Published online Nov 6, 2013. doi: 10.4292/wjgpt.v4.i4.97
Revised: September 12, 2013
Accepted: October 15, 2013
Published online: November 6, 2013
Processing time: 83 Days and 7 Hours
Metabolomics has increasingly been applied in addition to other “omic” approaches in the study of the pathophysiology of different gastrointestinal diseases. Metabolites represent molecular readouts of the cell status reflecting a physiological phenotype. In addition, changes in metabolite concentrations induced by exogenous factors such as environmental and dietary factors which do not affect the genome, are taken into account. Metabolic reactions initiated by the host or gut microbiota can lead to “marker” metabolites present in different biological fluids that allow differentiation between health and disease. Several lines of evidence implicated the involvement of intestinal microbiota in the pathogenesis of inflammatory bowel disease (IBD). Also in irritable bowel syndrome (IBS), a role of an abnormal microbiota composition, so-called dysbiosis, is supported by experimental data. These compositional alterations could play a role in the aetiology of both diseases by altering the metabolic activities of the gut bacteria. Several studies have applied a metabolomic approach to identify these metabolite signatures. However, before translating a potential metabolite biomarker into clinical use, additional validation studies are required. This review summarizes contributions that metabolomics has made in IBD and IBS and presents potential future directions within the field.
Core tip: Metabolic profiling is a powerful exploratory tool for understanding interactions between nutrients, the intestinal metabolism and the microbiota composition in health and disease and, to gain more insight in metabolic pathways. Metabolomics may advance our understanding, diagnosis and treatment of inflammatory bowel disease and irritable bowel syndrome. Metabolic reactions initiated by the host or gut microbiota can lead to “marker” metabolites present in different biological fluids that allow differentiation between health and disease. Disease-related mechanisms may be uncovered and verified, and candidate diagnostic biomarkers in biological samples are characterized.
- Citation: Preter VD, Verbeke K. Metabolomics as a diagnostic tool in gastroenterology. World J Gastrointest Pharmacol Ther 2013; 4(4): 97-107
- URL: https://www.wjgnet.com/2150-5349/full/v4/i4/97.htm
- DOI: https://dx.doi.org/10.4292/wjgpt.v4.i4.97
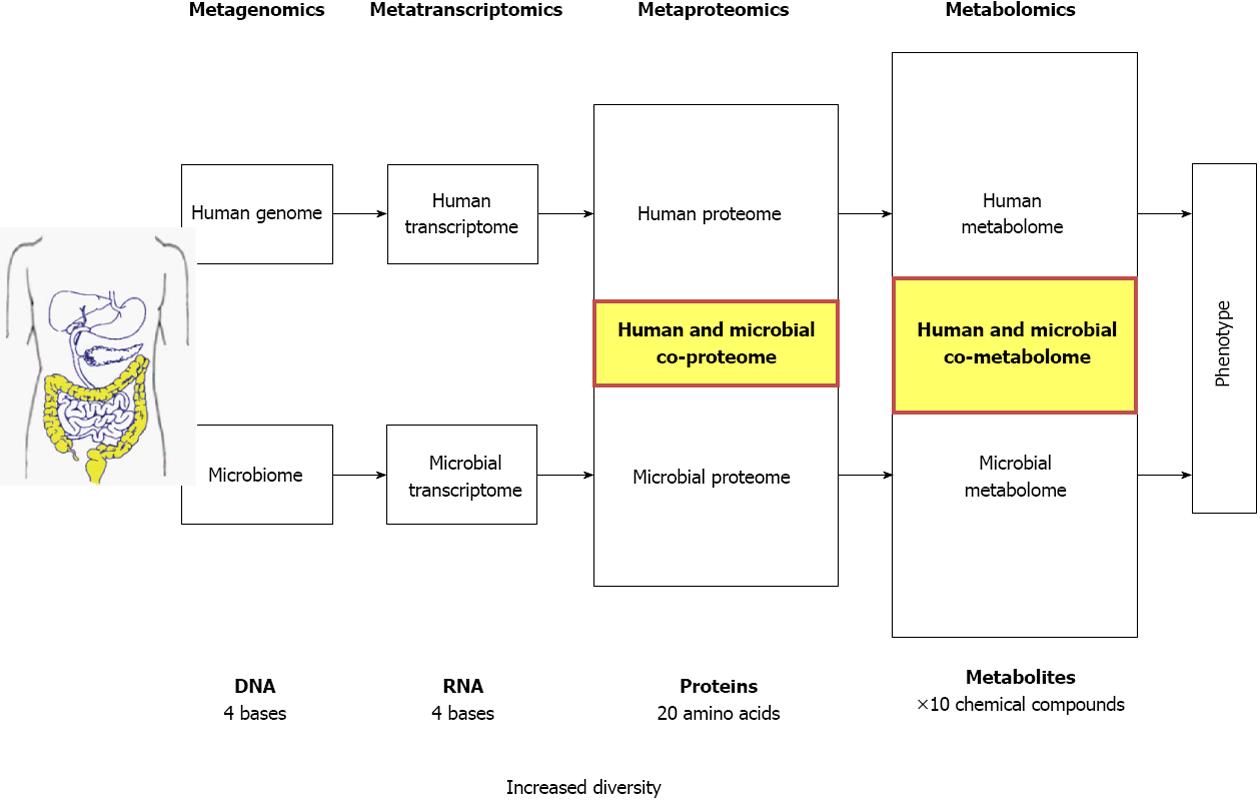
Different “Omic” approaches are currently applied to identify novel diagnostic targets and disease specific markers, and to characterize the link between gut microbiota or host metabolism and functional alterations in the pathophysiology of different diseases. Genomics, transcriptomics and proteomics provide extensive information regarding the genotype but convey limited information about the phenotype (Figure 1). Gene expression and protein data mainly indicate the potential for specific metabolic functions and do not always reflect the effective physiological processes as several downstream regulatory mechanisms are involved. As compared to other “omics”, metabolic profiling or metabolomics, integrates the effects of gene regulation, post-transcriptional regulation and pathway interactions. This downstream synthesis of diverse signals ultimately makes metabolites direct molecular readouts of cell status that reflect a meaningful physiological phenotype (Figure 1)[1,2]. In addition, changes in metabolite concentrations are also induced by exogenous factors such as environmental and dietary factors which do not affect the genome. Metabolomics is defined as “the non-biased identification and quantification of all metabolites in a biological system”[3]. For the quantitative analysis of metabolites in response to disease, Nicholson and colleagues introduced the term metabonomics or “the quantitative measurement of the multiparametric metabolic responses of a living system to pathophysiological stimuli or genetic modification”[4]. In practice, within human disease research, both terms are used indifferently.
