Published online Jun 20, 2025. doi: 10.5493/wjem.v15.i2.102395
Revised: January 5, 2025
Accepted: January 14, 2025
Published online: June 20, 2025
Processing time: 181 Days and 21.1 Hours
Hepatitis B virus (HBV) infection is still a major worldwide health concern, contributing to chronic liver disorders like hepatocellular carcinoma (HCC). This review comprehensively analyzes HBV genotype distribution, mutation patterns, and their clinical implications, focusing on diagnostic and therapeutic strategies for HBV-positive patients. The discussion begins with HBV virology, emphasizing its capacity for chronic hepatitis and its association with severe liver complications, notably HCC. Understanding HBV genotypes (A-J) and their distinct geo
Core Tip: Hepatitis B virus (HBV) genotypes and mutations significantly influence treatment responses, clinical outcomes, and disease etiology. Integrating genotyping and mutation studies improves diagnostic precision, treatment choices, and patient outcomes. Prioritizing genotype-specific management strategies will help healthcare practitioners maximize therapeutic efficacy and slow the course of disease. Future studies should concentrate on clarifying the molecular processes that underlie the pathophysiology and treatment outcomes specific to HBV genotypes. This thorough review highlights the significance of genotype-specific management approaches in the treatment of hepatitis B and offers an in-depth understanding of the intricate relationships between HBV genotypes, mutations, and clinical outcomes.
- Citation: Ratnaparkhi MM, Vyawahare CR, Gandham NR. Hepatitis B virus genotype distribution and mutation patterns: Insights and clinical implications for hepatitis B virus positive patients. World J Exp Med 2025; 15(2): 102395
- URL: https://www.wjgnet.com/2220-315x/full/v15/i2/102395.htm
- DOI: https://dx.doi.org/10.5493/wjem.v15.i2.102395
The DNA virus known as the hepatitis B virus (HBV) mainly attacks liver cells, which results in hepatitis. It is a major global health concern due to its ability to cause chronic infection, resulting in serious liver conditions such as hepatocellular carcinoma (HCC) and cirrhosis[1]. Understanding HBV's virology and genetic characteristics is crucial for developing effective diagnostic, therapeutic, and preventive strategies. HBV belongs to the Hepadnaviridae family and is recognized for its unique replication strategy, which involves reverse transcription of an RNA intermediate. The virus is highly infectious and can be transmitted through blood, sexual contact, and perinatally from mother to child. About 240 million people globally suffer from chronic HBV infection, which is a leading contributor toliver-related disease and mortality[2]. Even though a highly efficient vaccination is available, HBV continues to be a significant public health challenge, particularly in regions with limited access to vaccination and healthcare resources. The range of HBV infection symptoms is from acute to, self-limiting hepatitis inflammation to chronic infection, which can persist for decades. Chronic HBV infection is often asymptomatic in the early stages but can develop into cirrhosis, HCC, and liver fibrosis over time. Influential factors on the HBV infection's natural course include the age at infection, immune response, and viral genotype. Effective management of HBV requires a comprehensive understanding of these factors to optimize treatment and prevent long-term complications[1].
HBV exists as multiple genotypes (A-J) with distinct geographic distributions and clinical implications. Genotype variations influence disease progression, treatment responses, and the risk of developing complications like HCC[3]. Mutations within the HBV genome further complicate management by affecting viral replication, antigenicity, and resistance to antiviral therapies. Therefore, genotype and mutation analysis plays a pivotal role in customizing individual treatment regimens and enhancing the outcomes of the patients. Genotype analysis of HBV is essential for comprehending the natural history and epidemiology of the infection. Different genotypes are associated with varying clinical outcomes and responses to treatment. For instance, genotype C is linked to a reduced response to interferon therapy and an increased risk of HCC as compared to genotype B. In contrast, genotype A is often associated with a better response to antiviral treatment. Recognizing these genotype-specific characteristics enables clinicians to predict disease prognosis and select the most appropriate therapeutic approach. Mutations in the HBV genome can lead to the emergence of drug-resistant strains, complicating treatment and management. Common mutations, such as those in the promoter regions of the pre-core and basal core, can alter viral replication and antigen production, impacting disease severity and treatment efficacy. Additionally, mutations in the polymerase gene can confer resistance to nucleos(t)ide analogs, necessitating the use of alternative antiviral agents. Continuous monitoring of HBV genotypes and mutations is crucial for adapting treatment strategies and achieving sustained virological suppression, ultimately improving patient outcomes and reducing the burden of HBV-related liver disease[4].
One of the main risk factors for HCC is persistent HBV infection, accounting for a significant proportion of cases globally[5]. The virus integrates into the host genome, leading to genomic instability, oncogenic mutations, and dysregulation of cellular pathways involved in carcinogenesis. Long-term inflammation and immune responses against infected hepatocytes contribute to the development of liver cirrhosis from chronic hepatitis and ultimately HCC.
One crucial step in the pathogenesis of HCC is integrating HBV DNA into the host genome. This integration can disrupt host genes, including tumor suppressor genes, and activate oncogenes, which causes uncontrolled cell growth and malignancy. Moreover, integrated HBV DNA can produce truncated viral proteins with oncogenic potential, further promoting the development of HCC. The integration sites are often found near regions of the genome that are involved in cell cycle regulation and apoptosis, highlighting the direct HBV's part in liver cancer development. In addition to direct genetic effects, chronic HBV infection induces a persistent inflammatory response in the liver. The immune system's efforts to clear the virus result in repeated liver cell damage and regeneration cycles, creating an environment conducive to carcinogenesis[6]. Cytokines and other inflammatory mediators released during this process can induce oxidative stress, DNA damage, and fibrogenesis. As liver fibrosis progresses to cirrhosis, the risk of developing HCC increases significantly. Understanding these mechanisms underscores the importance of early detection and effective management of chronic HBV infection to prevent HCC development.
