2022;28:1520-1526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 9. | Zhang B, Wang HE, Bai YM, Tsai SJ, Su TP, Chen TJ, Wang YP, Chen MH. Inflammatory bowel disease is associated with higher dementia risk: a nationwide longitudinal study. Gut. 2021;70:85-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 218] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 10. | Rønnow Sand J, Troelsen FS, Horváth-Puhó E, Henderson VW, Sørensen HT, Erichsen R. Risk of dementia in patients with inflammatory bowel disease: a Danish population-based study. Aliment Pharmacol Ther. 2022;56:831-843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 11. | Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133-1163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3525] [Cited by in RCA: 3278] [Article Influence: 182.1] [Reference Citation Analysis (0)] |
| 12. | Smith GD, Ebrahim S. 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2788] [Cited by in RCA: 4158] [Article Influence: 180.8] [Reference Citation Analysis (0)] |
| 13. | Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017;26:2333-2355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 368] [Cited by in RCA: 1238] [Article Influence: 112.5] [Reference Citation Analysis (0)] |
| 14. | Zheng J, Baird D, Borges MC, Bowden J, Hemani G, Haycock P, Evans DM, Smith GD. Recent Developments in Mendelian Randomization Studies. Curr Epidemiol Rep. 2017;4:330-345. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 734] [Cited by in RCA: 818] [Article Influence: 90.9] [Reference Citation Analysis (0)] |
| 15. | Li H, Wen Z. Effects of ulcerative colitis and Crohn's disease on neurodegenerative diseases: A Mendelian randomization study. Front Genet. 2022;13:846005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 16. | Freuer D, Meisinger C. Association between inflammatory bowel disease and Parkinson's disease: A Mendelian randomization study. NPJ Parkinsons Dis. 2022;8:55. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 17. | Guo X, Chong L, Zhang X, Li R. Letter to the editor: Genetically determined IBD is associated with decreased risk of Alzheimer's disease: a Mendelian randomisation study. Gut. 2022;71:1688-1689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 18. | Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, Ripke S, Lee JC, Jostins L, Shah T, Abedian S, Cheon JH, Cho J, Dayani NE, Franke L, Fuyuno Y, Hart A, Juyal RC, Juyal G, Kim WH, Morris AP, Poustchi H, Newman WG, Midha V, Orchard TR, Vahedi H, Sood A, Sung JY, Malekzadeh R, Westra HJ, Yamazaki K, Yang SK; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium, Barrett JC, Alizadeh BZ, Parkes M, Bk T, Daly MJ, Kubo M, Anderson CA, Weersma RK. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979-986. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1898] [Cited by in RCA: 1986] [Article Influence: 180.5] [Reference Citation Analysis (0)] |
| 19. | Wu Y, Murray GK, Byrne EM, Sidorenko J, Visscher PM, Wray NR. GWAS of peptic ulcer disease implicates Helicobacter pylori infection, other gastrointestinal disorders and depression. Nat Commun. 2021;12:1146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 125] [Article Influence: 25.0] [Reference Citation Analysis (1)] |
| 20. | Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol. 2011;40:740-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 1408] [Article Influence: 88.0] [Reference Citation Analysis (0)] |
| 21. | Palmer TM, Lawlor DA, Harbord RM, Sheehan NA, Tobias JH, Timpson NJ, Davey Smith G, Sterne JA. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat Methods Med Res. 2012;21:223-242. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 956] [Cited by in RCA: 1008] [Article Influence: 72.0] [Reference Citation Analysis (0)] |
| 22. | Gill D, Efstathiadou A, Cawood K, Tzoulaki I, Dehghan A. Education protects against coronary heart disease and stroke independently of cognitive function: evidence from Mendelian randomization. Int J Epidemiol. 2019;48:1468-1477. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 119] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 23. | Levin MG, Judy R, Gill D, Vujkovic M, Verma SS, Bradford Y; Regeneron Genetics Center, Ritchie MD, Hyman MC, Nazarian S, Rader DJ, Voight BF, Damrauer SM. Genetics of height and risk of atrial fibrillation: A Mendelian randomization study. PLoS Med. 2020;17:e1003288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 140] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 24. | Esteban-Cornejo I, Ho FK, Petermann-Rocha F, Lyall DM, Martinez-Gomez D, Cabanas-Sánchez V, Ortega FB, Hillman CH, Gill JMR, Quinn TJ, Sattar N, Pell JP, Gray SR, Celis-Morales C. Handgrip strength and all-cause dementia incidence and mortality: findings from the UK Biobank prospective cohort study. J Cachexia Sarcopenia Muscle. 