Published online Dec 9, 2025. doi: 10.5409/wjcp.v14.i4.108733
Revised: May 12, 2025
Accepted: August 5, 2025
Published online: December 9, 2025
Processing time: 191 Days and 19.7 Hours
Autism spectrum disorders (ASD) represent a substantial social problem affecting at least 1 in 100 children worldwide. These conditions are very often accompanied by intellectual disability (ID) and speech delay; thus, they can be considered within a clinical continuum of neurodevelopmental disorders. Given the high heterogeneity of ASD, the subjective nature of diagnostic criteria, and the pre
To investigate the spectrum and frequency of rare genetic variants in genes with proven association with ASD in Russian children.
110 patients from 106 families were recruited into the study (mean age at diag
Pathogenic copy number variations were detected in three (7%) of the patients examined. Clinical exome sequ
These data confirm a tremendous diversity of genetic causes of ASD. Clinical exome sequencing may serve as a reasonable alternative to whole-exome sequencing.
Core Tip: While autism is a clinical diagnosis, genetic studies provide important clues on autism spectrum disorders (ASD) pathogenesis. From a practical point of view, DNA testing offers an opportunity to obtain valuable information on genetic risks and, sometimes, on the most effective treatment. The number of ASD-associated candidate genes exceeds 1000, yet often the causal role of a particular gene or allelic variant stays unproven. We utilized clinical exome sequencing for the DNA testing of ASD patients. The results obtained suggest that in children with ASD and developmental delay/mental retardation, the diagnostic yield of singleton clinical exome sequencing is comparable to that of singleton whole exome sequencing. Also, we assume that rare PCDH19 variants may play a role in causing autistic features in males.
- Citation: Suspitsin EN, Malysheva KS, Laptiev SA, Sharonova OS, Abuzova AS, Kuznitsyna AA, Melashenko TV, Efremova OV, Korzun PR, Binnatova JO, Gorgul YA, Syomina MV, Imyanitov EN. Monogenic defects in Russian children with autism spectrum disorders. World J Clin Pediatr 2025; 14(4): 108733
- URL: https://www.wjgnet.com/2219-2808/full/v14/i4/108733.htm
- DOI: https://dx.doi.org/10.5409/wjcp.v14.i4.108733
Autism spectrum disorders (ASD) affect at least 1% of children worldwide[1]. Heritability of these conditions is very high, being within the range 80%-90%[2,3]. ASD diagnosis requires the presence of deficits in social communication/interaction and restricted, repetitive patterns of behavior, interests or activities[4]. Patients with ASD often suffer from many comorbidities like intellectual disability (ID), speech delay, epilepsy, attention-deficit hyperactivity disorder, anxiety, sleep disorders, gastrointestinal problems, etc.[5]. Genomic architecture of autism is extremely complex and includes both rare events with strong effects [single nucleotide variations and copy number variations (CNV)] and multiple common variants providing mild predisposition[6]. Moreover, these events may have an additive effect when inherited together[7]. Many autism genes are shared with other neurodevelopmental disorders reflecting common pathogenetic mechanisms. These genes have important functions in neurogenesis, chromatin remodeling, and metabolic pathways. Most rare ASD-related genetic variants arise de novo, although some population-specific peculiarities of their spectrum may exist. While many countries have already done large-scale projects recruiting thousands of ASD patients, Russian population remains relatively understudied. Diagnosis of ASD is based on behavioral patterns, but DNA testing may provide valuable information on genetic risks and, sometimes, on effective treatment. It should be noted that there is a growing demand from families for more accurate evaluation of ASD recurrence risk in offspring. In a real-world setting this practical query is often far from being answered, facing ambiguous candidate genes, variants of unknown significance, incomplete penetrance, etc.
The list of the genes potentially associated with ASD is constantly expanding, yet there is little consensus on which genes should be tested routinely. There are ongoing efforts for selecting the genes demonstrating sufficient evidence for their causal role in ASD[8-10]. To help this curation, Simons Foundation Autism Research Initiative (SFARI) database provides ranking systems [SFARI Gene Score and Evaluation of Autism Gene Link Evidence (EAGLE) Score]. These scores allow genes to be ranked into several categories according to the strength of evidence for their link with autism.
Here we report our experience of detection of rare variants in genes with high SFARI and EAGLE integral scores (i.e., high confidence ASD-related genes) in predominantly simplex ASD families. We opted for clinical exome sequencing as a diagnostic method due to cost considerations, and this approach enabled us to focus on a relatively small number of genes with strong links to ASD.
The study included 110 children with ASD from 107 families. All patients had impaired social interaction and social communication, as well as stereotypical behavior, limited interests and hobbies. Patients were selected based on the results of consultation with a clinical geneticist and/or pediatric psychiatrist; the diagnosis was made in accordance with the standardized criteria provided by the American Psychiatric Association's Diagnostic and Statistical Manual, Fifth Edition[4].
The average age of patients was 6 years (range 2-18 years). 84 of the examined children (76.4%) were males, and 26 (23.6%) were females. Familial (multiplex) ASD cases included 6 patients from 3 families: One family had two affected monozygotic twins, one family had two siblings of different gender, and another one was characterized by two affected male first cousins. None of the parents reported consanguinity.
Slavic ethnic origin was reported by most of the study participants (96/110, 87%). In addition, this data set included Armenians (n = 3), Avars (n = 2), Dargins (n = 3), Chechens (n = 2), and Lezgins (n = 4).
17 patients (16%) had an isolated ASD, and 93 patients (84%) demonstrated a combination of ASD with developmental delay (DD, n = 82) or (ID, n = 11).
Informative facial dysmorphisms and/or other signs of a possible hereditary syndrome (micro- or macrocephaly, epilepsy, congenital heart disease, etc.) were observed in 18 patients (16%). Of those, Fragile X syndrome was suspected on clinical grounds in two male sibs; Fragile X Messenger Ribonucleoprotein 1 repeat analysis confirmed this diagnosis, therefore, these patients were excluded from further analysis.
All syndromic patients underwent standard karyotyping, as well as chromosomal microarray analysis (CMA; Genoscan3000, Genomed, Russia) with minimum resolution of 100 kb. Another 31 ASD patients underwent CMA test prior to recruitment to the study, so they were already known to have no relevant cytogenetic abnormalities. The results have been re-examined by a board-certified cytogeneticist.