The incidence of HBV-associated HCC varies geographically, with a high prevalence in regions including Sub-Saharan Africa and East Asia, where HBV infection is widespread. The risk of HCC is highest among individuals with chronic HBV infection, especially those who acquired the infection at birth or during early childhood[7].
In regions with high endemicity, vertical transmission (from mother to child) and horizontal transmission (among children) are the predominant modes of HBV spread[7]. These transmission routes contribute to a high rate of chronic infection, as the immune system is less likely to clear the virus when infection occurs early in life. As a result, individuals in these areas are more likely to develop long-term complications, including cirrhosis and HCC. Public health efforts, such as universal vaccination programs for newborns, have been implemented in many endemic regions to decrease the incidence of chronic HBV infection and subsequently lower the risk of HCC[7].
Socioeconomic factors and access to healthcare also play significant roles in determining the epidemiology of HBV-associated HCC. In low-resource settings, limited access to antiviral therapies and liver cancer screening programs contributes to higher rates of HCC[8]. Additionally, co-infections with more hepatitis viruses (like hepatitis C) or conditions like human immunodeficiency virus (HIV) may worsen liver damage and raise the chance of HCC in people with HBV infection. Addressing these disparities through improved healthcare infrastructure and targeted interventions is essential for reducing the global burden of HBV-associated HCC.
Public health and the economy are both greatly impacted by the HBV. About 820000 people die globally from liver disorders such cirrhosis and HCC each year as a result of it.
Due to its ability to transmit through blood, sexual contact, and pregnancy, HBV puts a strain on healthcare systems to establish extensive screening and immunization initiatives. Social and mental well-being are negatively impacted by the stigma and discrimination that people with HBV frequently experience. Programs for vaccination, diagnostic testing, antiviral drugs, liver transplants, and the management of problems associated to the liver are expensive.
While low-income nations struggle with underfunded healthcare systems, high-income nations spend millions each year handling issues related to HBV[8].
India is categorized as having intermediate HBV endemicity and there are about 40 million chronic HBV carriers in India. Significantly, prevalence rates are higher in some indigenous communities, suggesting that these communities require focused interventions. Both afflicted individuals and the healthcare system bear a heavy cost burden in managing HBV and its sequelae. Acknowledging the prevalence of HBV, the Indian government has taken a number of actions. In an effort to lower the disease burden and related mortality, the National Viral Hepatitis Control Program was established to offer free hepatitis B and C screening, diagnosis, and treatment[9].
Hepatitis B vaccination is used to prevent new infections, with a particular emphasis on birth-dose immunization to stop transmission during pregnancy.
The entire financial load could be lessened if scientific advances result in improved treatments, more affordable diagnostics, or even a cure. Making use of this knowledge of HBV's complex effects highlights the urgent need for more research into cutting-edge diagnostic, prophylactic, and treatment approaches to enhance individual and public health outcomes.
Several factors contribute to the progression of HCC in individuals with positive HBV, including high viral load, persistent viral replication, presence of specific HBV genotypes (e.g., genotype C), HIV and hepatitis C virus (HCV) co-infection, and underlying liver cirrhosis. Early detection of these risk factors through regular surveillance and screening is crucial for timely intervention and improved prognosis[10].
Among these risk factors, a high HBV DNA viral load is one significant predictor of HCC development. Research has indicated that individuals with higher levels of HBV DNA are at an increased possibility for liver cancer, irrespective of the stage of their liver disease. This underscores the importance of antiviral medication to suppress HBV replication and reduce viral load, which can subsequently lower the risk of HCC. Routine evaluation of HBV DNA concentrations in chronically infected patients is vital for assessing disease progression and how well the treatment plans work[10]. The likelihood of developing mutations varies throughout HBV genotypes. Precore (PC) and basal core promoter (BCP) mutations are more common in genotype C, which lowers hepatitis B e antigen (HBeAg) expression and causes immune evasion and chronic liver damage. The capacity of different HBV genotypes to create viral proteins, including HBV X protein (HBx), which is essential for carcinogenesis, varies. HBx can disrupt the function of the p53 tumor suppressor, encourage cell division, and prevent apoptosis.
HBx protein overexpression is more common in some genotypes, including genotype C, which increases the risk of HCC. Particularly in men, HBV genotype C is linked to delayed HBeAg seroconversion, which means that people stay in an active viral replication phase for longer. This is correlated with a higher risk of HCC.
The presence of liver cirrhosis, whether due to HBV infection alone or in combination with other factors, significantly elevates the risk of HCC. Cirrhosis represents the advanced stage of liver disease where impaired liver function results from the replacement of healthy liver tissue by scar tissue. Patients who have cirrhosis are more likely to develop malignant transformation of hepatocytes. Additionally, lifestyle factors such as obesity and alcohol use are two things that can make liver disease worse and further increase the risk of HCC in HBV-positive individuals[11]. Comprehensive management approaches that address risk factors that are both viral and non-viral are essential for reducing the occurrence of HCC in this particular population.
The HBV genome is compact and circular, approximately of size 3.2 kb consisting of four overlapping open reading frames (ORFs): (1) S region; (2) C region; (3) P region; and (4) X region. Each ORF encodes essential viral proteins involved in replication, assembly, and pathogenesis. Despite its small size, the HBV genome efficiently encodes multiple proteins through the use of overlapping reading frames and regulatory elements, allowing it to maintain its compact structure while carrying out complex functions necessary for its life cycle[1].