2022;13:1514-1525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 25. | Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, Reeve MP, Laivuori H, Aavikko M, Kaunisto MA, Loukola A, Lahtela E, Mattsson H, Laiho P, Della Briotta Parolo P, Lehisto AA, Kanai M, Mars N, Rämö J, Kiiskinen T, Heyne HO, Veerapen K, Rüeger S, Lemmelä S, Zhou W, Ruotsalainen S, Pärn K, Hiekkalinna T, Koskelainen S, Paajanen T, Llorens V, Gracia-Tabuenca J, Siirtola H, Reis K, Elnahas AG, Sun B, Foley CN, Aalto-Setälä K, Alasoo K, Arvas M, Auro K, Biswas S, Bizaki-Vallaskangas A, Carpen O, Chen CY, Dada OA, Ding Z, Ehm MG, Eklund K, Färkkilä M, Finucane H, Ganna A, Ghazal A, Graham RR, Green EM, Hakanen A, Hautalahti M, Hedman ÅK, Hiltunen M, Hinttala R, Hovatta I, Hu X, Huertas-Vazquez A, Huilaja L, Hunkapiller J, Jacob H, Jensen JN, Joensuu H, John S, Julkunen V, Jung M, Junttila J, Kaarniranta K, Kähönen M, Kajanne R, Kallio L, Kälviäinen R, Kaprio J; FinnGen, Kerimov N, Kettunen J, Kilpeläinen E, Kilpi T, Klinger K, Kosma VM, Kuopio T, Kurra V, Laisk T, Laukkanen J, Lawless N, Liu A, Longerich S, Mägi R, Mäkelä J, Mäkitie A, Malarstig A, Mannermaa A, Maranville J, Matakidou A, Meretoja T, Mozaffari SV, Niemi MEK, Niemi M, Niiranen T, O Donnell CJ, Obeidat ME, Okafo G, Ollila HM, Palomäki A, Palotie T, Partanen J, Paul DS, Pelkonen M, Pendergrass RK, Petrovski S, Pitkäranta A, Platt A, Pulford D, Punkka E, Pussinen P, Raghavan N, Rahimov F, Rajpal D, Renaud NA, Riley-Gillis B, Rodosthenous R, Saarentaus E, Salminen A, Salminen E, Salomaa V, Schleutker J, Serpi R, Shen HY, Siegel R, Silander K, Siltanen S, Soini S, Soininen H, Sul JH, Tachmazidou I, Tasanen K, Tienari P, Toppila-Salmi S, Tukiainen T, Tuomi T, Turunen JA, Ulirsch JC, Vaura F, Virolainen P, Waring J, Waterworth D, Yang R, Nelis M, Reigo A, Metspalu A, Milani L, Esko T, Fox C, Havulinna AS, Perola M, Ripatti S, Jalanko A, Laitinen T, Mäkelä TP, Plenge R, McCarthy M, Runz H, Daly MJ, Palotie A. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613:508-518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1241] [Cited by in RCA: 2813] [Article Influence: 937.7] [Reference Citation Analysis (0)] |
| 26. | Hartwig FP, Davies NM, Hemani G, Davey Smith G. Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int J Epidemiol. 2016;45:1717-1726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 243] [Cited by in RCA: 622] [Article Influence: 77.8] [Reference Citation Analysis (0)] |
| 27. | Ou YN, Yang YX, Shen XN, Ma YH, Chen SD, Dong Q, Tan L, Yu JT. Genetically determined blood pressure, antihypertensive medications, and risk of Alzheimer's disease: a Mendelian randomization study. Alzheimers Res Ther. 2021;13:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 28. | Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693-698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2495] [Cited by in RCA: 6212] [Article Influence: 776.5] [Reference Citation Analysis (0)] |
| 29. | Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377-389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1644] [Cited by in RCA: 3381] [Article Influence: 375.7] [Reference Citation Analysis (0)] |
| 30. | Greco M FD, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34:2926-2940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 1159] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 31. | Zhou S, Zhu G, Xu Y, Gao R, Zhang M, Zeng Q, Su W, Wang R. Mendelian randomization study on the causal effect of chickenpox on dementia. J Med Virol. 2023;95:e28420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 10] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 32. | Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1330] [Cited by in RCA: 4432] [Article Influence: 340.9] [Reference Citation Analysis (1)] |
| 33. | Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2275] [Cited by in RCA: 7164] [Article Influence: 651.3] [Reference Citation Analysis (1)] |
| 34. | Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4747] [Cited by in RCA: 5629] [Article Influence: 703.6] [Reference Citation Analysis (0)] |
| 35. | Kim JS, Chen MH, Wang HE, Lu CL, Wang YP, Zhang B. Inflammatory Bowel Disease and Neurodegenerative Diseases. Gut Liver. 2023;17:495-504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 29] [Reference Citation Analysis (0)] |
| 36. | Wang D, Zhang X, Du H. Inflammatory bowel disease: A potential pathogenic factor of Alzheimer's disease. Prog Neuropsychopharmacol Biol Psychiatry. 2022;119:110610. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 37. | Zingel R, Bohlken J, Kostev K. Association Between Inflammatory Bowel Disease and Dementia: A Retrospective Cohort Study. J Alzheimers Di. 80:1471-1478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 38. | Liu M, Li D, Hong X, Sun Z. Increased Risk for Dementia in Patients With Inflammatory Bowel Disease: A Systematic Review and Meta-Analysis of Population-Based Studies. Front Neurol. 