Metabolic profiling is a powerful exploratory tool for understanding interactions between nutrients, the intestinal metabolism and the microbiota composition in health and disease and, to gain more insight in metabolic pathways. Metabolomic studies allow evaluation of metabolites by a top-down approach bypassing the need for an a priori hypothesis. Generally, metabolomic analysis has in view two major opportunities. First, untargeted analysis of a large number of metabolites enhances the chance to discover metabolites that are associated with the disease and might serve as biomarkers. In this respect, biomarker models are designed to discriminate with optimal sensitivity/specificity between groups, but do not presume biological understanding as an absolute prerequisite for biomarker development. However, understanding of the biological pathways can certainly support an assay[5]. Second, the profile of metabolites affected by the disease may provide new insights into the pathogenesis and eventually reveal new therapeutic targets.
Until now, genomic and proteomic methodologies have often been applied to uncover gastrointestinal related pathophysiological processes[6-11]. However, currently, metabolomics technologies are increasingly used for discovery of gastrointestinal disease signatures and have been applied for the screening of different pathological conditions that are linked with a metabolic imbalance. This review focuses on the contribution of metabolic profiling in advancing research in the field of inflammatory bowel disease (IBD) and irritable bowel syndrome (IBS).
The microbiota residing in the human gastrointestinal tract, especially the large intestine, is recognized as one of the most metabolically active organs of the human body. This microbial ecosystem is extremely complex and dynamic with high densities of living bacteria consisting of approximately 500-1000 different species[12,13]. In healthy adults, 80% of the identified fecal microbiota can be classified into three dominant phyla: Firmicutes, Bacteroidetes and Actinobacteria, but there is substantial variation in the species composition between individuals[14]. A total of about 1014 bacterial cells are present in the adult intestine, which is ten times the number of cells in the human body[15]. This microbiome outnumbers the host’s genetic potential by two orders of magnitude[16] and provides a diverse range of biochemical and metabolic activities to complement the host’s physiology. The presence and metabolic activities of a specific bacterial community play an important role in maintaining the host’s overall health and well-being, and has been shown to respond to metabolic challenges and dietary factors. This complex microbial system varies with the host’s age, diet and health status[17].
Through the process of fermentation, colonic bacteria produce a wide range of compounds that may influence the physiological processes in the colon. The human microbiota is characterized by a significant degree of functional redundancy, meaning that different bacteria can perform similar functions and metabolize the same substrate, thereby producing similar metabolites[18]. Therefore, not only the composition but also the functional capacity of the intestinal microbiota is highly important regarding the clinical end points. Nevertheless, metabolic insights remain limited due to the inaccessibility of the intestinal habitat and the complexity of the microbiota composition[12]. A number of factors, such as nutrient availability, physicochemical nutrient properties, colonic transit time, and age of the host, influence the composition and the metabolic activity of the colonic microbiota. Nutrient availability is believed to be the most important regulator of bacterial metabolism. Especially the ratio of available carbohydrate to nitrogen determines the degree of saccharolytic vs proteolytic fermentation[19]. Colonic fermentation of carbohydrates results in the generation of short-chain fatty acids (SCFA) which are generally assumed to be beneficial for the host[20]. Protein fermentation gives rise to a variety of metabolites such as phenolic compounds, branched-chain fatty acids, S-containing compounds, amines and ammonia[21,22].
The intestinal microbiota produces a number of compounds during the metabolism of nutrients and xenobiotics (compounds of non-host origin that enter the gut with the diet or are produced by the microbiota). Some of these metabolites are excreted in feces whereas others are absorbed through the colonic mucosa and enter the systemic circulation where they can be further modified by human metabolism. For instance, p-cresol is a bacterial fermentation product produced in the colon from tyrosine that is effectively absorbed. It is conjugated in the colon mucosa or liver to p-cresol sulfate or p-cresol glucuronide which improves the water solubility and facilitates its urinary excretion[23]. These metabolites are called human commensal co-metabolites. Also the opposite occurs. A number of metabolites that are derived from host metabolism are returned to the gut via biliary excretion where they can be further metabolized by the microbiota. For instance, bile acids that have escaped absorption in the terminal ileum can be deconjugated and converted to secondary bile acids by microbial metabolism[24].
These host-microbiota metabolic interactions complicate the interpretation of metabolite profiles. In addition, this co-metabolism explains that the outcome of metabolome analyses clearly depends on the biomatrix chosen. The contribution of the microbial metabolism is more likely reflected in the fecal metabolome than in urinary, serum of breath profiles. Urinary profiles contain human and human-microbial co-metabolites whereas serum profiles seem less influenced by bacterial metabolism.