Massively parallel sequencing was performed on a Genolab M device (GeneMind, China) with at least 100-fold coverage. The sample preparation process included enzymatic DNA fragmentation and adapter ligation. For enrichment, the Clinical Exome Sequencing probe set (TruSight™ One Sequencing Panel, Illumina, 4813 genes) was utilized. Sequencing was performed using paired reads of 150 bp. Sequencing accuracy was assessed by the number of reads with a high-quality score (Phred Quality Score, or Q score). The BWA (Burrows-Wheeller Aligner) algorithm was used for the ali
Our analysis was intentionally limited to a list of highly confident ASD-associated genes. Gene selection was based on Simons Foundation Autism Research Initiative (SFARI) and EAGLE integral scores (https://gene.sfari.org/database/; latest Release 2024 Q4) reflecting the degree of evidence for association with ASD. When assessing the significance of variants, we considered information from publicly available databases (Franklin, OMIM, ClinVar). Combined annotation dependent depletion (CADD) score ≥ 20 was taken as a threshold for the prediction of functional significance of missense variants.
Variant prioritization was based on the following criteria: (1) Population frequency of ≤ 0.001 or not reported; (2) Variant affects ASD-associated gene and is classified as pathogenic or likely pathogenic according to the American College of Medical Genetics and Genomics (ACMG) and/or ClinVar criteria; (3) Missense variants affecting any of the 92 genes with EAGLE score of > 12 (high confidence) included into the diagnostic panel; (4) Likely gene damaging (LGD) event affecting any ASD-associated gene; and (5) Missense variants affecting any ASD-associated gene with integral CADD score of ≥ 30.
In addition, the possibility of a relationship between the phenotype and the presence of pathogenic/Likely pathogenic variants in genes that were not previously associated with ASD, was taken into account.
Whenever possible, the presence of the identified variants was analyzed in children`s parents by Sanger sequencing. Several possible mechanisms of ASD heritability were considered. In the most common scenarios, the disease is either associated with de novo variants absent in healthy parents or fits into an autosomal recessive or X-linked inheritance model. Alternatively, autosomal dominant inheritance with incomplete penetrance also cannot be completely excluded: Indeed, the presence of a rare heterozygous variant in a presumably healthy parent does not necessarily contradict with its potential causative role[11].
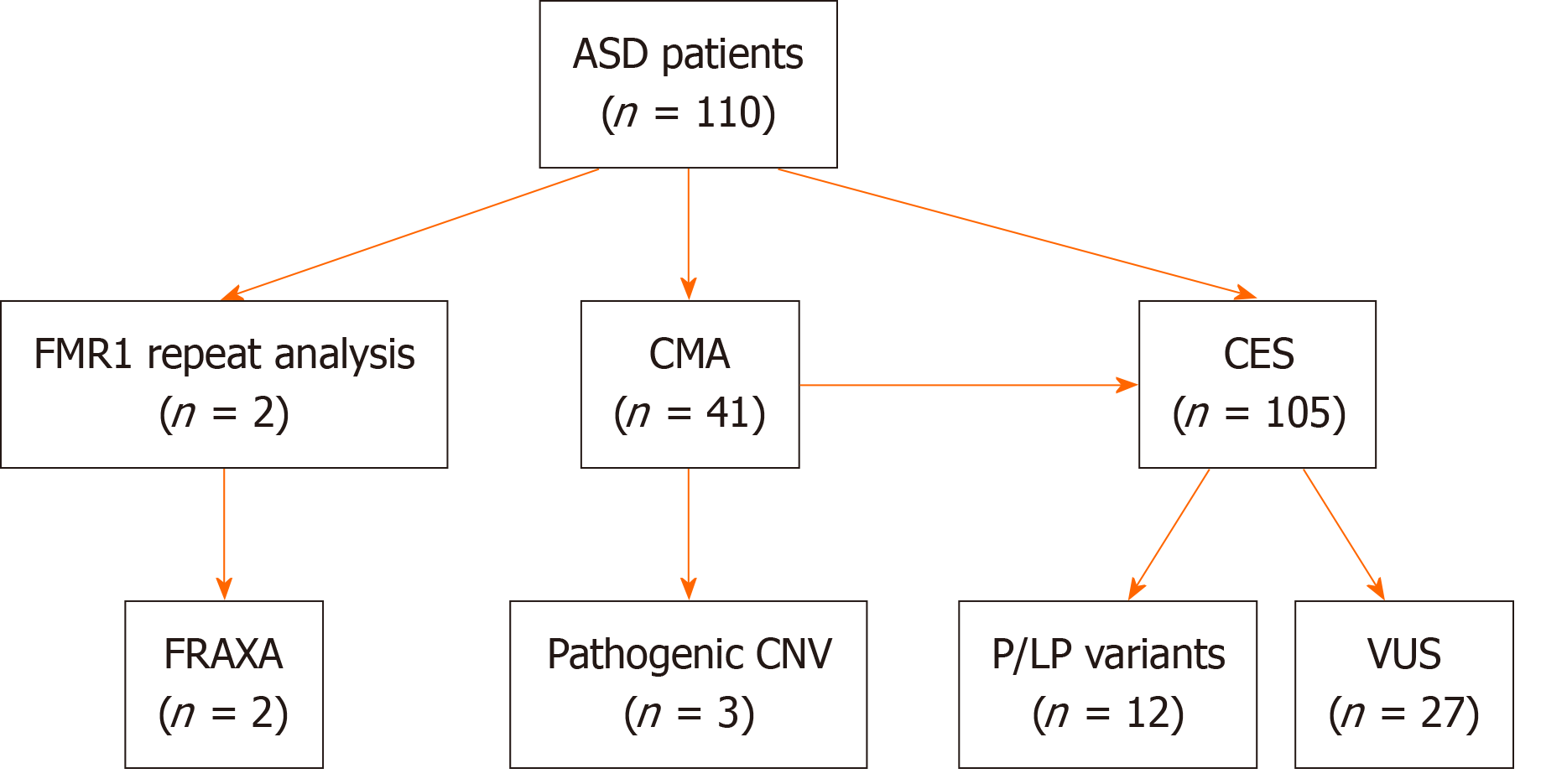
Key findings of the study are summarized in the Figure 1. Molecular cytogenetic analysis revealed pathogenic CNV in three patients (7% of 41 syndromic cases studied). A 2 years 7 months boy (karyotype 46, XY) and an 8-year-old girl [45, XX r(22), ring chromosome 22] were found to have heterozygous deletion 22q13.33, which includes the SHANK3 Locus and is associated with Phelan-McDermid syndrome. Along with severe DD and ASD, these patients had multiple minor congenital anomalies (deformed ears, short sunken nasal bridge, hypertelorism, epicanthus, fifth fingers clinodactyly, etc.). Muscular hypotonia and a high pain threshold for injuries were also noted. In addition, the girl had pronounced hypertrichosis of the back and limbs, and persistent self-harming behaviour (biting her wrists and fingers, resulting in chronic traumatization).
