Published online Sep 8, 2020. doi: 10.4292/wjgpt.v11.i4.69
Peer-review started: March 28, 2020
First decision: April 25, 2020
Revised: June 25, 2020
Accepted: August 15, 2020
Article in press: August 15, 2020
Published online: September 8, 2020
Processing time: 156 Days and 14.3 Hours
The existence of genetic anticipation has been long disputed in inflammatory bowel disease (IBD) in the absence of the explanatory mechanism.
To determine whether it was predictive of genetic anticipation, we evaluated telomere length in IBD. We hypothesized that multiplex IBD families exhibit a genetic defect impacting telomere maintenance mechanisms.
We studied three IBD families with multiple affected members in three successive generations. We determined telomere length (TL) in lymphocytes and granulocytes from peripheral blood of the affected members using flow cytometry and fluorescence in-situ hybridization (flow FISH). We also performed whole exome sequencing in the blood of all available family members and used PhenoDB to identify potential candidate gene variants with recessive or dominant modes of inheritance.
Out of twenty-four patients of European descent selected to participate in the study, eleven patients, eight parent-child pairs affected by IBD, were included in the genetic anticipation analysis. Median difference in age at diagnosis between two successive generations was 16.5 years, with earlier age at onset in the younger generations. In most of the affected members, the disease harbored similar gastrointestinal and extraintestinal involvement but was more aggressive among the younger generations. TL was not associated with earlier age at onset or more severe disease in members of successive generations affected by IBD. NOD2 gene mutations were present in the Crohn’s disease patients of one family. However, no gene variants were identified as potential candidates for inheritance.
Telomere shortening appears unlikely to be involved in mechanisms of possible genetic anticipation in IBD. Further studies using a larger sample size are required to confirm or refute our findings.
Core tip: This is a retrospective study to evaluate the role of telomere shortening in genetic anticipation of inflammatory bowel disease (IBD). Genetic anticipation is a long disputed concept in IBD and lacks an explanatory mechanism. We analyzed generational changes in telomere length of eight parent-child pairs of three generation IBD families with anticipation and performed whole exome sequencing to identify genetic variants for autosomal inheritance. Neither telomere shortening or autosomal inheritance was associated with anticipation in our three generation IBD families, suggesting other potential mechanisms underlie this phenomenon.
- Citation: Truta B, Wohler E, Sobreira N, Datta LW, Brant SR. Role of telomere shortening in anticipation of inflammatory bowel disease. World J Gastrointest Pharmacol Ther 2020; 11(4): 69-78
- URL: https://www.wjgnet.com/2150-5349/full/v11/i4/69.htm
- DOI: https://dx.doi.org/10.4292/wjgpt.v11.i4.69
Genetic anticipation is a phenomenon in which an inherited disorder manifests at a younger age and often more severely in offspring than in the affected parent[1]. This phenomenon has been reported in over twenty monogenetic degenerative neurological disorders[2]. In some of these disorders, such as Huntington’s disease, familial Parkinson’s disease, and fragile X syndrome, the molecular basis of anticipation has been linked to the expansion of unstable genomic triplet repeats, in step with the progression of the disease down through generations[3-7].
For inflammatory bowel disease (IBD) the involvement of genetic anticipation has been considered since the 1990’s[8,9] because of observed earlier ages at diagnosis and more severe disease among offspring of patients with IBD, mainly Crohn’s disease (CD). However, no mechanisms accounting for genetic anticipation have been identified, and many scientists have doubted genetic anticipation in IBD, attributing the observed differences in age at diagnosis and disease severity to biased observation[10,11].
The list of conditions exhibiting anticipation continues to grow. This list includes not only monogenic disease but also polygenic diseases such as bipolar disorder[12], schizophrenia[13], rheumatoid arthritis[14], Behcet’s disease[15], and chronic leukemia[16], in which the molecular mechanism is yet to be discovered.
One new cause of genetic anticipation is telomeropathies, which encompasses a wide variety of rare diseases caused by genetic defects in telomerase maintenance mechanisms[17,18]. Telomeres are nucleoproteins that protect chromosomal ends by counteracting the erosion caused by the problems inherent in terminal replication of the chromosomes. In telomeropathies, mutations in the telomerase reverse transcriptase, the specialized polymerase that synthesizes new telomere repeats, leads to progressive telomere shortening, causing earlier onset of disease in successive generations[18]. Families with telomere disorders, such as dyskeratosis congenita, Li-Fraumeni syndrome and Von-Hippel-Lindau disease display anticipation in generational changes, both in age at onset and in appearance of specific symptoms[17-20]. Although genetic causes have been discovered for a set of identified telomere disorders, a number of related syndromes may likewise be due to unrecognized telomeropathies[21].