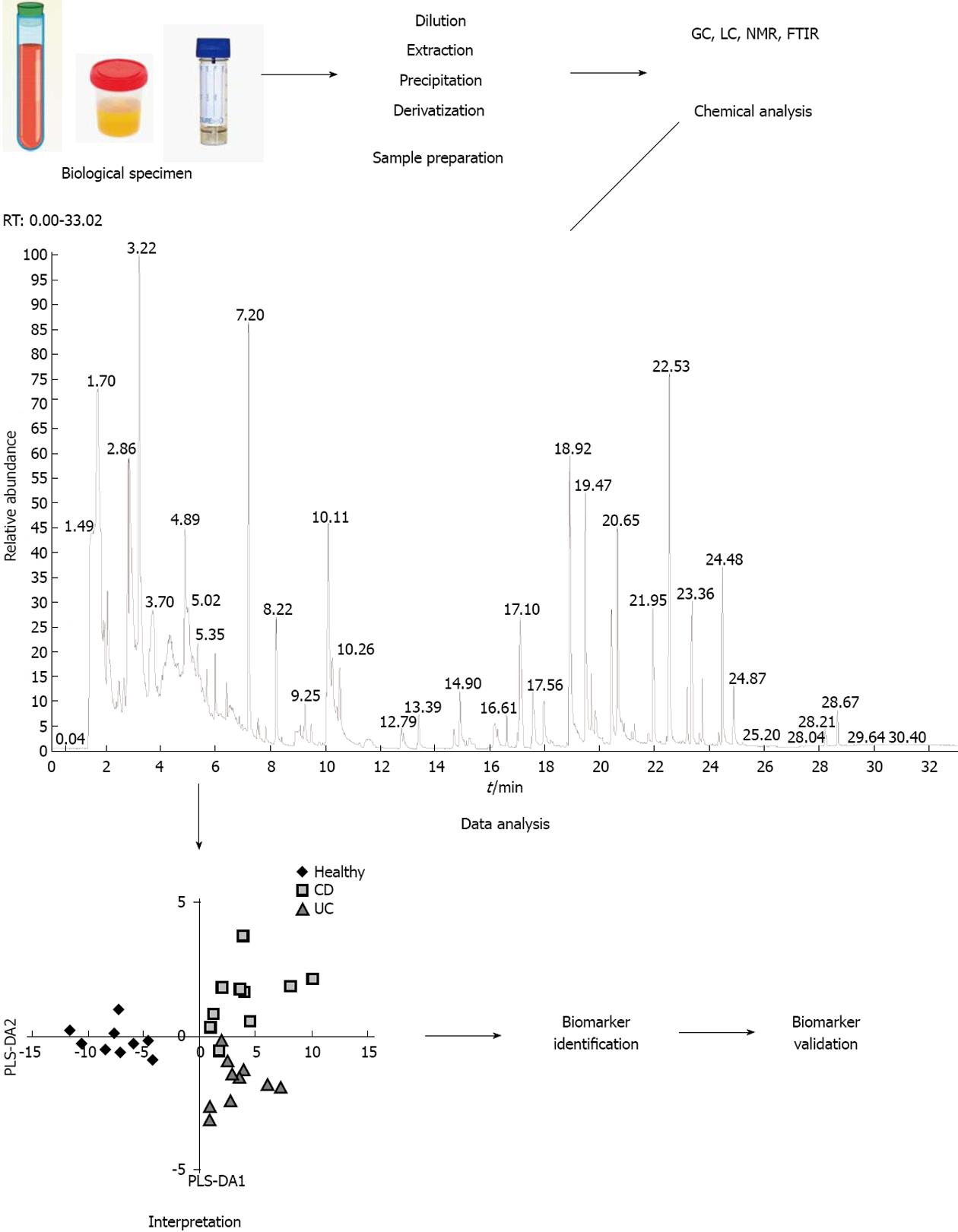
Fiehn et al[1] defined metabolomic analysis as “a comprehensive and quantitative analysis of the metabolome” with the metabolome defined as the whole of metabolites produced by an organism. However, due to the chemical diversity and different physicochemical properties of the metabolites and the large dynamic range of metabolite concentrations in different biological samples, it is virtually impossible to measure the complete metabolome. By selecting a specific analytical platform and a biofluid in which metabolites will be measured, the metabolome will be reduced to those specific conditions. Serum, plasma, urine, feces and tissue are the most studied biological matrices[25]. An overview of the different steps involved in the analytical process is shown in Figure 2.
Multiple analytical techniques have been used for the analysis of the metabolome. Gas chromatography (GC), liquid chromatography (LC) and high/ultra performance liquid chromatography (H/UPLC) coupled to mass spectrometry (MS), and nuclear magnetic resonance spectroscopy (NMR) enable detection, identification and quantification of metabolites[26,27]. Other analytical options consist of Fourier transform infrared spectroscopy (FTIS) or capillary electrophoresis (CE) coupled to MS[28,29].
The applicability of these analytical techniques differs. GC-MS provides an extraordinary resolution, permitting the separation of structurally similar compounds. However, this technique requires the compounds to be volatile and thermally stable. A chemical derivatization step can be applied prior to the chromatographic separation to render polar metabolites more volatile. Purge-and-trap and solid phase micro-extraction are sample preparation techniques often used in combination with GC. For metabolites that are not volatile and which cannot be derivatized, LC-MS is applied. LC-MS can detect a relatively broad spectrum of (polar and non-polar) metabolites with ample selectivity and sensitivity[30]. CE-MS is a rather new technique in metabolomics that is more sensitive and detects a wider spectrum of (polar) compounds than LC-MS[31,32]. 1H-NMR is a non-destructive technique that does not require prior separation of the compounds in the biofluid. It provides detailed information on molecular structure and requires only minimal sample preparation, but has a lower sensitivity than the MS based techniques[33]. Often, a combination of different techniques is applied as none of the individual methods will cover the full metabolome[27]. Several on-line databases for identifying metabolites from experimental NMR and/or MS data are available, as summarized in Table 1. These databases contain chemical, spectral, clinical and molecular data about metabolites found in different human biofluids.
| Database | URL or web address | Extra information |
| Human Metabolome Database (HMDB) | http://www.hmdb.ca/ | Wishart et al[34] |
| Madison Metabolomics Consortium (MMC) Database | http://mmcd.nmrfam.wisc.edu/ | Cui et al[35] |
| Biological Magnetic Resonance Data Bank (BMRB) | http://www.bmrb.wisc.edu/ | Ulrich et al[36] |
| Golm Metabolome Database | http://csbdb.mpimp-golm.mpg.de/csbdb/gmd/gmd.html | Kopka et al[37] |
| BiGG (a knowledgebase of Biochemically, Genetically and Genomically structured genome-scale metabolic network reconstructions) | http://bigg.ucsd.edu/ | Schellenberger et al[38] |
| SetupX and BinBase | http://fiehnlab.ucdavis.edu/ | Skogerson et al[39] |
| MassBank | http://www.massbank.jp/ | Horai et al[40] |
| METLIN | http://metlin.scripps.edu/ | Smith et al[41] |
Depending on the technique, detection limits vary: detection limits for NMR and FTIS (mM sensitivity) are much higher as compared to GC-MS (< mM sensitivity) and LC-MS (nM sensitivity). As a consequence, MS-based techniques are preferably applied for quantification of specific metabolites.
The analysis and interpretation of complex metabolomics data is facilitated by the application of chemometric and bio-statistical tools. Commonly used tests in metabolomic studies include principal component analysis (PCA) and partial-least squares discriminant analysis (PLS-DA). PCA is an unsupervised classification method, since the variation in the data is analyzed without a priori designation of samples into their classes. In contrast, PLS-DA is considered a supervised classification method, as the samples are designated into their classes for comparison.