A 16-year-old girl was found to have a combination of a pathogenic microduplication 1p36.33-p.36.32 of 2.6 Mbp with a 11p15.5 microdeletion of 424080 bp of unknown clinical significance. Multiple facial dysmorphisms were accompanied by autistic behavior and learning difficulties (difficulty expressing thoughts, attention deficit, poor memory, underdeveloped abstract thinking).
The remaining 105 patients underwent clinical exome sequencing. The results of the molecular testing are presented in Table 1.
| ID | Gender | Age at diagnosis | Phenotype | Mutation detected | Pathogenicity/disease | Segregation analysis | EAGLE gene score | SFARI gene score | Other rare variants detected |
| 2639 | Female | 2 years | ASD, DD; frontal bossing, flat nasal bridge, strabismus, arachnodactily | TCF4, NM_001083962.2 c.1841C>T, (p.Ala614Val), rs1568303352 | P (# 610954 PITT-HOPKINS SYNDROME) | De novo | 13.5 | 1s | - |
| 3942 | Female | 17 years | ASD, severe ID, epilepsy, CHD (pulmonary stenosis), smooth philtrum, broad nasal bridge; insensitivity to pain | ADNP NM_001282531.3 c.2157C>G (p.Tyr719*) rs587777526 | P (# 615873 HELSMOORTEL-VAN DER AA SYNDROME) | De novo | 41.5 | 1s | - |
| 4027 | Male | 3 years 10 months | ASD, DD; mild dysmorphic features | TRIP12 NM_001348323.3 c.3038_3041dup (p.Leu1014Phefs*37) | LP (# 617752 CLARK-BARAITSER SYNDROME) | De novo | 27 | 1s | - |
| 4071 | Male | 2 years 1 month | ASD, DD; congenital hypothyroidism, strabismus, muscular hypotonia, tall forehead, scaphocephaly, flat nasal bridge, short nose with anteverted nostrils, low-set ears | SON NM_138927.4 c.5753_5756 del (p.Val1918Glufs*87) | P (# 617140 ZTTK SYNDROME) | De novo | 11 | 1s | - |
| 4072 | Male | 3 years 9 months | ASD, DD | SLC9A9 NM_173653.4 c.8_9del (p.Arg3Thrfs*10) rs750792945 | LP (# 613410 AUTISM, SUSCEPTIBILITY TO, 16; AUTS16) | ND | - | 2 | - |
| 4217 | Female | 3 years 9 months | ASD, DD | ARID1B NM_001374828.1 c.1293_1311del (p.Gly434Alafs*12) rs943407609 | P (# 135900 COFFIN-SIRIS SYNDROME 1) | De novo | 34.75 | 1s | TRIP12 NM_001348323.3 c.4426G>A (p.Val1476Met) rs781311402 |
| 4232 | Female | 7 years 3 months | ASD, DD; strabismus, low-set ears, micrognathia, low anterior hairline | SERPINI1 NM_001122752.2 c.553T>G (p.Tyr185Asp) | LP (# 604218 ENCEPHALOPATHY, FAMILIAL, WITH NEUROSERPIN INCLUSION BODIES) | ND | - | - | DIP2A NM_015151.4 c.2626C>T (p.Arg876Cys) rs199807759 |
| 4259 | Male | 6 years 6 months | ASD, ID; cerebellar vermis hypoplasia, tetraventricular dilatation | TUBB3 NM_006086.4 c.1172G>A (p.Arg391His) rs886039497 | P/LP (# 614039 CORTICAL DYSPLASIA, COMPLEX, WITH OTHER BRAIN MALFORMATIONS 1) | ND | - | - | CACNA1D NM_001128840.3 c.5017G>A (p.Glu1673 Lys) rs147973409 CACNA1D NM_001128840.3 c.5377C>T (p.Arg1793Trp) rs555675934 |
| 4386 | Male | 6 years | ASD, ID; epilepsy, hydrocephalus.Triangular face, beaked nose, protruding ears, thin upper lip | OPHN1 NM_002547.3 c.644_645del (p.Val215Glyfs*35) rs1569244467 hemizygous | P (# 300486 INTELLECTUAL DEVELOPMENTAL DISORDER, X-LINKED, SYNDROMIC, BILLUART TYPE) | Maternal | - | 2 | - |
| 4436 | Male | 16 years | ASD, ID; hypertelorism | EHMT1 NM_024757.5 c.732del (p.Phe244 Leufs*38) | LP (# 610253 KLEEFSTRA SYNDROME 1) | De novo | 13.5 | 1s | EP300 NM_001429.4 c.4256T>C (p.Ile1419Thr) rs1278019392 paternal. RAI1 NM_030665.4 c.4340G>C (p.Arg1447Thr) rs767484843 maternal |
| KM | Male | 15 years | ASD, ID; macrocephaly, frontal bossing hypertelorism | CHD3 NM_001005273.3 c.5642G>T (p.Arg1881 Leu) rs1567877108 | P/LP (#618205 SNIJDERS-BLOC-CAMPEAU SYNDROME) | De novo | - | 1s | - |
| ShV | Male | 5 years | ASD, DD; macrocephaly, epilepsy | PIK3CA NM_006218.4 c.23G>A (p.Gly8Asp) | LP (# 615108 COWDEN SYNDROME 5) | De novo | - | 3 | - |
In total, pathogenic/Likely pathogenic variants were found in 12 of 105 (11%) patients assessed by clinical exome sequencing. Notably, 4 patients also carried additional rare alleles of unknown significance in high-confidence ASD genes (Table 1). Pathogenic/Likely pathogenic variants were detected in the majority of syndromic (9 of 13, 69%) children compared to only 3 of 92 (3%) nonsyndromic patients (P = 0.0000).
In 6 cases, the detected variants were clearly considered pathogenic by ACMG criteria, which made it possible to confidently establish the diagnosis of a monogenic condition (Pitt-Hopkins syndrome, ZTTK syndrome, syndromic X-linked ID, Billuart type, Snijders-Blok-Campeau, Helsmoortel-van der Aa, and Coffin-Siris syndrome).