S region: Encodes the surface antigens [hepatitis B surface antigen (HBsAg)] responsible for virus entry and immune recognition. The S region includes pre-S1, pre-S2, and S genes, which are critical for the formation of the viral envelope and binding itself to receptors on host cells. The accurate expression of these antigens is crucial for the ability of the virus to infect the hepatocytes and elicit an immune response.
C region: Produces core proteins and HBeAg involved in viral assembly and immune modulation. The core protein forms the nucleocapsid, which houses the viral DNA, while HBeAg modulates the host immune response, helping the virus evade detection and establish persistent infection.
P region: Encodes the viral DNA polymerase essential for genome replication. The polymerase has multiple functions, including priming, reverse transcription, and DNA synthesis, making it a vital component for the replication of the viral genome. Mutations in this region can lead to resistance to antiviral therapies.
X region: Codes for the X protein, which regulates viral transcription, host cell survival pathways, and immune evasion strategies. The X protein is a multifunctional regulator that can modulate cellular signalling pathways, promote cell survival, and contribute to the oncogenic potential of HBV by interfering with cell cycle control and apoptosis.
Mutations within these genomic regions can alter viral fitness, antigenicity, and susceptibility to antiviral drugs. For example, the pre-core and core promoter region mutations can lead to HBeAg-negative chronic hepatitis B, which is linked to liver disease that is more severe and reduced treatment response[12]. Additionally, the polymerase gene mutations can result in resistance to the nucleos(t)ide analogs, complicating treatment efforts. Understanding these mutations and their impacts on the viral lifecycle and host interactions is essential for developing targeted therapies and effective management strategies for HBV infection.
HBV genotypes exhibit different geographical distributions and clinical outcomes. Prevalence of genotype A is observed in Europe and Africa, while genotypes B and C predominates the East Asia[13]. Genotype D is common in the Mediterranean region, and genotype E is found predominantly in West Africa. Genotypes F and H are mainly seen in Central and South America, whereas genotype G is less common and observed sporadically in the United States and France. This global distribution pattern is influenced by historical human migration, socio-economic factors, and regional healthcare practices, reflecting the complex epidemiology of HBV.
Worldwide, five of the nine genotypes account for almost 96% of chronic HBV infections: Mostly found in East Asia, genotype C is the most prevalent, making up 26% of infections. With 22% of the population, genotype D is common in the Middle East, India, and the Mediterranean region. With 18% of infections are caused by genotype E, mostly in sub-Saharan Africa. Common across North America, Europe, and Africa, genotype A accounts for 17%. In Southeast Asia, genotype B accounts for 14% of the population.
The understanding of global HBV genotype distribution is crucial for public health planning and resource allocation. For instance, vaccination programs can be tailored to target predominant genotypes in different regions, enhancing their effectiveness. Additionally, epidemiological studies focusing on genotype distribution can help identify at-risk popu
The distribution of HBV genotypes influences disease progression and treatment responses. For instance, the risk of having HCC is increased in those with genotype C compared to other genotypes, whereas genotype A is linked to a better reaction to interferon-based treatments[14]. In areas where genotypes B and C are more common, such as East Asia, the incidence of liver cirrhosis and HCC is notably higher. Conversely, in regions dominated by genotype A, patients tend to exhibit a more favourable prognosis with a higher likelihood of achieving sustained virological response[15].
Understanding regional variations in genotype prevalence is vital for the development of region-specific clinical guidelines and therapeutic protocols. Healthcare providers can leverage this knowledge to offer personalized treatment plans that account for genotype-related risks and benefits, thereby improving patient outcomes. Moreover, regional genotype data can inform public health policies, including vaccination and screening programs, tailored to the unique epidemiological landscape of each area.
In India, HBV genotypes show considerable diversity, with genotypes A and D being the most prevalent. Genotype D is the dominant strain found across various regions of the country, while genotype A is more commonly observed in the northern parts. The presence of genotypes B and C is relatively rare but has been documented in certain populations, reflecting the influence of migration and ethnic diversity.
Understanding the genotype distribution in India is essential for optimizing treatment strategies and public health interventions. For instance, genotype D, which is more common in India, is associated with a poorer response to interferon-based therapies compared to genotype A. This information can guide clinicians in selecting appropriate antiviral therapies and help in developing targeted vaccination programs to control the spread of HBV. Furthermore, recognizing regional genotype variations can aid in identifying high-risk groups and tailoring screening efforts to mitigate the impact of HBV-related liver diseases in the country[16,17].
Different HBV genotypes exhibit varying degrees of pathogenicity and responsiveness to antiviral therapies. Genotype-specific mutations can affect treatment outcomes by influencing drug resistance profiles and viral replication dynamics. For example, patients having genotype C are more prone to develop drug-resistant mutations, complicating treatment regimens. In contrast, genotype B is linked to a lower incidence of resistance to the drug but a higher likelihood of liver inflammation and fibrosis. Understanding these genotype-specific differences is critical for optimizing therapeutic strategies and improving patient care[16].
Research has shown that certain genotypes, such as genotype C, have a higher propensity for chronicity and liver damage, leading to more severe clinical outcomes. Conversely, genotype A patients often experience better responses to pegylated interferon treatment, making them suitable candidates for this therapeutic approach. These insights into genotype-disease relationships underscore the importance of genotype testing in clinical practice. By incorporating genotype analysis into routine diagnostics, healthcare providers can make more informed decisions regarding treatment options, monitor disease progression more accurately, and ultimately enhance the quality of care for HBV-infected individuals[16].