Interestingly, short telomeres were found to be associated with genetic anticipations in hereditary breast cancer (in women carrying a mutation in BRCA1 or BRCA2 genes, and a subset of BRCAX families)[22] and thyroid cancer[23]. There is increasing evidence suggesting that short telomeres and subsequent genomic instability contribute to malignant transformation[24,25]. Telomere shortening has been associated with dysplasia and neoplasia in the early onset of ulcerative colitis (UC)[26-28].
These findings generate thought-provoking questions about the possibility that telomere shortening may be the cause of genetic anticipation in other familial disorders associated with increased risk of malignancies such as IBD (which has an increased risk of cancer) - and a subset of these familial patients might result from unrecognized telomeropathies, particularly if there is also an accompanying identifiable genetic mutation related to telomere biology.
In our study, the primary objective was to evaluate generational changes in telomere length in parent-child pairs of familial IBD, to determine the role of telomere shortening, and its potential implication as a mechanism of anticipation in familial IBD. The secondary objective focused on investigating potential candidate gene variants for autosomal inheritance in these multi-generation IBD families.
Study design: We selected three multi-generation IBD families included in the Johns Hopkins IBD Family Study. Each selected family had three or more first-degree relatives affected by IBD: Either CD, UC or IBD-unclassified (IBD-U). Affected and non-affected individuals completed a multiple-choice questionnaire. This questionnaire interrogated patient demographics, family history of IBD, household specifics to childhood such as water source, number of people in the house, birth order, passive and active smoking; questions also inquired about disease onset, disease location, extraintestinal manifestations and complications, as well as medical and surgical therapy for IBD. We selected those families where all three affected generations had access to similar therapy to avoid the confounding factor of different classes of therapy on severity of the disease. Therefore, most of the patients were treated with mesalamine, thiopurine and steroids. None of the patients were treated with methotrexate. The patients were treated before biologics were commonly used as part of the IBD therapy.
Blood samples also obtained from participants were used to isolate lymphocytes for storage, serum for serological analysis of antibodies and other proteins relevant to IBD and DNA for genotyping or sequencing.
The Johns Hopkins IBD Family Study, conducted with the overall goal of identifying IBD susceptibility genes, was initially approved in 1996 and later registered with clinical trials in 2010. The review of electronic and paper records and the genetic analysis for the current study was performed in 2019.
IBD diagnosis: To confirm the diagnosis of IBD, define the severity of the disease, and determine current and past therapies, we reviewed electronic and paper medical records including consult notes, imaging tests (computer tomography, upper gastrointestinal, small bowel and barium enemas studies), endoscopic (upper endoscopy, colonoscopy and capsule endoscopy) and pathology reports of all available family members. We used the National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) IBD Genetic Consortium phenotype operating manual (version May 10, 2006) and forms to classify of disease as either CD, UC or IBD-U (https://repository.niddk.nih.gov/media/studies/ibd/ibd_phenotyping-manual.pdf).
Telomere length measurement: DNA was purified from whole blood using Qiagen Puregene kit. To measure average length of telomere repeats in cells, we used fluorescent in situ hybridization with labeled peptide nucleic acid probes specific for telomere repeats in combination with fluorescence measurements by flow cytometry (flow FISH) as previously described[29,30]. This method was developed to measure single-cell Telomere length (TL) using a fluorescently labeled probe that hybridizes to telomere DNA and has the unique advantage over other methods for measuring telomere length by providing multi-parameter information on the length of telomere repeats in thousands of individual cells. The accuracy and reproducibility of the measurements are augmented by the automation of most pipetting (aspiration and dispensing) steps and by including an internal standard (control cells) with a known TL in every tube. Individuals were diagnosed with short telomeres if TL was shorter than the 10th percentile of the age-matched control population.