In another 6 cases, likely pathogenic variants in TRIP12, SLC9A9, SERPINI1, TUBB3, EHMT1, and PIK3CA genes were detected. In four patients (4027, 4072, 4259, 4436) molecular findings corresponded well to the phenotype, leading to a diagnosis of Clark-Baraitser syndrome, Autism susceptibility type 16, Complex cortical dysplasia with other brain malformations type 1, and Kleefstra syndrome type 1, respectively. A 5-year-old boy (Patient ShV) with macrocephaly and epilepsy had germline PIK3CA variant. Analysis of the DNA obtained from his parents revealed that this variant arose de novo; this fact, together with clinical findings, let us reclassify this variants with unknown clinical significance (VUS) to LP variant and make a diagnosis of Cowden syndrome type 5. Of note, PIK3CA gene has SFARI gene score 3 (Suggestive Evidence).
A 7-year-old girl (Patient 4232) carried likely pathogenic allele c.553T>G in the SERPINI1 gene. The associated neurodegenerative condition, familial encephalopathy with neuroserpin inclusion bodies, typically manifests from the third to fifth decade of life; thus, presymptomatic diagnosis of this condition cannot be excluded. SERPINI1 defects have never been described in association with ASD.
Segregation analysis was performed in 9 families, in 8 of which the de novo status of the mutation was proven, and in one case (Patient 4386) an X-linked variant was inherited from a healthy mother.
In another 27 patients (27/105, 26%), 37 VUS in DSCAM, SHANK2, AUTS2, ADNP, ANKRD11, APBA2, ARID1B, ASTN2, ATRX, SCN1A, CHD2, DEAF1, EHMT1, GRIN2B, MBD5, NBEA, NR4A2, TRIO, TRIP12, POGZ, EP300, FOXP1, PCDH19, GRIN2A, NCKAP1, and CHD8 genes were detected. 8 patients carried two, and 1 patient had 3 rare VUS. Details of the identified variants are provided in Supplementary Table 1.
Three LGD variants, which are not currently classified as pathogenic/Likely pathogenic, were identified. These variants affected YY1, ATRX and GRIN2A genes, being found in 1 patient each. 23 missense variants in genes with EAGLE score of > 12 with and integral CADD predictive score of ≥ 20 were detected in 18 patients. 6 missense variants with CADD score of ≥ 30 detected in 6 patients; they involved SCN1A, APBA2, ASTN2, NR4A2, and NBEA genes.
No specific variant was detected more than once in unrelated patients. Among the genes with rare variants found in 2 or more patients were TRIP12 (n = 4), AUTS2 (n = 3), ARID1B (n = 3), PCDH19 (n = 3), EP300 (n = 3), TRIO (n = 2), ASTN2 (n = 2), EHMT1 (n = 2), and CHD2 (n = 2).
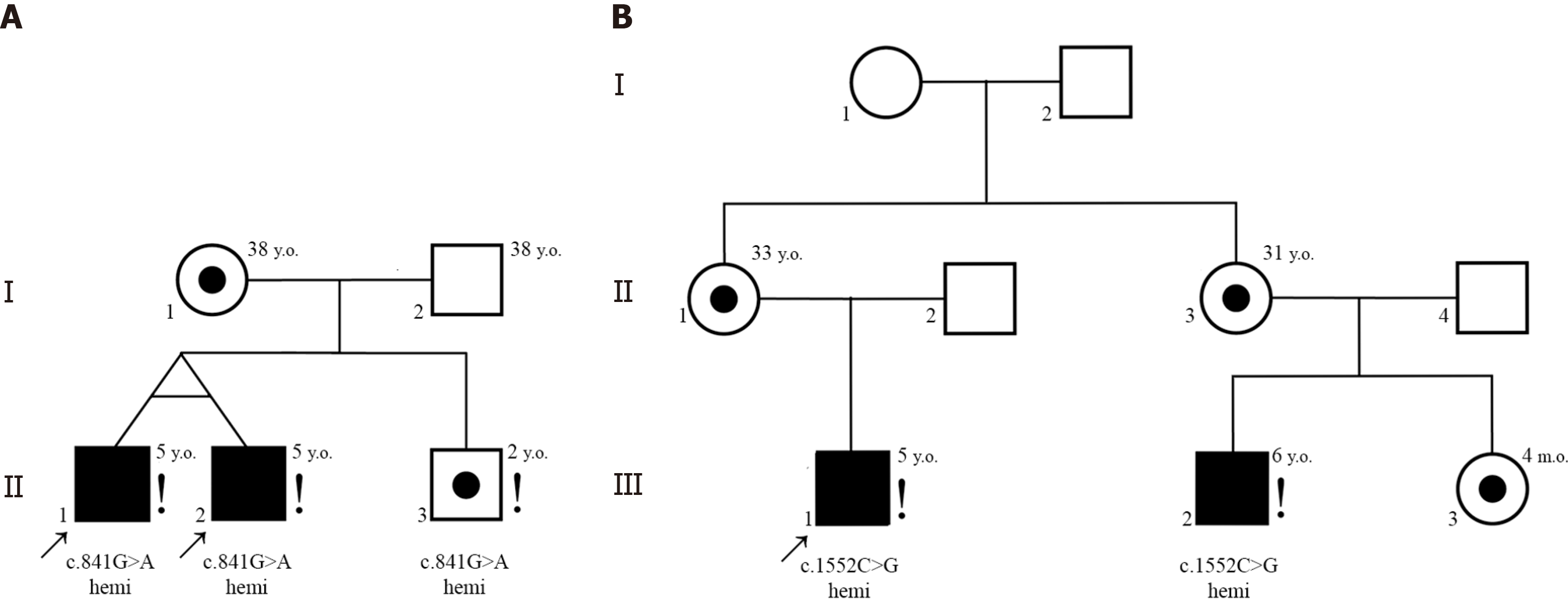
Interestingly, non-mosaic hemizygous missense variants of PCDH19 were identified in five male ASD patients (mean age at diagnosis 6 years). In particular, such variants were detected in two unrelated families (two first cousins and two monozygotic twins, respectively) as well as in one simplex case (Patient 4123). All these patients had both ASD and DD. All PCDH19 candidate variants were maternally inherited and located within extracellular (EC) domains EC3 or EC5 (exon 1).
Family 1. Monozygotic twin brothers (patients 4448, II-1, and 4449, II-2), aged 5 years, were diagnosed with ASD and DD. Patient II-2 also had a history of epileptiform activity on EEG. Genetic testing revealed a hemizygous PCDH19 variant NM_001184880.2 c.841G>A (p.Val281Ile) classified as a VUS. Their mother was a healthy heterozygous carrier of the same variant. Their younger male sibling also carried the hemizygous PCDH19 c.841G>A variant but exhibited no neurological or psychiatric symptoms (Figure 2). He was considered as having low ASD risk upon M-CHAT-R testing. The variant has not been reported in gnomAD.