HBV undergoes frequent mutations due to its high replication rate and error-prone viral polymerase. Common mutations occur in the pre-core, core promoter, and polymerase regions, affecting viral replication efficiency, antigen expression, and drug susceptibility. In the pre-core region, the G1896A mutation leads to the production of a defective HBeAg, resulting in HBeAg-negative chronic hepatitis B[18]. The core promoter region mutations, such as A1762T/G1764A, are associated with higher viral replication and increased risk of liver disease progression. Polymerase region mutations, such as rtM204V/I, A181T/V and N236T are linked to resistance against antiviral drugs like lamivudine, adefovir and entecavir respectively making treatment more challenging. K130M and V131I X gene mutations enhance oncogenic potential.
In addition to these, pre-S region mutations causes oxidative stress and liver damage. Antigenic epitope changes lessen immune recognition. The accumulation of pre-S protein mutations raises the risk of HCC and adds to genomic instability. It connection with the development of HCC and severe liver disease. Itassociated with decreased detection of surface antigens, which leads to diagnostic failures. S region can undergo mutations that alter the structure of the HBsAg-HBsAg, impacting vaccine efficacy and diagnostic test accuracy. These mutations can lead to "escape mutants" that are not detected by standard assays or are less responsive to vaccine-induced immunity. Therefore, continuous monitoring and updating of diagnostic tools and vaccines are essential to manage these evolving viral forms effectively.
Mutations in HBV are driven by selective pressure of the host's immune system responses and antiviral therapies. Escape mutations allow the virus to evade immune detection and persist in the host, contributing to chronic infection and treatment resistance. The immune system exerts selective pressure by targeting viral antigens, leading to the emergence of variants that can avoid immune recognition. Antiviral therapies, particularly nucleos(t)ide analogs, impose selective pressure by inhibiting viral replication, which can result in the selection of resistant strains. The error-prone nature of the HBV polymerase further facilitates the accumulation of these mutations[19]. Reverse transcription, the method by which HBV replicates, is prone to errors due to its lack of proofreading skills. A high rate of mutation results from this, contributing to the development of diverse virus populations within the host. Genetic recombination events can occasionally occur across distinct HBV genotypes, producing novel variations with distinctive characteristics. These recombination events can affect virulence and add to the genomic diversity of the virus.
Environmental factors, such as co-infections with other hepatitis viruses or HIV, can also influence mutation patterns by altering the immune landscape and therapeutic responses. Additionally, host genetic factors, including poly
Certain mutations confer resistance to nucleos(t)ide analogs used in HBV treatment, such as lamivudine, entecavir, and tenofovir. These resistance-associated mutations can emerge during treatment and necessitate alternative therapeutic approaches to achieve viral suppression and prevent disease progression. For example, the rtM204V/I mutation in the polymerase gene leads to resistance to lamivudine and may reduce the efficacy of other antiviral drugs. The emergence of such mutations during therapy can result in viral rebound and an increased risk of liver disease progression[20].
For example, the rtM204V/I mutation in the polymerase gene leads to resistance to lamivudine and may reduce the efficacy of other antiviral drugs. Although lamivudine is not preferred nowadays for treatment due to high resistance rates, it remains important to understand its resistance mechanisms. The rtM204V/I mutation causes a significant viral rebound and increases the risk of liver disease progression when resistance develops during therapy.
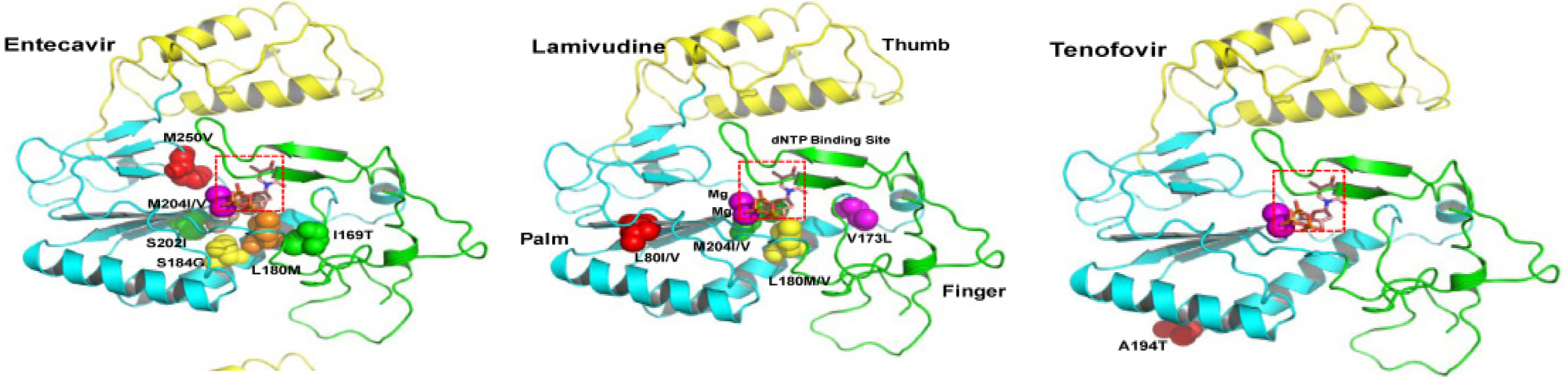
In the case of entecavir, the rtI169T, rtT184G, rtS202I, and rtM250V mutations have been identified as key resistance-associated mutations. These mutations can reduce the efficacy of entecavir, necessitating a switch to alternative therapies to maintain viral suppression and prevent disease progression[21,22].