Whole exome sequencing: Performed on eight individuals from the three families as part of the Baylor-Hopkins Center for Mendelian Genomics. Whole exome sequencing (WES) was performed using Agilent SureSelect HumanAllExonV5Clinicalon the Illumina HiSeq2000 platform. Libraries were sequenced on the HiSeq2500 platform with onboard clustering using 125 bp-paired end runs and sequencing chemistry kits HiSeq PE Cluster Kit v4 and HiSeq SBS Kit v4. Fastq files were aligned with BWA mem[31] version 0.7.8 to the 1000 genomes phase 2 (GRCh37) human genome reference[32,33]. Duplicate molecules were flagged with Picard version 1.109(1716). Local realignment around indels and base call quality score recalibration were performed using the Genome Analysis Toolkit 26 version v3.1-1- g07a4bf8 or v3.3-0-g37228af. Variant filtering was done using the Variant Quality Score Recalibration (VQSR) method[34]. Variants that passed VQSR filtering were annotated using Annovar (version 2013_09_11) against a variety of data sources. Ultimately, PhenoDB[35] was used to analyze the data for coding, frameshifting, nonsense, or missense, rare (minor allele frequency > 1% in ExAC, gnomAD, 1000 Genomes, or Exome Variant Server) variants in genes that were shared between the three affected families based on recessive or dominant modes of inheritance.
Results are expressed as mean ± SD for n number of samples. Analysis of difference between groups was conducted using an analysis of variance, t test, or Fisher exact test, as appropriate. A two-sided P value < 0.05 was used for statistical significance. All analysis was done using GraphPad Prism software (GraphPad 5.0).
Approval of the study was obtained from the Institutional Review Board of Johns Hopkins University.
There were twenty-four individuals selected to participate in the study. Only eleven out of the twenty-four were eligible (successive generations affected by IBD) for genetic anticipation analysis: Five patients with UC, four patients with CD and two patients with IBD-U. These represent eight parent-child pairs from the three families studied (Table 1).
| ID | Member | IBD | Tobacco | AOD | NOD2 | Extent | Behavior | Medications | Surgery |
| 1-01 | Mother | CD | None | 21 | + | Ileum, colon | B2 | Steroids 5-ASA | Yes |
| 1-02 | Proband/F | CD | Former | 9 | + | Ileum, jejunum, colon | B3p | Steroids 5-ASA | No |
| 1-05 | Sister | CD | None | 14 | + | Ileum, colon | B2 | Steroids 5-ASA thiopurine | No |
| 1-07 | Grandmother | CD | Passive | 42 | + | Ileum, colon | B2 | 5-ASA | Yes |
| 2-01 | Father | IBD-U | Active | 21 | _ | Left colon | NA | 5-ASA, thiopurine | Yes |
| 2-02 | Proband/F | UC | Active | 4 | _ | Left colon | NA | Steroids 5-ASA, thiopurine | Yes |
| 2-03 | Grandfather | UC | Former | 63 | _ | Left colon | NA | Steroids 5-ASA | Yes |
| 3-01 | Mother | UC | None | 40 | _ | Left colon | NA | Steroids 5-ASA | Yes |
| 3-03 | Proband/M | UC | None | 19 | _ | Left colon | NA | Steroids | No |
| 3-04 | Uncle | UC | Active | 41 | _ | Left colon | NA | Steroids 5-ASA | No |
| 3-05 | Grandfather | IBD-U | Former | 56 | _ | Pancolitis | NA | Steroids | No |
IBD diagnosis confirmation: Through electronic and paper medical records review and the IBD phenotype was defined per NIDDK IBD Genetic Consortium phenotype operating manual (version May 10, 2006) to classify disease as either CD, UC or IBD-U.
Children’s age at diagnosis: The median age of parents at diagnosis was 16.5 years older than that of the children affected by IBD and the median age at blood draw was 39.6 ± 23 years for the entire group. The mean age of parents at diagnosis was 40 ± 17 and the mean age of children was 21 ± 13, P < 0.0014. The UC families were of mixed descent Italian/Irish/Japanese and Poland/Russian/Puerto Rican while the CD family was of European descent (Polish/Russian). The later two families were also identified as being of Ashkenazi Jewish descent.
Disease extent and severity in parent-child pairs: Table 1 shows the extent of disease involvement in the CD, UC, and mixed families. Similar extent of the disease was noted in either parents or their children. In the family with CD, the disease extended into the large and small intestine in all three generations of affected individuals. All four affected members had a penetrating form (entero-enteral fistula, anal fistulas) of the disease. Arthropathy was the most common extraintestinal manifestation of IBD. In UC patients, the disease involved mainly the rectum and sigmoid colon (left side disease) and showed increasing severity as the disease was passed to the subsequent generation. All the affected members of the second family underwent total colectomy for disease refractory to medical therapy with the youngest member requiring surgery at only 6 years of age.