In metabolomics, typically the number of variables or metabolites largely exceeds the number of samples measured. This can lead to the discovery of a number of variables that randomly, i.e., by chance, correlate to the outcome variable and in this way give the impression of a good predictive ability. However, if such a set of variables is chosen and the model is applied to new samples, the predictive ability might be very poor. This is known as over-fitting or fitting to the noise and can be avoided by careful cross-validation of the model. Cross-validation implicates that the data set is split in a training set and a test set. The biomarker model is discovered using the training set and the performance of the model is evaluated using the test set. In case of a relatively low sample number (< 100), multiple rounds of cross validation are performed using different partitions of the data in training set/test set and the performance results are averaged[5].
In gastrointestinal diseases such as IBD and IBS, there is an emerging consensus hypothesis that a dysbiosis of the microbiota is involved in initiating the disease or maintaining it. Several studies identified a disproportion of the predominant bacteria in fecal samples of IBD patients[18,42] and IBS patients[43]. For example, a reduction in the abundance and diversity of Firmicutes is frequently associated with IBD and IBS. At present, studies comparing the metabolic activity of the microbiota of IBD and IBS patients as compared to normal individuals are emerging thereby investigating whether eventual differences could be related to the pathogenesis of the disease[44,45] or whether they could be used as a classification tool in clinical diagnosis.
Inflammatory bowel diseases comprise Crohn’s disease and ulcerative colitis as the two major phenotypes. Although both phenotypes share similar pathophysiological and clinical features, they require different therapeutic management and display different prognosis. Both manifestations are influenced by genetic predispositions as well as microbial and environmental factors. At present, the diagnosis of IBD mainly relies on clinical, endoscopic, radiologic and histologic examination which implicates that diagnosis is only possible at a relatively advanced stage of the disease. Less invasive methods such as analysis of biomarkers from urine, serum, or feces, however, would be of significant advantage and useful for primary diagnosis, surveillance, and early detection of relapses.
Several biomarkers or sets of biomarkers have been tested in clinical trials including acute phase proteins such as C-reactive protein, fecal markers (lactoferrin, calprotectin, and PMN-elastase) and serological markers (antibodies against luminal antigens and anti-glycan antibodies)[46]. Recently, the exploration of metabolomics in IBD rose from the need to improve diagnosis and to allow better stratification of patients into IBD subtypes.
Several studies have applied a metabolomic approach to discriminate IBD patients from healthy controls, to discriminate CD from UC and patients with active disease from patients in remission. An overview of the studies in humans is presented in Table 2.
| Reference | Analytical platform | Biofluid | Samples | Observations |
| Marchesi et al[47] | 1HNMR | Faecal extracts | CD (n = 10), UC (n = 10), HC (n = 13) | Depletion of SCFA and methylamine and trimethylamine in CD patients Higher amounts of amino acids in UC and CD compared to healthy controls |
| Jansson et al[64] | ICR-FT/MS | Faecal water | 10 twin pairs with CD, 7 healthy twin pairs | Discrimination based on disease location (ileal or colonic CD) significant differences in the types and number of metabolites within specific pathways, including tyrosine and phenyl-alanine metabolism and bile acid and fatty acid biosynthesis |
| Le Gall et al[49] | 1HNMR | Faecal water | UC (n = 13; 31 samples), IBS (n = 10; 21 samples), HC (n = 22; 72 samples) | Discrimination between UC and HC; no classification of IBS Increased taurine and cadaverine in UC |
| Walton et al[50] | GC-MS | Faeces | UC (n = 20), CD (n = 22), IBS (n = 26), HC (n = 19) | Increased concentrations of ester and alcohol derivates of short-chain fatty acids and indole in CD After treatment, metabolite patterns are more similar to those of HC |
| Williams et al[52] | 1HNMR | Urine | CD (n = 86), UC (n = 60), HC (n = 60) | Discrimination between CD, UC and HC Significantly different metabolites include hippurate, p-cresol sulfate and formate Clustering independent of diet and medication |
| Schicho et al[53] | 1HNMR | Urine, serum, plasma | CD (n = 20), UC (n = 20), HC (n = 40) | IBD patients could be discriminated from HC, differences between CD and UC less pronounced Discriminating metabolites include amino acids, creatine, creatinine, metabolites of urea cycle, monosaccharides, hippurate (urine) |
| Stephens et al[54] | 1HNMR | Urine | CD (n = 30), UC (n = 30), HC (n = 60) | Metabolites for distinguishing IBD from HC: TCA cycle intermediates, amino acids metabolites derived from gut microflora (methanol, formate, hippurate, acetate, and methylamine); as well as the other metabolites trigonelline, creatine, urea, and taurine No discrimination between UC and CD after removal of patients with surgical intervention confounder |
| Ooi et al[58] | GC-MS | Colonic biopsies, serum | Colonic biopsies: UC (n = 22), serum: UC (n = 13), CD (n = 21), HC (n = 17) | Reduced levels of amino acids resulting in reduced levels of TCA cycle related downstream molecules in colonic tissue of UC Serum amino acid profiling enabled discrimination between UC and CD |
| Bjerrum et al[60] | 1HNMR | Colonic biopsies, colonocytes, lymphocytes, urine | Active UC (n = 35), quiescent UC (n = 33), HC (n = 25) | No discrimination between active UC, inactive UC and HC based on urine or lymphocyte profiles Inactive UC could not be differentiated from HC Active UC characterized by higher antioxidants and amino acids and lower levels of lipid, myo-inositol, betaine and glycerophosphoglycine 20% of inactive UC had similar profile as active UC |
| Bezabeh et al[63] | 1HNMR | Colonic biopsies | UC (n = 26; 45 samples), CD (n = 21; 31 samples), controls (38 non-inflamed IBD, 25 cancer patients) | Accurate classification of UC vs CD Some non-inflamed tissues from IBD had abnormal NMR-spectra |
| Balasubramanian et al[61] | 1HNMR | Colonic biopsies | Active UC (n = 20), Inactive UC (n = 11), Active CD (n = 20), Inactive CD (n = 6), HC (n = 26) | Higher α-glucose and lower amino acids, membrane components, lactate and succinate in active UC and CD compared to HC Lower lactate, glycerophosphorylcholine and myo-inositol in inactive UC and lower lactate in inactive CD compared to HC Lower formate in active UC vs active CD |
| Sharma et al[62] | 1HNMR | Colonic biopsies (inflamed and non-inflamed) | UC (n = 12), CD (n = 9), controls (n = 25) | No differentiation between inflamed and non-inflamed samples Lower levels of amino acids, membrane components, lactate and formate in IBD vs controls and higher levels of glucose |
| Hisamatsu et al[66] | AA analyzer | plasma | CD (n = 165), UC (n = 222), HC (n = 210) | Multivariate indexes established from plasma aminograms distinguish CD or UC from HC Other indexes distinguish active UC and CD from each remission patients and correlate with disease activity indices |
| Zhang et al[67] | 1HNMR | Serum | Active UC (n = 20), HC (n = 19) | Active UC displayed increased 3-hydroxybutyrate, β-glucose, α-glucose and phenylalanine and decreased lipid compared to healthy controls |
| Ponnusamy et al[71] | GC-MS | Faeces | IBS (n = 11) vs non-IBS (n = 8) | Elevated levels of amino acids and phenolic compounds that were highly correlated with abundance of lactobacilli and Clostridium |
| Ohman et al[69] | GC-MS | Faeces | IBS-D (n = 30), CD (n = 62), UC (n = 48), HC (n = 109) | Significantly more esters in IBS-D, association of aldehydes with IBD Accurate separation of IBS-D from active CD, UC and HC |
The growing acceptance of the involvement of the gut microbiota in the chronic mucosal immune activation underlying the pathogenesis of IBD has led to an interest in the use of fecal extracts or fecal samples as biofluids to apply metabolite profiling.