2022;13:813266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 39. | Zhang MN, Shi YD, Jiang HY. The risk of dementia in patients with inflammatory bowel disease: a systematic review and meta-analysis. Int J Colorectal Dis. 2022;37:769-775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 40. | Zuin M, De Giorgio R, Capatti E, Boschetti E, Zuliani G. Inflammatory bowel disease as a new risk factor for dementia. Aging Clin Exp Res. 2022;34:1725-1728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 41. | Zhou M, Xu R, Kaelber DC, Gurney ME. Tumor Necrosis Factor (TNF) blocking agents are associated with lower risk for Alzheimer's disease in patients with rheumatoid arthritis and psoriasis. PLoS One. 2020;15:e0229819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 42. | Cooper J, Pastorello Y, Slevin M. A meta-analysis investigating the relationship between inflammation in autoimmune disease, elevated CRP, and the risk of dementia. Front Immunol. 2023;14:1087571. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 44] [Reference Citation Analysis (0)] |
| 43. | Xing Y, Li P, Jia Y, Zhang K, Liu M, Jiang J. Association of inflammatory bowel disease and related medication exposure with risk of Alzheimer's disease: An updated meta-analysis. Front Aging Neurosci. 2022;14:1082575. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 44. | Kaneko R, Matsui A, Watanabe M, Harada Y, Kanamori M, Awata N, Kawazoe M, Takao T, Kobayashi Y, Kikutake C, Suyama M, Saito T, Saido TC, Ito M. Increased neutrophils in inflammatory bowel disease accelerate the accumulation of amyloid plaques in the mouse model of Alzheimer's disease. Inflamm Regen. 2023;43:20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 45. | He XF, Li LL, Xian WB, Li MY, Zhang LY, Xu JH, Pei Z, Zheng HQ, Hu XQ. Chronic colitis exacerbates NLRP3-dependent neuroinflammation and cognitive impairment in middle-aged brain. J Neuroinflammation. 2021;18:153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 87] [Article Influence: 17.4] [Reference Citation Analysis (1)] |
| 46. | Liu N, Wang Y, He L, Sun J, Wang X, Li H. Inflammatory bowel disease and risk of dementia: An updated meta-analysis. Front Aging Neurosci. 2022;14:962681. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 47. | Huang J, Su B, Karhunen V, Gill D, Zuber V, Ahola-Olli A, Palaniswamy S, Auvinen J, Herzig KH, Keinänen-Kiukaanniemi S, Salmi M, Jalkanen S, Lehtimäki T, Salomaa V, Raitakari OT, Matthews PM, Elliott P, Tsilidis KK, Jarvelin MR, Tzoulaki I, Dehghan A. Inflammatory Diseases, Inflammatory Biomarkers, and Alzheimer Disease: An Observational Analysis and Mendelian Randomization. Neurology. 2023;100:e568-e581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 48. | Saji N, Niida S, Murotani K, Hisada T, Tsuduki T, Sugimoto T, Kimura A, Toba K, Sakurai T. Analysis of the relationship between the gut microbiome and dementia: a cross-sectional study conducted in Japan. Sci Rep. 2019;9:1008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 160] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 49. | Chauhan N, Tay ACY, Marshall BJ, Jain U. Helicobacter pylori VacA, a distinct toxin exerts diverse functionalities in numerous cells: An overview. Helicobacter. 2019;24:e12544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 102] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 50. | Gorospe EC, Dave JK. The risk of dementia with increased body mass index. Age Ageing. 2007;36:23-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 106] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 51. | Papazoglou A, Arshaad MI, Henseler C, Daubner J, Broich K, Haenisch B, Weiergräber M. The Janus-like Association between Proton Pump Inhibitors and Dementia. Curr Alzheimer Res. 2021;18:453-469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 52. | Watad A, McGonagle D, Anis S, Carmeli R, Cohen AD, Tsur AM, Ben-Shabat N, Luigi Bragazzi N, Lidar M, Amital H. TNF inhibitors have a protective role in the risk of dementia in patients with ankylosing spondylitis: Results from a nationwide study. Pharmacol Res. 2022;182:106325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 53. | Dong L, Shen Y, Li H, Zhang R, Yu S, Wu Q. Shared Genes of PPARG and NOS2 in Alzheimer's Disease and Ulcerative Colitis Drive Macrophages and Microglia Polarization: Evidence from Bioinformatics Analysis and Following Validation. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 18] [Reference Citation Analysis (0)] |
| 54. | Adewuyi EO, O'Brien EK, Nyholt DR, Porter T, Laws SM. A large-scale genome-wide cross-trait analysis reveals shared genetic architecture between Alzheimer's disease and gastrointestinal tract disorders. Commun Biol. 2022;5:691. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 55. | Adewuyi EO, O'Brien EK, Porter T, Laws SM. Relationship of Cognition and Alzheimer's Disease with Gastrointestinal Tract Disorders: A Large-Scale Genetic Overlap and Mendelian Randomisation Analysis. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 22] [Reference Citation Analysis (0)] |
Open-Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Psychiatry
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: M'Koma AE, United States; Yau PTO, United Kingdom S-Editor: Qu XL L-Editor: A P-Editor: Zhao S