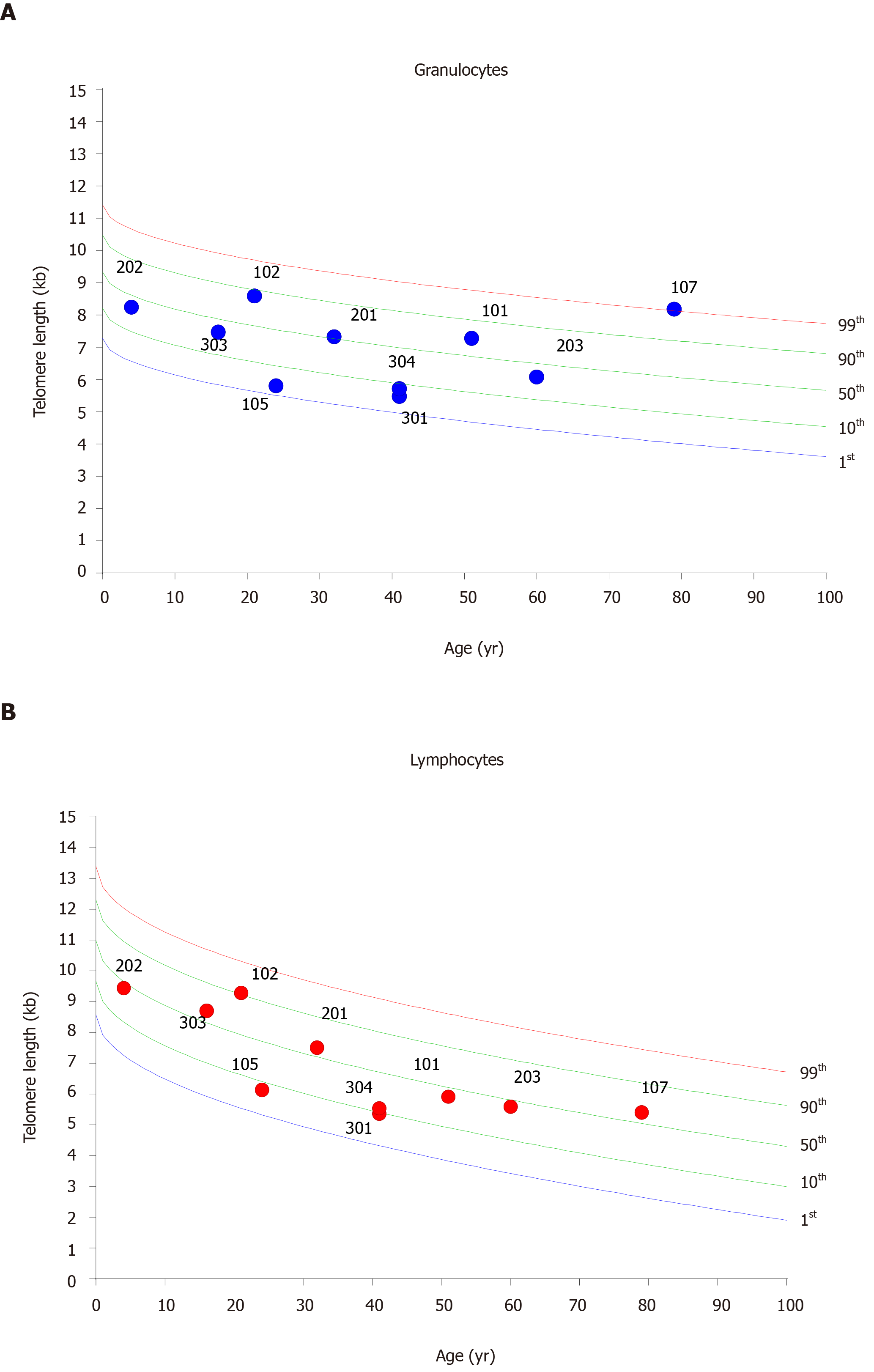
TL in successive generations: In order to evaluate the hypothesis that telomere shortening may be associated with the earlier onset of the disease, we investigated the mean relative TL of blood lymphocytes and granulocytes (Figure 1). Individuals were diagnosed with short telomeres if their telomere length was shorter than the 10th percentile of the age matched control population[17]. No associations were found between TL in lymphocytes and granulocytes and anticipation in the age at onset observed in successive generations.
WES for genetic inheritance: Finally, we explored the possibility of a gene variant with potential for autosomal inheritance in these Mendelian families. We used WES and PhenoDB to identify a genetic variant common to all three IBD families. NOD2 gene mutations were present in the CD affected patients of one family. WES did not detect any rare frameshift, nonsense or missense coding variants in genes shared by these three families for either dominant or recessive modes of inheritance. NOD2 gene mutations were present in the Crohn’s affected patients of one family.