Marchesi et al[47] was the first to differentiate IBD patients from healthy controls based on 1H-NMR analysis of fecal water extracts and to discriminate CD patients from UC patients. Fecal water extracts from IBD patients were characterized by a depletion of bacterial degradation products such as SCFA, dimethylamine and trimethylamine suggesting a disruption of the normal bacterial ecology, called dysbiosis, either as the cause or consequence of the disease. In a study in identical twins including healthy twins and twins with inactive CD, either concordant or discordant, fecal extracts were analyzed using ICR-FT/MS with ultrahigh mass resolution. Healthy subjects could nicely be discriminated from CD patients. In addition, it was possible to separate patients with predominantly ileal involvement from patients with predominantly colonic involvement of the disease. Interestingly, the separation of the groups was higher for the metabolite profiles than for the microbial community profiles, analyzed on the same samples with T-RFLP fingerprints generated using general bacterial and Bacteroides group-specific primers[48]. The higher discrimination of the metabolite data was attributed to a direct link of the metabolites to function.
In another study, 1H-NMR profiling of fecal extract allowed to discriminate patients with UC from healthy controls. Elevated levels of taurine and cadaverine in UC patients were the major discriminative findings. Samples were also analyzed for microbiota composition using DGGE. Canonical correlation analysis between NMR and DGGE data sets, based on PC scores accounting for 90% of the original variance, revealed a good correlation (r = 0.85, P < 0.002) between gut microbiota profile and metabolite composition suggesting a direct link between both parameters[49].
A recent study analyzed metabolite profiles in feces of chronic gastrointestinal disorders using GC-MS with headspace sample preparation. Samples of CD patients showed significant higher levels of ester and alcohol derivatives of SCFA and indole as compared to healthy controls, UC and IBS patients. Following treatment, the metabolite profile was altered to more closely resemble that of healthy volunteers[50]. As many microbial metabolites are absorbed and excreted in urine, either as such or after further metabolism by human enzymes, metabolite profiles of urine samples may also reflect the impact of the microbiota composition[51]. Several studies were able to discriminate IBD patients from healthy controls based on metabolite profiling of urine samples[52-54]. In all these studies, hippurate levels were lower in IBD patients as compared to controls suggesting hippurate as a biomarker of IBD. Hippurate or N-benzoylglycine is a mammalian-microbial co-metabolite that originates from bacterial fermentation of dietary aromatic compounds (polyphenols, purines or aromatic amino acids) to benzoic acid which is further conjugated to glycine in the liver[55]. Remarkably, urinary metabolite profiling allowed to differentiate between UC and CD in only one study[51], whereas two other studies failed to do so[53,54]. This discrepancy may highlight the fact that IBD is a multifactorial disease with a high variety in phenotypes and severity[56]. Indeed, the notion that IBD is actually a syndrome comprising several disease subtypes, is gaining more and more acceptance[46].
Metabolite profiles in serum or plasma or in colonic mucosal biopsies rather reflect changes in the host’s metabolism and provide less information on the impact of the gut microbiota composition and/or activity. As compared to urinary and fecal profiles, metabolite profiles in serum or plasma may be less affected by environmental factors and are subject to less inter-individual variation[57].
Results from studies that analyzed serum/plasma or colonic mucosa cells indicate that both CD and UC have an impact on the amino acid metabolism[53,58-63]. Several amino acids occurred in lower levels in colonic mucosal cells from IBD patients as compared to controls. As higher amino acid levels were found in fecal extracts[47,64], this may be the result of malabsorption due to inflammation. An alternative explanation is that inflammatory conditions induce large energy requirements to repair the damaged mucosa leading to enhanced protein catabolism. Specifically, the role of decreased levels of glutamine in the pathogenesis of IBD has been studied. Besides butyrate, glutamine is an important energy source for the colonocytes and accounts for about 30% of their energy needs. In a mouse model of DSS-induced colitis, similar reductions in glutamine levels in serum and colonic tissue were observed and supplementation with glutamine attenuated the DSS-induced colitis[65].
In a recent study, Hisamatsu et al[66] calculated an AminoIndex based on multivariate analysis of amino acid profiles in serum of IBD patients which allowed to distinguish between CD and UC and also reflected disease activity.
Zhang et al[67] specifically recruited patients with active UC and short disease duration (2.7 years) and evaluated the efficacy of 1H-NMR metabolic profiling of serum for early stage diagnosis of UC. Although active UC patients could be discriminated from healthy controls in a multivariate OPLS-DA model, the number of altered metabolites in this study was rather limited.
Irritable bowel syndrome is a multifactorial functional disorder of the gastrointestinal tract that affects about 10%-15% of the adult population. IBS patients have symptoms of pain and bowel dysfunction. A subset of patients that developed IBS after an infection, so-called post-infectious IBS, may have microscopic inflammation but with normal mucosal appearance on endoscopy. Recently, a new IBS entity, IBD-IBS, has been described: these patients have pain and diarrhea similar to IBS in association with minimal or no evident intestinal inflammation[68]. The exact etiology has not been identified. Yet, environmental, psycho-social, physiological and genetic factors are believed to play a role. Also the role of an abnormal microbiota composition is supported by clinical and experimental data[69]. Disruption of the balance between the host and the intestinal microbiota results in changes in the mucosal immune system that range from overt inflammation, as seen in Crohn’s disease, to low-grade inflammation without tissue injury, as seen in a subset of IBS patients[70].