Family 2. Two male first cousins (Patients 3897, III-1 and 3851, III-2), aged 5 and 6 years, respectively, presented with ASD and DD. Both carried rare hemizygous PCDH19 variant NM_001184880.2 c.1552C>G (p.Leu518Val) classified as a VUS. Their mothers (II-1 and II-3) and the younger sister of the patient 3851, III-3, were heterozygous carriers but exhibited no neurological or psychiatric symptoms (Figure 2). This variant is reported in gnomAD Exomes with frequency of 0.0000055 (one heterozygote found in 114184 females; no hemizygotes reported).
Simplex case 4123. Patient 4123, an 8-year-old male, was diagnosed with moderate ID, structural epilepsy, and ASD-like symptoms. Genetic analysis identified a previously undescribed PCDH19 variant c.1651G>T (p.Val551Phe) in hemizygous state. The variant was also detected in the patient’s mother and sister, none of whom ever exhibited neu
Besides the abovementioned two multiplex families with PCDH19 variants, the study included only one family with two siblings affected. Discordance between the siblings was found: The 5-year boy had MBD5 c.83G>A (p.Arg28His) variant, which was absent in his 7-year-old sister.
This is the first systematic next generation sequencing-based study of the genetic causes of ASD in Russian patients. It confirms a striking diversity of ASD-associated alleles and suggests a role for the PCDH19 gene in causing autistic features in males.
The trajectory of the genetic testing of subjects with ASD is a subject of debate. While most professional societies suggest starting analysis of ASD genetics from CMA[12,13], some researchers advocate using whole exome sequencing (WES) as a first-tier test[14]. The efficiency of detecting genetic causes of ASD by this method falls within 8%-26%[15]. Diagnostic efficiency may be substantially increased with the use of WES in the trio format[16], however, this approach is associated with additional costs.
The diagnostic yield of DNA testing varies greatly depending on the characteristics of the patients. Overall, causative genetic defects are more often detected in patients with a combination of ASD and ID[17,18]. While the probability of finding a disease-causing variant is generally higher in WES compared with targeted panels, recent meta-analysis suggests that this difference is not significant[19].
Although the total number of ASD candidate genes is rapidly growing and approaches up to 1000[20], reliable data on ASD-causality are often lacking. So we decided to limit our search by a relatively humble list of high-confidence ASD genes that may be used in clinical rather than research settings. This restriction was aimed at more accurate risk assessment and making genetic counseling more understandable for the families.
Our results confirm that causative variants are more likely to be detected in syndromic than in nonsyndromic ASD, as demonstrated by others[21,22]. The yield of clinical exome sequencing in our study (11% of pathogenic/Likely pathogenic variants) may be directly compared with the one obtained by Chérot et al[23] who detected pathogenic/Likely pathogenic variants in 4% of ASD patients using the same enrichment kit. Moreover, our data demonstrate approximately the same yield as singleton exome sequencing[22,24].
Of three families with multiplex ASD patients, two, including one with monozygotic twins, demonstrated concordance of genetic events; brother and sister from another family were discordant. Contrary to conventional wisdom, non-sharing of putatively causative SNVs between affected siblings has been reported by several researchers[25,26]. For example, data obtained by Yuen et al[26] demonstrated that 69% of affected siblings did not share the same ASD-related variant(s).
Rare hemizigous variants of the PCDH19 were found in two monozygous twin boys and two male first cousins with ASD as well as in one simplex case. Although in one family a hemizygous boy was unaffected, this finding remains interesting. The PCDH19 gene encodes protocadherin-19, a protein primarily expressed in the brain. It plays a crucial role in cell adhesion through homophilic binding, i.e., interacting only with other protocadherin-19 molecules on adjacent cells. This process is crucial for the proper formation of neuronal connections and the stability of neural networks. Conversely, dysfunction of PCDH19 can lead to the disorganization of neural networks[27]. Heterozygous mutations in PCDH19 are known to cause severe X-linked epilepsy restricted to females (EFMR). This condition follows an unusual X-linked inheritance pattern, where clinical manifestations are observed in heterozygous females while hemizygous males are described as “asymptomatic carriers”[28]. Severe phenotypic expression in females is mainly attributed to cellular interference resulting from random X-chromosome inactivation. A similar effect occurs in mosaic males, where both mutant and wild-type cells coexist in the brain causing symptoms resembling those in females[29]. Hemizygous non-mosaic males were long thought to be unaffected, but recent findings suggest that rare PCDH19 variants may be associated with ASD[30–32] and intellectual deficiency/mental retardation[33]. Moreover, Pcdh19 knock-out mouse models exhibited autism-like behaviors in males, including impaired social interaction, repetitive behaviors, and heightened anxiety[34].
Previously, variants in non-mosaic affected males were found both in EC[30] and in the cytoplasmic domain[31,32]. All the variants detected in affected boys included in this study were located in the EC domains EC3 and EC5 of exon 1. Notably, most pathogenic PCDH19 variants associated with epilepsy are also found in exon 1[35,36], almost half of those being located in the EC3 and EC4 domains[37].
Our findings confirm the observations that at least some hemizygous males with non-mosaic PCDH19 variants may present with neurobehavioral abnormalities. Evidently, such variants do not cause EFMR phenotype in patients’ mothers or sisters.
Our study has some limitations. Firstly, a targeted singleton approach has been used instead of whole-exome trio sequencing, thus restricting the opportunity for discovery of new ASD candidates and putting aside the majority of non-coding variants.
Secondly, CNV analysis was performed only in a minor fraction of our ASD patients; thus, we focused our efforts exclusively on rare monogenic/oligogenic events. In contrast, the only study of Russian ASD patients available in PubMed[38] is based on case-control study of CNV burden and common SNP. We are aware of the possibility of CNV and SNV co-occurrence in the same individual, providing additive effect[39]. Data on rare SNVs detected in Russian patients are currently limited to a single disease entity[40] or to a clinically special subgroup, e.g., autism with regression[41]. Thirdly, the study involved a substantial number of hospitalized patients, i.e. subjects with severe clinical manifestations of the disease. As a result, there was a recruitment bias towards patients with both ASD and ID, which may have affected the diagnostic yield.
It is highly likely that in the near future, whole-genome trio sequencing will largely replace other approaches to the genetic examination of patients with suspected monogenic disease. However, this approach is not compatible with resources currently available, and the search for cost-efficient alternatives remains a valuable avenue for clinical investigations.