In addition to drug resistance, mutations can enhance HBV virulence by increasing viral replication and persistence. The BCP mutations (A1762T/G1764A) not only enhance viral replication but also raises the HCC development risk[19]. Similarly, pre-core mutations that inhibit HBeAg production are associated with more aggressive liver disease. These mutations can also affect the clinical management of HBV infection, necessitating regular monitoring and potentially more aggressive treatment strategies. Understanding the role of these mutations is essential for optimizing treatment regimens and improving patient outcomes in HBV management. Table 1 shows mutation impact on HBV and its clinical consequences. Table 2 shows common mutations in HBV genome. Figure 1 shows the ribbon diagram for HBV drugs resistance[23].
| Region | Effect on hepatitis B virus | Clinical consequences |
| Pre-S region | Impaired secretion, immune escape | Increased HCC risk, occult infections |
| S region | Altered hepatitis B surface antigen structure | Diagnostic failures, vaccine escape |
| Basalcore promoter region | Reduced HBeAg, increased replication | Severe liver disease, HCC |
| Precore region | Loss of HBeAg | Silent chronic infection, higher liver inflammation |
| Polymerase region | Antiviral resistance | Treatment failure, persistent viremia |
| X gene | Enhanced oncogenesis | Direct contribution to HCC |
| No. | Mutation | Location | Effect | Clinical significance | Impact on treatment |
| 1 | G1896A | Pre-core | Defective HBeAg production | HBeAg-negative chronic hepatitis B | May affect serological monitoring |
| 2 | G145R, T126I | S region | Immuneand diagnostic escape | Occult infections, vaccine breakthrough | May affect serological monitoring |
| 3 | A1762T/G1764A | Core promoter | Higherviral replication | Increased risk of liverdisease progression | Requires close monitoring |
| 4 | RtM204V/I | Polymerase | Lamivudine resistance | Viral rebound and liver disease progression | Reduced efficacy of lamivudine and potential cross-resistance |
| 5 | RtI169T, rtT184G, rtS202I, rtM250V | Polymerase | Entecavir resistance | Necessitates alternative therapies | Reduced efficacy of entecavir |
| 6 | RtA194T | Polymerase | Tenofovir resistance | Complicates treatment strategies | Reduced susceptibility to tenofovir, especially with priorlamivudine exposure |
| 7 | K130M, V131I | X gene | Adefovir resistance | Enhances viral replication and transcription | These mutations stabilizes the virus under drug pressure |
HBV genotypes significantly influence the natural history and therapeutic outcomes of infection. Genotypes B and C are associated with a higher likelihood of experiencing liver problems, including HCC and cirrhosis, compared to genotypes A and D. For instance, genotype C is particularly linked to a higher incidence of HCC due to its association with pro
Mutations within the HBV genome can alter the efficacy of antiviral drugs, leading to genotype-specific treatment considerations. For example, mutations in the polymerase gene (e.g., rtM204V/I) confer resistance to drugs like lamivudine and require alternative therapies such as entecavir or tenofovir. Genotypic testing is crucial for identifying these drug-resistant mutations early in treatment to optimize therapeutic outcomes and prevent treatment failure[25,26].
Genotype and mutation analysis provides valuable prognostic information that helps predict disease progression and treatment responses in HBV-infected patients. Patients with specific HBV genotypes or drug-resistant mutations may have different clinical trajectories and treatment needs[27]. For instance, individuals with genotype C and certain mutations may require more aggressive monitoring for liver complications and personalized treatment regimens to achieve sustained viral suppression and prevent disease progression[28-30].
HBV escape mutants pose significant challenges in both diagnostics and clinical management. These mutants can evade detection by standard serological assays, leading to false-negative results in diagnostic testing. Moreover, they can evade vaccine-induced immune responses, compromising the effectiveness of HBV vaccination programs. Understanding the prevalence and impact of escape mutants is crucial for developing improved diagnostic assays and enhancing vaccine efficacy to control HBV transmission effectively[31,32].
By integrating genotype and mutation analysis into clinical practice, healthcare professionals can enhance risk stratification, optimize therapeutic approaches, and improve patient outcomes for those infected with HBV. Continued research into HBV genotypes, mutations, and their clinical implications is essential for advancing therapeutic approaches and reducing the global burden of HBV-related liver diseases[33-35].
Occult HBV infection (OBI) is defined as the presence of HBV DNA in the liver (and sometimes detectable in serum) without detectable HBsAg in the blood. Its function in the course of liver disease and HCC, as well as its potential for reactivation and transmission (for example, through organ transplants or blood transfusions), make it a significant clinical entity. OBI can result from various factors such as structural alterations in the HBsAg caused by mutations in the pre-S or S sections of HBV can decrease its detectability in conventional techniques. HBeAg expression is decreased or eliminated by mutations in the BCP and PC, which also aid in immune evasion.
Mutations in the "a-determinant" of HBsAg and other surface gene (S gene) mutations affect antigenicity and diagnostic test recognition. The occult condition may be exacerbated by extremely low viral loads brought on by strong immune-mediated inhibition of HBV replication. Co-infections with HIV or the HCV can change the clinical appearance of HBV or inhibit its replication. Chemotherapy and organ transplantation are examples of immunosuppression that might reactivate OBI. Common HBV mutations linked to OBI are deletions or insertions in the pre-S1 or pre-S2 regions, reducing HBsAg production. Mutations in the S gene (e.g., G145R, D144A/E, or T126I) affect the antigenicity of HBsAg. BCP mutations A1762T/G1764A are common and reduce the expression of HBeAg, which helps HBV evade immune detection. PC G1896A mutation creates a premature stop codon in the precore region, leading to the absence of HBeAg production. X Gene HBx protein mutations (e.g., K130M/V131I) may impact viral replication and persistence. Diagnosis requires sensitive HBV DNA assays, as HBsAg is not detectable. Adverse consequences can be avoided by detecting and monitoring OBI early, especially in high-risk populations[36-38].