Surprisingly, very little work has been conducted on metabolite profiling of IBS patients. 1H-NMR metabolite profiling of fecal extracts allowed to separate IBS patients from healthy controls with moderate success (sensitivity = 57%, specificity = 76%)[49]. Two other studies applied GC/MS to analyze fecal samples although with different sample preparation. Ahmed et al[71] analyzed the volatile organic compounds (VOC) in the headspace of the fecal samples, i.e., compounds with a vapour pressure that is sufficiently high to enable them to move from the solid or liquid phase into the gaseous phase. Those VOC comprise hydrocarbons, alcohols, aldehydes, ketones, esters and organic acids[72]. In contrast, the volatility of polar compounds in the fecal samples was increased by trimethylsilyl derivatization in the study by Ponnusamy et al[73]. Although both studies were able to discriminate IBS patients from healthy controls, the metabolites responsible for discrimination were different which is inherent to the different sample preparation. Ahmed et al[71] found increased abundance of esters in diarrhea predominant IBS-patients compared with healthy controls. Of the 28 VOCs positively associated with IBS, 22 belonged to the class of esters. Ponnusamy et al[73] highlighted higher levels of specific amino acids (alanine and pyroglutamate) and phenolic compounds (hydroxyphenyl acetate and hydroxyphenyl propionate) and associated these metabolic changes to alterations of specific gut microbial populations including lactobacilli and Clostridium.
Metabolites are promising biomarkers because they can be easily measured from non-invasive breath, urine, feces or blood samples. Several studies have identified such metabolite signatures that should allow classification of samples as healthy or diseased. To translate these models into the clinic, additional validation studies are required. First of all, experiments to validate the biomarker model need to be performed. This level of validation includes (1) lab repeatability studies where the same samples are analyzed in the same laboratory by the same observer; (2) lab replication studies, where independent samples are analyzed in the same lab by the same observer; (3) inter-lab repeatability studies, were the original samples are analyzed in a different laboratory by a different observer; and (4) inter-lab replication studies, were independent samples are analyzed in a different laboratory.
Secondly, most studies mentioned above have compared metabolite profiles obtained from patients with a distinct diagnosis of either IBD or IBS to healthy subjects. To ensure clinical utility, it might be necessary to include additional control groups including patients with other gastrointestinal or inflammatory disorders like patients with infectious GI disease or neoplastic disease.
Thirdly, the influence of potential confounders on the performance of the models needs to be established. Confounding factors might be related to the subject (gender, age, disease location, comorbidities) or might be of environmental origin (diet, diurnal variation, medication, smoking).
Metabolomics may advance our understanding, diagnosis and treatment of inflammatory bowel disease and irritable bowel syndrome. Using this approach, disease-related mechanisms may be uncovered and verified, and candidate diagnostic biomarkers in biological samples are characterized. Before usage as clinical diagnostics, metabolites must be verified and validated in large clinical trials. To translate metabolomics data into a more profound biological understanding of the disease, more knowledge on the relevance of a decrease or increase in certain metabolites is warranted. Metabolomic profiling mainly detects associations between profiles and specific phenotypes which may not always be meaningful. In addition, it often remains unknown whether changes in metabolites are the cause or a consequence of the disease. Integration of other “Omics” with metabolomic may enable a further understanding of gastrointestinal related pathophysiological processes.
| 1. | Fiehn O. Metabolomics--the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155-171. [PubMed] |
| 2. | Assfalg M, Bertini I, Colangiuli D, Luchinat C, Schäfer H, Schütz B, Spraul M. Evidence of different metabolic phenotypes in humans. Proc Natl Acad Sci USA. 2008;105:1420-1424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 192] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 3. | Ellis DI, Dunn WB, Griffin JL, Allwood JW, Goodacre R. Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics. 2007;8:1243-1266. [PubMed] |
| 4. | Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181-1189. [PubMed] |
| 5. | Xia J, Broadhurst DI, Wilson M, Wishart DS. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013;9:280-299. [PubMed] |
| 6. | Berndt U, Bartsch S, Philipsen L, Danese S, Wiedenmann B, Dignass AU, Hämmerle M, Sturm A. Proteomic analysis of the inflamed intestinal mucosa reveals distinctive immune response profiles in Crohn’s disease and ulcerative colitis. J Immunol. 2007;179:295-304. [PubMed] |
| 7. | Olsen J, Gerds TA, Seidelin JB, Csillag C, Bjerrum JT, Troelsen JT, Nielsen OH. Diagnosis of ulcerative colitis before onset of inflammation by multivariate modeling of genome-wide gene expression data. Inflamm Bowel Dis. 2009;15:1032-1038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 107] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 8. | Arijs I, Quintens R, Van Lommel L, Van Steen K, De Hertogh G, Lemaire K, Schraenen A, Perrier C, Van Assche G, Vermeire S. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm Bowel Dis. 2010;16:2090-2098. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 140] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 9. | Lamendella R, VerBerkmoes N, Jansson JK. ‘Omics’ of the mammalian gut--new insights into function. Curr Opin Biotechnol. 2012;23:491-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 10. | Yau Y, Leong RW, Zeng M, Wasinger VC. Proteomics and metabolomics in inflammatory bowel disease. J Gastroenterol Hepatol. 2013;28:1076-1086. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 11. | Granlund Av, Flatberg A, Østvik AE, Drozdov I, Gustafsson BI, Kidd M, Beisvag V, Torp SH, Waldum HL, Martinsen TC. Whole genome gene expression meta-analysis of inflammatory bowel disease colon mucosa demonstrates lack of major differences between Crohn’s disease and ulcerative colitis. PLoS One. 2013;8:e56818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 92] [Cited by in RCA: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 12. | Tuohy KM, Gougoulias C, Shen Q, Walton G, Fava F, Ramnani P. Studying the human gut microbiota in the trans-omics era--focus on metagenomics and metabonomics. Curr Pharm Des. 2009;15:1415-1427. [PubMed] |
| 13. | Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207-214. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9292] [Cited by in RCA: 8375] [Article Influence: 598.2] [Reference Citation Analysis (4)] |