Awareness of the changing of disease pattern in IBD families with multiply affected generations not only has implications for genetic counseling and surveillance of at-risk members but also for implementation of correct treatment strategies. Genetic anticipation in IBD, although observed in most studies, has been questioned by many scientists due to lack of evidence for a molecular mechanism to explain this phenomenon. It has been suggested that anticipation may result from observation bias[10,11]. Recently, a Bayesian method (that corrects for random effects, isolating the confounding effect of changes in secular trends, screening and medical practices, and adjusts for changes in age-specific incidence across birth cohorts), confirmed anticipation observed among successive generations of Lynch Syndrome families, a syndrome that likewise had controversy regarding genetic anticipation[36]. Nevertheless, the molecular mechanism of genetic anticipation in Lynch Syndrome has not been identified. It is therefore possible, that genetic anticipation in IBD may be demonstrated using an optimal study design and statistical methods and/or evidence of a molecular mechanism[37].
In several monogenetic diseases, including Huntington disease[3], myotonic dystrophy[38], and Friedreich ataxia[39], the biological basis for genetic anticipation resides in expansion of unstable genomic triple repeats. In polygenic diseases, the complexity of multifactorial inheritance makes the identification of such molecular mechanism very challenging. In IBD, genomic imprinting of age at diagnosis has been proposed but when tested in 137 affected parent-child pairs from 96 families, it was not observed[40].
The patterns of genetic anticipation discovered for some telomeropathies generated the question of whether a similar defect in telomere maintenance mechanisms may have a role in IBD. The TL has been studied in IBD in association with colon cancer and severity of inflammation in UC but not in the context of genetic anticipation[26-28,40]. In particular, we have questioned if these familial forms of IBD with observed anticipation might be due to unrecognized telomerophaties.
In our study, we examined three IBD families with clinical features of genetic anticipation. In all three families, the disease onset was reported at an early age (median 16.5) and became more severe in successive generations. In order to encompass three successive generations, we reviewed the disease history of families who received care at our center as early as 1996. Confusion related to the potential impact of newer therapy on the severity of the disease was avoided since all the recruited affected members had to receive treatment before the introduction of the biologics. Since population genetics has temporal stability, the recruitment methodology did not influence the results of the study which remain valid today despite the age of our families.
We have shown that when controlling for age, no association exists between TL and age at disease onset among members of successive generations affected by IBD. Genetic anticipation is unlikely to be the result of telomere shortening and other mechanism should be explored. We also investigated the potential for these IBD families with Mendelian pattern of inheritance to represent a monogenic form of IBD similar to Very Early Onset IBD[41]. In particular, we wanted to learn if there is an accompanying identifiable genetic mutation related to telomere biology[32] in these families and if its effects were overlooked due to our study sample size. Moreover, in some monogenic diseases, genetic anticipation has been linked to the expansion of unstable genomic triplet repeats[3], a mechanism that we would have explored in IBD had it been monogenetic. However, besides the NOD2 mutations identified among the affected members one of the CD families, WES did not detect any rare frameshift, nonsense or missense coding variants in genes shared by these three families for either dominant or recessive modes of inheritance. In adults, NOD2 is typically the most highly penetrant established gene associated with CD but the population attributable risk of NOD2 variance to CD is only 26%[43]. No mutation in genes involved in the telomere maintenance mechanism were detected.
The study represents the first evaluation of TL defects in genetic anticipation of IBD. Even though no association was found between TL and age at disease onset among members of successive generations, this pilot study is very valuable as an attempt to find the molecular mechanism of genetic anticipation in IBD. While we have shown that genetic anticipation is unlikely to be the result of telomere shortening, our results do point the way to explore other mechanisms. Nonetheless, the sample size may be too small to rule out whether telomeropathies may play a role in other families with similar strong evidence for anticipation. Further studies to confirm our findings should address a larger population of familial IBD that likewise have strong evidence for anticipation among successive generations.
The existence of genetic anticipation, a phenomenon in which an inherited disorder manifests at a younger age and often more severely in offspring than in the affected parent, has been long disputed in inflammatory bowel disease (IBD). In the absence of the explanatory mechanism, it has been suggested that anticipation may result from observation bias. In monogenetic degenerative neurological disorders the mechanism for genetic anticipation was identified as unstable genomic triple repeats. In polygenic diseases, such as IBD, the complexity of multifactorial inheritance makes the identification of genetic mechanism very challenging. I Most recently, mutations in the telomerase reverse transcriptase, have been shown to lead to progressive telomere shortening, causing earlier onset of disease in successive generations with telemeropahities. Telomere length (TL) has also been found to be associated with severity of inflammation and colon cancer in IBD.