Laboratory techniques play a crucial role in detecting HBV genotypes and mutations, providing essential information for clinical management. Polymerase chain reaction (PCR), direct sequencing, and hybridization assays are among the primary methods used for genotype and mutation detection. PCR amplifies specific HBV DNA sequences, allowing for precise identification of viral genotypes[39]. Direct sequencing provides detailed information about genetic variations within the HBV genome, including mutations that influence treatment responses. Hybridization assays, such as line probe assays, offer rapid and accurate detection of specific mutations associated with drug resistance. The Xpert® HBV Viral Load test [Cepheid, Sunnyvale, CA, United States; CE-IVD (in vitro diagnostic medical devices)] is a real-time PCR assay for HBV quantification run on the automated GeneXpert® systems is also available[40]. These techniques enable healthcare providers to customize therapy plans according to unique virus traits and enhance clinical outcomes[41].
But there are certain limitations associated with these techniques like in patients with very low viral load or when the virus is latent, it may be challenging to detect HBV. Particularly in patients receiving antiviral medication or those with an occult HBV infection, false negative results might happen. Not all HBV genotypes or mutations may be efficiently amplified by PCR since it requires particular primers and probes. A diagnosis that is not correct may arise from mispriming or from the failure to identify certain genotypes or mutations. Because of its great sensitivity and susceptibility to contamination, PCR might provide false positive results.
Repetitive testing and strict laboratory controls are necessary, which raises the expense and complexity[42,43].
Personalized medicine approaches in HBV treatment leverage genotype and mutation status to tailor therapeutic strategies to individual patient needs[26]. By identifying specific HBV genotypes and drug-resistant mutations, clinicians can select antiviral agents with optimal efficacy and safety profiles. For example, patients with genotype C and mutations conferring resistance to lamivudine may benefit from first-line therapy with tenofovir or entecavir, which have demonstrated efficacy against drug-resistant variants. Personalized treatment aims to achieve sustained virological suppression, minimize treatment-related adverse effects, and prevent disease progression. Close monitoring of viral load and liver function markers helps assess treatment response and guide adjustments in therapy when necessary[44].
Future research directions in HBV management focus on advancing diagnostic technologies and developing novel therapeutic approaches. Emerging technologies such as next-generation sequencing offer enhanced sensitivity and resolution in detecting HBV genotypes and mutations, facilitating early detection of drug-resistant variants and guiding personalized treatment decisions[45]. By using small interfering RNAs to break down HBV RNA transcripts, RNAi-based therapies lower the synthesis and replication of viral proteins. Clinical investigations have indicated that this strategy has the potential to lower HBsAg levels. Novel antiviral agents targeting specific viral proteins or pathways aim to overcome existing treatment challenges, including drug resistance and incomplete viral suppression. Additionally, research efforts are directed towards immunotherapeutic strategies that enhance host immune responses against HBV-infected hepatocytes, potentially achieving functional cure or long-term virological remission. By using gene editing technologies HBV DNA can be directly targeted and disrupted within infected cells using methods like clustered regularly interspaced short palindromic repeats-Cas9, which may eradicate the virus's genetic material. This strategy has the potential to be a permanent cure, even if it is still in its early phases. Initiatives for collaboration between researchers, physicians, and pharmaceutical companies are critical for accelerating the translation of these innovations into clinical practice, improving patient outcomes, and ultimately reducing the global burden of HBV-related liver diseases.
Continued investment in research, innovation, and collaborative efforts is essential for advancing HBV management strategies, enhancing treatment efficacy, and ultimately achieving the goal of global HBV elimination[46-48].
HBV genotypes and mutations play critical roles in disease pathogenesis, clinical outcomes, and treatment responses. Understanding the genetic diversity of HBV is essential for implementing effective management strategies and improving patient care.
Incorporating genotype and mutation analysis into routine clinical practice enhances diagnostic accuracy, informs treatment decisions, and improves patient outcomes. Healthcare providers should prioritize genotype-specific mana
Future research should focus on elucidating the molecular mechanisms underlying HBV genotype-specific pathogenesis and treatment responses. Addressing the challenges posed by HBV escape mutants through innovative vaccine strategies and antiviral therapies is essential for achieving global hepatitis B control and elimination goals.
This detailed overview provides a comprehensive understanding of the complex interactions between HBV genotypes, mutations, and clinical outcomes, highlighting the importance of genotype-specific management strategies in hepatitis B management.
The authors are grateful to Dr. D. Y. Patil Medical College, Hospital and Research Centre and Dr. D. Y. Patil Vidyapeeth, for providing necessary facility and support.