Even the use of relatively small gene panels reveals many VUS which can rarely be reliably reclassified by bio
Clinical exome sequencing may serve as a more accessible alternative to WES in diagnosis of well-established genetic causes of ASD. Mutation-negative patients could be further analyzed by WES or whole-genome sequencing to reveal new ASD candidates.
| 1. | Genovese A, Butler MG. Clinical Assessment, Genetics, and Treatment Approaches in Autism Spectrum Disorder (ASD). Int J Mol Sci. 2020;21:4726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 146] [Article Influence: 24.3] [Reference Citation Analysis (11)] |
| 2. | Tick B, Bolton P, Happé F, Rutter M, Rijsdijk F. Heritability of autism spectrum disorders: a meta-analysis of twin studies. J Child Psychol Psychiatry. 2016;57:585-595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 772] [Cited by in RCA: 656] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 3. | Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, Reichenberg A. The Heritability of Autism Spectrum Disorder. JAMA. 2017;318:1182-1184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 466] [Article Influence: 51.8] [Reference Citation Analysis (0)] |
| 4. | American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 2013. [RCA] [DOI] [Full Text] [Cited by in Crossref: 66101] [Cited by in RCA: 61183] [Article Influence: 3599.0] [Reference Citation Analysis (5)] |
| 5. | Khachadourian V, Mahjani B, Sandin S, Kolevzon A, Buxbaum JD, Reichenberg A, Janecka M. Comorbidities in autism spectrum disorder and their etiologies. Transl Psychiatry. 2023;13:71. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 125] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 6. | Iakoucheva LM, Muotri AR, Sebat J. Getting to the Cores of Autism. Cell. 2019;178:1287-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 188] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 7. | Antaki D, Guevara J, Maihofer AX, Klein M, Gujral M, Grove J, Carey CE, Hong O, Arranz MJ, Hervas A, Corsello C, Vaux KK, Muotri AR, Iakoucheva LM, Courchesne E, Pierce K, Gleeson JG, Robinson EB, Nievergelt CM, Sebat J. A phenotypic spectrum of autism is attributable to the combined effects of rare variants, polygenic risk and sex. Nat Genet. 2022;54:1284-1292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 136] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 8. | Sanders SJ, He X, Willsey AJ, Ercan-Sencicek AG, Samocha KE, Cicek AE, Murtha MT, Bal VH, Bishop SL, Dong S, Goldberg AP, Jinlu C, Keaney JF 3rd, Klei L, Mandell JD, Moreno-De-Luca D, Poultney CS, Robinson EB, Smith L, Solli-Nowlan T, Su MY, Teran NA, Walker MF, Werling DM, Beaudet AL, Cantor RM, Fombonne E, Geschwind DH, Grice DE, Lord C, Lowe JK, Mane SM, Martin DM, Morrow EM, Talkowski ME, Sutcliffe JS, Walsh CA, Yu TW; Autism Sequencing Consortium, Ledbetter DH, Martin CL, Cook EH, Buxbaum JD, Daly MJ, Devlin B, Roeder K, State MW. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215-1233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 952] [Cited by in RCA: 1042] [Article Influence: 94.7] [Reference Citation Analysis (0)] |
| 9. | C Yuen RK, Merico D, Bookman M, L Howe J, Thiruvahindrapuram B, Patel RV, Whitney J, Deflaux N, Bingham J, Wang Z, Pellecchia G, Buchanan JA, Walker S, Marshall CR, Uddin M, Zarrei M, Deneault E, D'Abate L, Chan AJ, Koyanagi S, Paton T, Pereira SL, Hoang N, Engchuan W, Higginbotham EJ, Ho K, Lamoureux S, Li W, MacDonald JR, Nalpathamkalam T, Sung WW, Tsoi FJ, Wei J, Xu L, Tasse AM, Kirby E, Van Etten W, Twigger S, Roberts W, Drmic I, Jilderda S, Modi BM, Kellam B, Szego M, Cytrynbaum C, Weksberg R, Zwaigenbaum L, Woodbury-Smith M, Brian J, Senman L, Iaboni A, Doyle-Thomas K, Thompson A, Chrysler C, Leef J, Savion-Lemieux T, Smith IM, Liu X, Nicolson R, Seifer V, Fedele A, Cook EH, Dager S, Estes A, Gallagher L, Malow BA, Parr JR, Spence SJ, Vorstman J, Frey BJ, Robinson JT, Strug LJ, Fernandez BA, Elsabbagh M, Carter MT, Hallmayer J, Knoppers BM, Anagnostou E, Szatmari P, Ring RH, Glazer D, Pletcher MT, Scherer SW. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat Neurosci. 2017;20:602-611. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 502] [Cited by in RCA: 608] [Article Influence: 67.6] [Reference Citation Analysis (0)] |
| 10. | Schaaf CP, Betancur C, Yuen RKC, Parr JR, Skuse DH, Gallagher L, Bernier RA, Buchanan JA, Buxbaum JD, Chen CA, Dies KA, Elsabbagh M, Firth HV, Frazier T, Hoang N, Howe J, Marshall CR, Michaud JL, Rennie O, Szatmari P, Chung WK, Bolton PF, Cook EH, Scherer SW, Vorstman JAS. A framework for an evidence-based gene list relevant to autism spectrum disorder. Nat Rev Genet. 2020;21:367-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 92] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 11. | Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He ZX, Leal SM, Bernier R, Eichler EE. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582-588. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 413] [Cited by in RCA: 448] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 12. | Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, Martin CL, Ledbetter DH. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749-764. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2229] [Cited by in RCA: 1958] [Article Influence: 122.4] [Reference Citation Analysis (0)] |