| 14. | Ventura M, Turroni F, Canchaya C, Vaughan EE, O’Toole PW, van Sinderen D. Microbial diversity in the human intestine and novel insights from metagenomics. Front Biosci (Landmark Ed). 2009;14:3214-3221. [PubMed] |
| 15. | Xu J, Gordon JI. Honor thy symbionts. Proc Natl Acad Sci USA. 2003;100:10452-10459. [PubMed] |
| 16. | Human Microbiome Project Consortium. A framework for human microbiome research. Nature. 2012;486:215-221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2152] [Cited by in RCA: 1862] [Article Influence: 133.0] [Reference Citation Analysis (0)] |
| 17. | Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O’Sullivan O. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1993] [Cited by in RCA: 2390] [Article Influence: 170.7] [Reference Citation Analysis (0)] |
| 18. | Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci USA. 2009;106:5859-5864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 557] [Article Influence: 32.8] [Reference Citation Analysis (0)] |
| 19. | De Preter V, Hamer HM, Windey K, Verbeke K. The impact of pre- and/or probiotics on human colonic metabolism: does it affect human health? Mol Nutr Food Res. 2011;55:46-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 107] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 21. | Smith EA, Macfarlane GT. Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol. 1996;81:288-302. [PubMed] |
| 22. | Fooks LJ, Fuller R, Gibson GR. Prebiotics, probiotics and human gut microbiology. Int Dairy J. 1999;9:53-61. [RCA] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 209] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 23. | Evenepoel P, Meijers BK, Bammens BR, Verbeke K. Uremic toxins originating from colonic microbial metabolism. Kidney Int Suppl. 2009;S12-S19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 341] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 24. | Bajor A, Gillberg PG, Abrahamsson H. Bile acids: short and long term effects in the intestine. Scand J Gastroenterol. 2010;45:645-664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 107] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 25. | Issaq HJ, Abbott E, Veenstra TD. Utility of separation science in metabolomic studies. J Sep Sci. 2008;31:1936-1947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 41] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 26. | De Preter V, Van Staeyen G, Esser D, Rutgeerts P, Verbeke K. Development of a screening method to determine the pattern of fermentation metabolites in faecal samples using on-line purge-and-trap gas chromatographic-mass spectrometric analysis. J Chromatogr A. 2009;1216:1476-1483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 54] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 27. | Weckwerth W, Morgenthal K. Metabolomics: from pattern recognition to biological interpretation. Drug Discov Today. 2005;10:1551-1558. [PubMed] |
| 28. | Dunn WB, Bailey NJ, Johnson HE. Measuring the metabolome: current analytical technologies. Analyst. 2005;130:606-625. [PubMed] |
| 29. | van der Greef J, Stroobant P, van der Heijden R. The role of analytical sciences in medical systems biology. Curr Opin Chem Biol. 2004;8:559-565. [PubMed] |
| 30. | Barderas MG, Laborde CM, Posada M, de la Cuesta F, Zubiri I, Vivanco F, Alvarez-Llamas G. Metabolomic profiling for identification of novel potential biomarkers in cardiovascular diseases. J Biomed Biotechnol. 2011;2011:790132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 31. | Soga T, Ishikawa T, Igarashi S, Sugawara K, Kakazu Y, Tomita M. Analysis of nucleotides by pressure-assisted capillary electrophoresis-mass spectrometry using silanol mask technique. J Chromatogr A. 2007;1159:125-133. [PubMed] |
| 32. | Ramautar R, Somsen GW, de Jong GJ. CE-MS for metabolomics: developments and applications in the period 2010-2012. Electrophoresis. 2013;34:86-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 33. | Ala-Korpela M. Critical evaluation of 1H NMR metabonomics of serum as a methodology for disease risk assessment and diagnostics. Clin Chem Lab Med. 2008;46:27-42. [PubMed] |
| 34. | Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801-D807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2109] [Cited by in RCA: 2206] [Article Influence: 157.6] [Reference Citation Analysis (0)] |
| 35. | Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol. 2008;26:162-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 511] [Cited by in RCA: 442] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 36. | Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z. BioMagResBank. Nucleic Acids Res. 2008;36:D402-D408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1295] [Cited by in RCA: 1279] [Article Influence: 71.1] [Reference Citation Analysis (0)] |
| 37. | Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmüller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635-1638. [PubMed] |
| 38. | Schellenberger J, Park JO, Conrad TM, Palsson BØ. BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics. 2010;11:213. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 400] [Cited by in RCA: 389] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 39. | Skogerson K, Wohlgemuth G, Barupal DK, Fiehn O. The volatile compound BinBase mass spectral database. BMC Bioinformatics. 2011;12:321. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 146] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 40. | Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K. MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom. 2010;45:703-714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1496] [Cited by in RCA: 1726] [Article Influence: 107.9] [Reference Citation Analysis (0)] |
| 41. | Smith CA, O’Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747-751. [PubMed] |
| 42. | Joossens M, Huys G, Cnockaert M, De Preter V, Verbeke K, Rutgeerts P, Vandamme P, Vermeire S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 692] [Cited by in RCA: 819] [Article Influence: 54.6] [Reference Citation Analysis (0)] |
| 43. | Krogius-Kurikka L, Lyra A, Malinen E, Aarnikunnas J, Tuimala J, Paulin L, Mäkivuokko H, Kajander K, Palva A. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9:95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 201] [Cited by in RCA: 241] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 44. | McNiven EM, German JB, Slupsky CM. Analytical metabolomics: nutritional opportunities for personalized health. J Nutr Biochem. 2011;22:995-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 39] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 45. | Olivares M, Laparra JM, Sanz Y. Host genotype, intestinal microbiota and inflammatory disorders. Br J Nutr. 2013;109 Suppl 2:S76-S80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 46. | Dotan I. New serologic markers for inflammatory bowel disease diagnosis. Dig Dis. 2010;28:418-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 47. | Marchesi JR, Holmes E, Khan F, Kochhar S, Scanlan P, Shanahan F, Wilson ID, Wang Y. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546-551. [PubMed] |