| 1. | Liang TJ. Hepatitis B: the virus and disease. Hepatology. 2009;49:S13-S21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 747] [Cited by in RCA: 666] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 2. | Burns GS, Thompson AJ. Viral hepatitis B: clinical and epidemiological characteristics. Cold Spring Harb Perspect Med. 2014;4:a024935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 87] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 3. | Croagh CM, Desmond PV, Bell SJ. Genotypes and viral variants in chronic hepatitis B: A review of epidemiology and clinical relevance. World J Hepatol. 2015;7:289-303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 102] [Article Influence: 9.3] [Reference Citation Analysis (1)] |
| 4. | Nguyen MH, Wong G, Gane E, Kao JH, Dusheiko G. Hepatitis B Virus: Advances in Prevention, Diagnosis, and Therapy. Clin Microbiol Rev. 2020;33:e00046-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 347] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 5. | Pandyarajan V, Govalan R, Yang JD. Risk Factors and Biomarkers for Chronic Hepatitis B Associated Hepatocellular Carcinoma. Int J Mol Sci. 2021;22:479. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 28] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 6. | Cho HJ, Cheong JY. Role of Immune Cells in Patients with Hepatitis B Virus-Related Hepatocellular Carcinoma. Int J Mol Sci. 2021;22:8011. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 7. | Kim DY. Changing etiology and epidemiology of hepatocellular carcinoma: Asia and worldwide. J Liver Cancer. 2024;24:62-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 89] [Reference Citation Analysis (0)] |
| 8. | Gnyawali B, Pusateri A, Nickerson A, Jalil S, Mumtaz K. Epidemiologic and socioeconomic factors impacting hepatitis B virus and related hepatocellular carcinoma. World J Gastroenterol. 2022;28:3793-3802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 9. | National guidelines for diagnosis and management of viral hepatitis in India, NVHCP. Available from: http://nvhcp.mohfew.gov.in. |
| 10. | Mak LY, Cruz-Ramón V, Chinchilla-López P, Torres HA, LoConte NK, Rice JP, Foxhall LE, Sturgis EM, Merrill JK, Bailey HH, Méndez-Sánchez N, Yuen MF, Hwang JP. Global Epidemiology, Prevention, and Management of Hepatocellular Carcinoma. Am Soc Clin Oncol Educ Book. 2018;38:262-279. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 170] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 11. | Li S, Saviano A, Erstad DJ, Hoshida Y, Fuchs BC, Baumert T, Tanabe KK. Risk Factors, Pathogenesis, and Strategies for Hepatocellular Carcinoma Prevention: Emphasis on Secondary Prevention and Its Translational Challenges. J Clin Med. 2020;9:3817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 38] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 12. | Kim H, Lee SA, Kim BJ. X region mutations of hepatitis B virus related to clinical severity. World J Gastroenterol. 2016;22:5467-5478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 36] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 13. | Sunbul M. Hepatitis B virus genotypes: global distribution and clinical importance. World J Gastroenterol. 2014;20:5427-5434. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 264] [Cited by in RCA: 305] [Article Influence: 25.4] [Reference Citation Analysis (8)] |
| 14. | Shoraka S, Hosseinian SM, Hasibi A, Ghaemi A, Mohebbi SR. The role of hepatitis B virus genome variations in HBV-related HCC: effects on host signaling pathways. Front Microbiol. 2023;14:1213145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 15. | Ashtari S, Pourhoseingholi MA, Sharifian A, Zali MR. Hepatocellular carcinoma in Asia: Prevention strategy and planning. World J Hepatol. 2015;7:1708-1717. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 84] [Cited by in RCA: 92] [Article Influence: 8.4] [Reference Citation Analysis (2)] |
| 16. | Lin CL, Kao JH. Hepatitis B virus genotypes and variants. Cold Spring Harb Perspect Med. 2015;5:a021436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 193] [Article Influence: 17.5] [Reference Citation Analysis (1)] |
| 17. | Pacholi S, Bishwal SC, Barde PV. Identification of hepatitis B virus genotypes detected in Lahaul & Spiti district of Himachal Pradesh, India. Indian J Med Res. 2022;156:779-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 18. | Zhand S, Karami C, Hosseinzadeh Adli A, Tabarraei A, Khodabakhshi B, Moradi A. Correlation Between Hepatitis B G1896A Precore Mutations and HBeAg in Chronic HBV Patients. Jundishapur J Microbiol. 2015;8:e17126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Carabelli AM, Peacock TP, Thorne LG, Harvey WT, Hughes J; COVID-19 Genomics UK Consortium, Peacock SJ, Barclay WS, de Silva TI, Towers GJ, Robertson DL. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol. 2023;21:162-177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 286] [Cited by in RCA: 525] [Article Influence: 175.0] [Reference Citation Analysis (0)] |
| 20. | Mokaya J, McNaughton AL, Bester PA, Goedhals D, Barnes E, Marsden BD, Matthews PC. Hepatitis B virus resistance to tenofovir: fact or fiction? A systematic literature review and structural analysis of drug resistance mechanisms. Wellcome Open Res. 2020;5:151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 25] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 21. | Lau KCK, Burak KW, Coffin CS. Impact of Hepatitis B Virus Genetic Variation, Integration, and Lymphotropism in Antiviral Treatment and Oncogenesis. Microorganisms. 2020;8:1470. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 22. | Tenney DJ, Levine SM, Rose RE, Walsh AW, Weinheimer SP, Discotto L, Plym M, Pokornowski K, Yu CF, Angus P, Ayres A, Bartholomeusz A, Sievert W, Thompson G, Warner N, Locarnini S, Colonno RJ. Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to Lamivudine. Antimicrob Agents Chemother. 2004;48:3498-3507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 427] [Article Influence: 19.4] [Reference Citation Analysis (1)] |
| 23. | Kim KH, Kim ND, Seong BL. Discovery and development of anti-HBV agents and their resistance. Molecules. 2010;15:5878-5908. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 24. | McMahon BJ, Nolen LD, Snowball M, Homan C, Negus S, Roik E, Spradling PR, Bruden D. HBV Genotype: A Significant Risk Factor in Determining Which Patients With Chronic HBV Infection Should Undergo Surveillance for HCC: The Hepatitis B Alaska Study. Hepatology. 2021;74:2965-2973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 25. | Caligiuri P, Cerruti R, Icardi G, Bruzzone B. Overview of hepatitis B virus mutations and their implications in the management of infection. World J Gastroenterol. 2016;22:145-154. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 102] [Cited by in RCA: 111] [Article Influence: 11.1] [Reference Citation Analysis (2)] |