| 13. | Srivastava S, Cole JJ, Cohen JS, Chopra M, Smith HS, Deardorff MA, Pedapati E, Corner B, Anixt JS, Jeste S, Sahin M, Gurnett CA, Campbell CA; Intellectual and Developmental Disabilities Research Center (IDDRC) Workgroup on Advocating for Access to Genomic Testing. Survey of the Landscape of Society Practice Guidelines for Genetic Testing of Neurodevelopmental Disorders. Ann Neurol. 2024;96:900-913. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 14. | Arteche-López A, Gómez Rodríguez MJ, Sánchez Calvin MT, Quesada-Espinosa JF, Lezana Rosales JM, Palma Milla C, Gómez-Manjón I, Hidalgo Mayoral I, Pérez de la Fuente R, Díaz de Bustamante A, Darnaude MT, Gil-Fournier B, Ramiro León S, Ramos Gómez P, Sierra Tomillo O, Juárez Rufián A, Arranz Cano MI, Villares Alonso R, Morales-Pérez P, Segura-Tudela A, Camacho A, Nuñez N, Simón R, Moreno-García M, Alvarez-Mora MI. Towards a Change in the Diagnostic Algorithm of Autism Spectrum Disorders: Evidence Supporting Whole Exome Sequencing as a First-Tier Test. Genes (Basel). 2021;12:560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 15. | Savatt JM, Myers SM. Genetic Testing in Neurodevelopmental Disorders. Front Pediatr. 2021;9:526779. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 137] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 16. | Wu Q, Morrow EM, Gamsiz Uzun ED. A deep learning model for prediction of autism status using whole-exome sequencing data. PLoS Comput Biol. 2024;20:e1012468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 17. | Feliciano P, Zhou X, Astrovskaya I, Turner TN, Wang T, Brueggeman L, Barnard R, Hsieh A, Snyder LG, Muzny DM, Sabo A; SPARK Consortium, Gibbs RA, Eichler EE, O'Roak BJ, Michaelson JJ, Volfovsky N, Shen Y, Chung WK. Exome sequencing of 457 autism families recruited online provides evidence for autism risk genes. NPJ Genom Med. 2019;4:19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 170] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 18. | Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, Yuen RK, Uddin M, Roberts W, Weksberg R, Woodbury-Smith M, Zwaigenbaum L, Anagnostou E, Wang Z, Wei J, Howe JL, Gazzellone MJ, Lau L, Sung WW, Whitten K, Vardy C, Crosbie V, Tsang B, D'Abate L, Tong WW, Luscombe S, Doyle T, Carter MT, Szatmari P, Stuckless S, Merico D, Stavropoulos DJ, Scherer SW, Fernandez BA. Molecular Diagnostic Yield of Chromosomal Microarray Analysis and Whole-Exome Sequencing in Children With Autism Spectrum Disorder. JAMA. 2015;314:895-903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 307] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 19. | Stefanski A, Calle-López Y, Leu C, Pérez-Palma E, Pestana-Knight E, Lal D. Clinical sequencing yield in epilepsy, autism spectrum disorder, and intellectual disability: A systematic review and meta-analysis. Epilepsia. 2021;62:143-151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 114] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 20. | SPARK Consortium. SPARK Consortium. SPARK: A US Cohort of 50,000 Families to Accelerate Autism Research. Neuron. 2018;97:488-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 326] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 21. | Postma JK, Harrison MA, Kutcher S, Webster RJ, Cloutier M, Bourque DK, Yu AC, Carter MT. The diagnostic yield of genetic and metabolic investigations in syndromic and nonsyndromic patients with autism spectrum disorder, global developmental delay, or intellectual disability from a dedicated neurodevelopmental disorders genetics clinic. Am J Med Genet A. 2024;194:e63791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 22. | Sánchez Suárez A, Martínez Menéndez B, Escolar Escamilla E, Martínez Sarries FJ, Esparza Garrido MI, Gil-Fournier B, Ramiro León S, Rubio Gribble B, Quesada Espinosa JF, Alcaraz Romero AJ. Whole Exome Sequencing and Panel-Based Analysis in 176 Spanish Children with Neurodevelopmental Disorders: Focus on Autism Spectrum Disorder and/or Intellectual Disability/Global Developmental Delay. Genes (Basel). 2024;15:1310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 23. | Chérot E, Keren B, Dubourg C, Carré W, Fradin M, Lavillaureix A, Afenjar A, Burglen L, Whalen S, Charles P, Marey I, Heide S, Jacquette A, Heron D, Doummar D, Rodriguez D, Billette de Villemeur T, Moutard ML, Guët A, Xavier J, Périsse D, Cohen D, Demurger F, Quélin C, Depienne C, Odent S, Nava C, David V, Pasquier L, Mignot C. Using medical exome sequencing to identify the causes of neurodevelopmental disorders: Experience of 2 clinical units and 216 patients. Clin Genet. 2018;93:567-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 86] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 24. | Tal-Ben Ishay R, Shil A, Solomon S, Sadigurschi N, Abu-Kaf H, Meiri G, Flusser H, Michaelovski A, Dinstein I, Golan H, Davidovitch N, Menashe I. Diagnostic Yield and Economic Implications of Whole-Exome Sequencing for ASD Diagnosis in Israel. Genes (Basel). 2021;13:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 25. | Miyake N, Tsurusaki Y, Fukai R, Kushima I, Okamoto N, Ohashi K, Nakamura K, Hashimoto R, Hiraki Y, Son S, Kato M, Sakai Y, Osaka H, Deguchi K, Matsuishi T, Takeshita S, Fattal-Valevski A, Ekhilevitch N, Tohyama J, Yap P, Keng WT, Kobayashi H, Takubo K, Okada T, Saitoh S, Yasuda Y, Murai T, Nakamura K, Ohga S, Matsumoto A, Inoue K, Saikusa T, Hershkovitz T, Kobayashi Y, Morikawa M, Ito A, Hara T, Uno Y, Seiwa C, Ishizuka K, Shirahata E, Fujita A, Koshimizu E, Miyatake S, Takata A, Mizuguchi T, Ozaki N, Matsumoto N. Molecular diagnosis of 405 individuals with autism spectrum disorder. Eur J Hum Genet. 2024;32:1551-1558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 9] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 26. | Yuen RK, Thiruvahindrapuram B, Merico D, Walker S, Tammimies K, Hoang N, Chrysler C, Nalpathamkalam T, Pellecchia G, Liu Y, Gazzellone MJ, D'Abate L, Deneault E, Howe JL, Liu RS, Thompson A, Zarrei M, Uddin M, Marshall CR, Ring RH, Zwaigenbaum L, Ray PN, Weksberg R, Carter MT, Fernandez BA, Roberts W, Szatmari P, Scherer SW. Whole-genome sequencing of quartet families with autism spectrum disorder. Nat Med. 2015;21:185-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 370] [Cited by in RCA: 378] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 27. | Park J, Lee E, Kim CH, Ohk J, Jung H. Mosaicism-independent mechanisms contribute to Pcdh19-related epilepsy and repetitive behaviors in Xenopus. Proc Natl Acad Sci U S A. 2024;121:e2321388121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 28. | Dell'Isola GB, Vinti V, Fattorusso A, Tascini G, Mencaroni E, Di Cara G, Striano P, Verrotti A. The Broad Clinical Spectrum of Epilepsies Associated With Protocadherin 19 Gene Mutation. Front Neurol. 2021;12:780053. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 29. | Terracciano A, Trivisano M, Cusmai R, De Palma L, Fusco L, Compagnucci C, Bertini E, Vigevano F, Specchio N. PCDH19-related epilepsy in two mosaic male patients. Epilepsia. 2016;57:e51-e55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 30. | Piton A, Gauthier J, Hamdan FF, Lafrenière RG, Yang Y, Henrion E, Laurent S, Noreau A, Thibodeau P, Karemera L, Spiegelman D, Kuku F, Duguay J, Destroismaisons L, Jolivet P, Côté M, Lachapelle K, Diallo O, Raymond A, Marineau C, Champagne N, Xiong L, Gaspar C, Rivière JB, Tarabeux J, Cossette P, Krebs MO, Rapoport JL, Addington A, Delisi LE, Mottron L, Joober R, Fombonne E, Drapeau P, Rouleau GA. Systematic resequencing of X-chromosome synaptic genes in autism spectrum disorder and schizophrenia. Mol Psychiatry. 2011;16:867-880. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 265] [Cited by in RCA: 249] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 31. | van Harssel JJ, Weckhuysen S, van Kempen MJ, Hardies K, Verbeek NE, de Kovel CG, Gunning WB, van Daalen E, de Jonge MV, Jansen AC, Vermeulen RJ, Arts WF, Verhelst H, Fogarasi A, de Rijk-van Andel JF, Kelemen A, Lindhout D, De Jonghe P, Koeleman BP, Suls A, Brilstra EH. Clinical and genetic aspects of PCDH19-related epilepsy syndromes and the possible role of PCDH19 mutations in males with autism spectrum disorders. Neurogenetics. 2013;14:23-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 32. | Chouery E, Makhlouf J, Daoud Khatoun W, Mehawej C, Megarbane A. PCDH19 in Males: Are Hemizygous Variants Linked to Autism? Genes (Basel). 2023;14:598. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 33. | Tarpey PS, Smith R, Pleasance E, Whibley A, Edkins S, Hardy C, O'Meara S, Latimer C, Dicks E, Menzies A, Stephens P, Blow M, Greenman C, Xue Y, Tyler-Smith C, Thompson D, Gray K, Andrews J, Barthorpe S, Buck G, Cole J, Dunmore R, Jones D, Maddison M, Mironenko T, Turner R, Turrell K, Varian J, West S, Widaa S, Wray P, Teague J, Butler A, Jenkinson A, Jia M, Richardson D, Shepherd R, Wooster R, Tejada MI, Martinez F, Carvill G, Goliath R, de Brouwer AP, van Bokhoven H, Van Esch H, Chelly J, Raynaud M, Ropers HH, Abidi FE, Srivastava AK, Cox J, Luo Y, Mallya U, Moon J, Parnau J, Mohammed S, Tolmie JL, Shoubridge C, Corbett M, Gardner A, Haan E, Rujirabanjerd S, Shaw M, Vandeleur L, Fullston T, Easton DF, Boyle J, Partington M, Hackett A, Field M, Skinner C, Stevenson RE, Bobrow M, Turner G, Schwartz CE, Gecz J, Raymond FL, Futreal PA, Stratton MR. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41:535-543. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 490] [Cited by in RCA: 476] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 34. | Lim J, Ryu J, Kang S, Noh HJ, Kim CH. Autism-like behaviors in male mice with a Pcdh19 deletion. Mol Brain. 2019;12:95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 20] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 35. | Depienne C, Trouillard O, Bouteiller D, Gourfinkel-An I, Poirier K, Rivier F, Berquin P, Nabbout R, Chaigne D, Steschenko D, Gautier A, Hoffman-Zacharska D, Lannuzel A, Lackmy-Port-Lis M, Maurey H, Dusser A, Bru M, Gilbert-Dussardier B, Roubertie A, Kaminska A, Whalen S, Mignot C, Baulac S, Lesca G, Arzimanoglou A, LeGuern E. Mutations and deletions in PCDH19 account for various familial or isolated epilepsies in females. Hum Mutat. 2011;32:E1959-E1975. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 36. | Zhao X, Wang Y, Mei S, Kong X. A novel PCDH19 missense mutation, c.812G>A (p.Gly271Asp), identified using whole-exome sequencing in a Chinese family with epilepsy female restricted mental retardation syndrome. Mol Genet Genomic Med. 2020;8:e1234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 37. | Kolc KL, Sadleir LG, Scheffer IE, Ivancevic A, Roberts R, Pham DH, Gecz J. A systematic review and meta-analysis of 271 PCDH19-variant individuals identifies psychiatric comorbidities, and association of seizure onset and disease severity. Mol Psychiatry. 2019;24:241-251. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 90] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 38. | Gibitova EA, Dobrynin PV, Pomerantseva EA, Musatova EV, Kostareva A, Evsyukov I, Rychkov SY, Zhukova OV, Naumova OY, Grigorenko EL. A Study of the Genomic Variations Associated with Autistic Spectrum Disorders in a Russian Cohort of Patients Using Whole-Exome Sequencing. Genes (Basel). 2022;13:920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 39. | Pizzo L, Jensen M, Polyak A, Rosenfeld JA, Mannik K, Krishnan A, McCready E, Pichon O, Le Caignec C, Van Dijck A, Pope K, Voorhoeve E, Yoon J, Stankiewicz P, Cheung SW, Pazuchanics D, Huber E, Kumar V, Kember RL, Mari F, Curró A, Castiglia L, Galesi O, Avola E, Mattina T, Fichera M, Mandarà L, Vincent M, Nizon M, Mercier S, Bénéteau C, Blesson S, Martin-Coignard D, Mosca-Boidron AL, Caberg JH, Bucan M, Zeesman S, Nowaczyk MJM, Lefebvre M, Faivre L, Callier P, Skinner C, Keren B, Perrine C, Prontera P, Marle N, Renieri A, Reymond A, Kooy RF, Isidor B, Schwartz C, Romano C, Sistermans E, Amor DJ, Andrieux J, Girirajan S. Rare variants in the genetic background modulate cognitive and developmental phenotypes in individuals carrying disease-associated variants. Genet Med. 2019;21:816-825. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 132] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 40. | Parfenenko MA, Dantsev IS, Bochenkov SV, Kuramagomedova RG, Vinogradova NV, Afanaseva MP, Groznova OS, Voinova VI. Expansion of phenotypic and genotypic data in autism spectrum disorders due to variants in the CHD8 gene. Neurogenetics. 2024;26:4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 41. | Yin J, Chun CA, Zavadenko NN, Pechatnikova NL, Naumova OY, Doddapaneni HV, Hu J, Muzny DM, Schaaf CP, Grigorenko EL. Next Generation Sequencing of 134 Children with Autism Spectrum Disorder and Regression. Genes (Basel). 2020;11:853. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
Open Access: This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: https://creativecommons.org/Licenses/by-nc/4.0/