| 48. | Dicksved J, Halfvarson J, Rosenquist M, Järnerot G, Tysk C, Apajalahti J, Engstrand L, Jansson JK. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J. 2008;2:716-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 335] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 49. | Le Gall G, Noor SO, Ridgway K, Scovell L, Jamieson C, Johnson IT, Colquhoun IJ, Kemsley EK, Narbad A. Metabolomics of fecal extracts detects altered metabolic activity of gut microbiota in ulcerative colitis and irritable bowel syndrome. J Proteome Res. 2011;10:4208-4218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 284] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 50. | Walton C, Fowler DP, Turner C, Jia W, Whitehead RN, Griffiths L, Dawson C, Waring RH, Ramsden DB, Cole JA. Analysis of volatile organic compounds of bacterial origin in chronic gastrointestinal diseases. Inflamm Bowel Dis. 2013;19:2069-2078. [PubMed] |
| 51. | Nicholls AW, Mortishire-Smith RJ, Nicholson JK. NMR spectroscopic-based metabonomic studies of urinary metabolite variation in acclimatizing germ-free rats. Chem Res Toxicol. 2003;16:1395-1404. [PubMed] |
| 52. | Williams HR, Cox IJ, Walker DG, North BV, Patel VM, Marshall SE, Jewell DP, Ghosh S, Thomas HJ, Teare JP. Characterization of inflammatory bowel disease with urinary metabolic profiling. Am J Gastroenterol. 2009;104:1435-1444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 53. | Schicho R, Shaykhutdinov R, Ngo J, Nazyrova A, Schneider C, Panaccione R, Kaplan GG, Vogel HJ, Storr M. Quantitative Metabolomic Profiling of Serum, Plasma, and Urine by (1)H NMR Spectroscopy Discriminates between Patients with Inflammatory Bowel Disease and Healthy Individuals. J Proteome Res. 2012;Epub ahead of print. [PubMed] |
| 54. | Stephens NS, Siffledeen J, Su X, Murdoch TB, Fedorak RN, Slupsky CM. Urinary NMR metabolomic profiles discriminate inflammatory bowel disease from healthy. J Crohns Colitis. 2013;7:e42-e48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 55. | Williams HR, Cox IJ, Walker DG, Cobbold JF, Taylor-Robinson SD, Marshall SE, Orchard TR. Differences in gut microbial metabolism are responsible for reduced hippurate synthesis in Crohn’s disease. BMC Gastroenterol. 2010;10:108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 92] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 56. | Strober W, Fuss I, Mannon P. The fundamental basis of inflammatory bowel disease. J Clin Invest. 2007;117:514-521. [PubMed] |
| 57. | Lenz EM, Bright J, Wilson ID, Morgan SR, Nash AF. A 1H NMR-based metabonomic study of urine and plasma samples obtained from healthy human subjects. J Pharm Biomed Anal. 2003;33:1103-1115. [PubMed] |
| 58. | Ooi M, Nishiumi S, Yoshie T, Shiomi Y, Kohashi M, Fukunaga K, Nakamura S, Matsumoto T, Hatano N, Shinohara M. GC/MS-based profiling of amino acids and TCA cycle-related molecules in ulcerative colitis. Inflamm Res. 2011;60:831-840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 134] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 59. | Yoshida M, Hatano N, Nishiumi S, Irino Y, Izumi Y, Takenawa T, Azuma T. Diagnosis of gastroenterological diseases by metabolome analysis using gas chromatography-mass spectrometry. J Gastroenterol. 2012;47:9-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 60. | Bjerrum JT, Nielsen OH, Hao F, Tang H, Nicholson JK, Wang Y, Olsen J. Metabonomics in ulcerative colitis: diagnostics, biomarker identification, and insight into the pathophysiology. J Proteome Res. 2010;9:954-962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 128] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 61. | Balasubramanian K, Kumar S, Singh RR, Sharma U, Ahuja V, Makharia GK, Jagannathan NR. Metabolism of the colonic mucosa in patients with inflammatory bowel diseases: an in vitro proton magnetic resonance spectroscopy study. Magn Reson Imaging. 2009;27:79-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 90] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 62. | Sharma U, Singh RR, Ahuja V, Makharia GK, Jagannathan NR. Similarity in the metabolic profile in macroscopically involved and un-involved colonic mucosa in patients with inflammatory bowel disease: an in vitro proton ((1)H) MR spectroscopy study. Magn Reson Imaging. 2010;28:1022-1029. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 63. | Bajpai J, Sinha BN, Srivastava AN. Clinical study of Volkmann’s ischemic contracture of the upper limb. Int Surg. 1975;60:162-164. [PubMed] |
| 64. | Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, Tysk C, Schmitt-Kopplin P. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS One. 2009;4:e6386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 372] [Cited by in RCA: 412] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 65. | Shiomi Y, Nishiumi S, Ooi M, Hatano N, Shinohara M, Yoshie T, Kondo Y, Furumatsu K, Shiomi H, Kutsumi H. GCMS-based metabolomic study in mice with colitis induced by dextran sulfate sodium. Inflamm Bowel Dis. 2011;17:2261-2274. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 66. | Hisamatsu T, Okamoto S, Hashimoto M, Muramatsu T, Andou A, Uo M, Kitazume MT, Matsuoka K, Yajima T, Inoue N. Novel, objective, multivariate biomarkers composed of plasma amino acid profiles for the diagnosis and assessment of inflammatory bowel disease. PLoS One. 2012;7:e31131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 155] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 67. | Zhang Y, Lin L, Xu Y, Lin Y, Jin Y, Zheng C. 1H NMR-based spectroscopy detects metabolic alterations in serum of patients with early-stage ulcerative colitis. Biochem Biophys Res Commun. 2013;433:547-551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 68. | Grover M, Herfarth H, Drossman DA. The functional-organic dichotomy: postinfectious irritable bowel syndrome and inflammatory bowel disease-irritable bowel syndrome. Clin Gastroenterol Hepatol. 2009;7:48-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 85] [Article Influence: 5.0] [Reference Citation Analysis (1)] |
| 69. | Ohman L, Simrén M. Intestinal microbiota and its role in irritable bowel syndrome (IBS). Curr Gastroenterol Rep. 2013;15:323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 70. | Collins SM, Denou E, Verdu EF, Bercik P. The putative role of the intestinal microbiota in the irritable bowel syndrome. Dig Liver Dis. 2009;41:850-853. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 86] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 71. | Ahmed I, Greenwood R, Costello Bde L, Ratcliffe NM, Probert CS. An investigation of fecal volatile organic metabolites in irritable bowel syndrome. PLoS One. 2013;8:e58204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 72. | Wang S, Ang HM, Tade MO. Volatile organic compounds in indoor environment and photocatalytic oxidation: state of the art. Environ Int. 2007;33:694-705. [PubMed] |
| 73. | Ponnusamy K, Choi JN, Kim J, Lee SY, Lee CH. Microbial community and metabolomic comparison of irritable bowel syndrome faeces. J Med Microbiol. 2011;60:817-827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 176] [Article Influence: 11.7] [Reference Citation Analysis (1)] |
P- Reviewers: Hasanein P, Kanazawa M, Munoz M S- Editor: Qi Y L- Editor: A E- Editor: Liu XM