| 26. | Gopalakrishna H, Ghany MG. Perspective on Emerging Therapies to Achieve Functional Cure of Chronic Hepatitis B. Curr Hepatol Rep. 2024;23:241-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 27. | Zhang Q, Cao G. Genotypes, mutations, and viral load of hepatitis B virus and the risk of hepatocellular carcinoma: HBV properties and hepatocarcinogenesis. Hepat Mon. 2011;11:86-91. [PubMed] |
| 28. | Zhao Q, Liu H, Tang L, Wang F, Tolufashe G, Chang J, Guo JT. Mechanism of interferon alpha therapy for chronic hepatitis B and potential approaches to improve its therapeutic efficacy. Antiviral Res. 2024;221:105782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 48] [Reference Citation Analysis (0)] |
| 29. | Ray G. Functional cure of chronic hepatitis B-hope or hype? World J Hepatol. 2024;16:1199-1205. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 30. | Zhang S, Chau HT, Tun HM, Huang FY, Wong DK, Mak LY, Yuen MF, Seto WK. Virological response to nucleos(t)ide analogues treatment in chronic hepatitis B patients is associated with Bacteroides-dominant gut microbiome. EBioMedicine. 2024;103:105101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 3] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 31. | Lazarevic I, Banko A, Miljanovic D, Cupic M. Immune-Escape Hepatitis B Virus Mutations Associated with Viral Reactivation upon Immunosuppression. Viruses. 2019;11:778. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 78] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 32. | Bhosale S, Tripathi S, Fernandez K. Knowledge attitude and practices regarding hepatitis ′B′ among class IV hospital workers of tertiary care teaching hospital in Pune, India. Med J Dr D Y Patil Univ. 2014;7:96. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 33. | Patel A, Dossaji Z, Gupta K, Roma K, Chandler TM, Minacapelli CD, Catalano K, Gish R, Rustgi V. The Epidemiology, Transmission, Genotypes, Replication, Serologic and Nucleic Acid Testing, Immunotolerance, and Reactivation of Hepatitis B Virus. Gastro Hep Adv. 2024;3:139-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 34. | Singh KP, Avihingsanon A, Zerbato JM, Zhao W, Braat S, Tennakoon S, Rhodes A, Matthews GV, Fairley CK, Sasadeusz J, Crane M, Audsley J, Lewin SR. Predictors of liver disease progression in people living with HIV-HBV co-infection on antiretroviral therapy. EBioMedicine. 2024;102:105054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 35. | Zhang B, Han H, Zhao X, Li AN, Wang Y, Yuan W, Yang Z, Li MD. An HBV susceptibility variant of KNG1 modulates the therapeutic effects of interferons α and λ1 in HBV infection by promoting MAVS lysosomal degradation. EBioMedicine. 2023;94:104694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 36. | Saitta C, Pollicino T, Raimondo G. Occult Hepatitis B Virus Infection: An Update. Viruses. 2022;14:1504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 86] [Reference Citation Analysis (0)] |
| 37. | Besharat S, Katoonizadeh A, Moradi A. Potential mutations associated with occult hepatitis B virus status. Hepat Mon. 2014;14:e15275. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 38. | Sun H, Chang L, Yan Y, Ji H, Jiang X, Song S, Xiao Y, Lu Z, Wang L. Naturally occurring pre-S mutations promote occult HBV infection by affecting pre-S2/S promoter activity. Antiviral Res. 2022;208:105448. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 39. | Datta S, Chatterjee S, Veer V. Recent advances in molecular diagnostics of hepatitis B virus. World J Gastroenterol. 2014;20:14615-14625. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (2)] |
| 40. | Marcuccilli F, Chevaliez S, Muller T, Colagrossi L, Abbondanza G, Beyser K, Wlassow M, Ortonne V, Perno CF, Ciotti M. Multicenter Evaluation of the Cepheid Xpert(®) HBV Viral Load Test. Diagnostics (Basel). 2021;11:297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 41. | Farshadpour F, Taherkhani R, Saberi F. Molecular evaluation of hepatitis B virus infection and predominant mutations of pre-core, basal core promoter and S regions in an Iranian population with type 2 diabetes mellitus: a case-control study. BMC Infect Dis. 2022;22:553. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 42. | Chang ML, Liaw YF. Emerging Therapies for Chronic Hepatitis B and the Potential for a Functional Cure. Drugs. 2023;83:367-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 43. | Tsertsvadze T, Sharvadze L, Chkhartishvili N. Cost-effectiveness of PCR-based diagnostic methods for hepatitis B detection in resource-limited settings. J Med Virol. 2014;86:1396-1402. |
| 44. | Lok AS. Personalized treatment of hepatitis B. Clin Mol Hepatol. 2015;21:1-6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 45. | Hebeler-Barbosa F, Wolf IR, Valente GT, Mello FCDA, Lampe E, Pardini MIMC, Grotto RMT. A New Method for Next-Generation Sequencing of the Full Hepatitis B Virus Genome from A Clinical Specimen: Impact for Virus Genotyping. Microorganisms. 2020;8:1391. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 46. | Howell J, Pedrana A, Schroeder SE, Scott N, Aufegger L, Atun R, Baptista-Leite R, Hirnschall G, 't Hoen E, Hutchinson SJ, Lazarus JV, Olufunmilayo L, Peck R, Sharma M, Sohn AH, Thompson A, Thursz M, Wilson D, Hellard M. A global investment framework for the elimination of hepatitis B. J Hepatol. 2021;74:535-549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 73] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 47. | Wei N, Zheng B, Cai H, Li N, Yang J, Liu M. Systematic review and meta-analysis: de novo combination of nucleos(t)ide analogs and pegylated interferon alpha versus pegylated interferon alpha monotherapy for the functional cure of chronic hepatitis B. Front Pharmacol. 2024;15:1403805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (3)] |
| 48. | Zhang X, Yang X, Tan L, Tian Y, Zhao Z, Ru S. The efficacy and safety of addition of pegylated interferon to long-term nucleos(t)ide analogue therapy on functional cure of chronic hepatitis B patient: a systematic review and meta-analysis. Front Pharmacol. 2024;15:1474342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/