The role of TL in genetic anticipation of patients with IBD has not been studied.
In our study, the primary objective was to evaluate generational changes in TL in parent-child pairs of familial IBD, to determine the role of telomere shortening, and its potential implication as a mechanism of anticipation in familial IBD. The secondary objective focused on investigating potential candidate gene variants for autosomal inheritance in these multi-generation IBD families.
We studied three IBD families with multiple affected members in three successive generations. We determined TL in lymphocytes and granulocytes from peripheral blood of the affected members using flow cytometry and fluorescence in-situ hybridization (flow FISH). We also performed whole exome sequencing (WES) in the blood of all available family members and used PhenoDB to identify potential candidate gene variants for recessive or dominant modes of inheritance.
Out of twenty-four patients of European descent selected to participate in the study, eleven patients, eight parent-child pairs affected by IBD, were included in the genetic anticipation analysis. Median difference in age at diagnosis between two successive generations was 16.5 years, with earlier age at onset in the younger generations. Five patients were affected by ulcerative colitis, four patients by Crohn’s disease and two patients by IBD-Unclassified. In most of the affected members, the disease harbored similar gastrointestinal and extraintestinal involvement but was more aggressive among the younger generations. TL was not associated with earlier age at onset or with more severe disease in members of successive generations affected by IBD. NOD2 gene mutations were present in the Crohn’s affected patients of one family. However, no gene variants were identified as potential candidates for Mendelian inheritance.
The study represents the first evaluation of TL defects in genetic anticipation of IBD. Even though no association was found between TL and age at disease onset among members of successive generations, this pilot study is very valuable as an attempt to find the molecular mechanism of genetic anticipation in IBD.
While we have shown that genetic anticipation is unlikely to be the result of telomere shortening, our results do point the way to exploration of other mechanisms. Further studies to confirm our findings should address a larger population of familial IBD that likewise have strong evidence for anticipation among successive generations.
The authors thank Professor Mary Armanios for assistance with telomere analysis. The authors would like to dedicate this paper in memoriam to Dr. Theodore M. Bayless, a great mentor and a believer in genetic anticipation in inflammatory bowel disease.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Serban ED, Triantafillidis J S-Editor: Liu M L-Editor: A P-Editor: Wang LL
| 1. | McInnis MG. Anticipation: an old idea in new genes. Am J Hum Genet. 1996;59:973-979. [PubMed] |
| 2. | McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786-799. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 403] [Cited by in RCA: 369] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 3. | Khristich AN, Mirkin SM. On the wrong DNA track: Molecular mechanisms of repeat-mediated genome instability. J Biol Chem. 2020;295:4134-4170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 224] [Article Influence: 37.3] [Reference Citation Analysis (0)] |
| 4. | Rosenberg RN. DNA triplet repeats and neurologic disease. N Engl J Med. 1996;335:1222-1224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 26] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 5. | Liu Y, Wilson SH. DNA base excision repair: a mechanism of trinucleotide repeat expansion. Trends Biochem Sci. 2012;37:162-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 6. | Everett CM, Wood NW. Trinucleotide repeats and neurodegenerative disease. Brain. 2004;127:2385-2405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 120] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 7. | Ashley CT, Warren ST. Trinucleotide repeat expansion and human disease. Annu Rev Genet. 1995;29:703-728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 285] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 8. | Bayless TM, Picco MF, LaBuda MC. Genetic anticipation in Crohn's disease. Am J Gastroenterol. 1998;93:2322-2325. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 9. | Polito JM 2nd, Rees RC, Childs B, Mendeloff AI, Harris ML, Bayless TM. Preliminary evidence for genetic anticipation in Crohn's disease. Lancet. 1996;347:798-800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 45] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 10. | Picco MF, Goodman S, Reed J, Bayless TM. Methodologic pitfalls in the determination of genetic anticipation: the case of Crohn disease. Ann Intern Med. 2001;134:1124-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 11. | Lee JC, Bridger S, McGregor C, Macpherson AJ, Jones JE. Why children with inflammatory bowel disease are diagnosed at a younger age than their affected parent. Gut. 1999;44:808-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 34] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 12. | Faybush EM, Blanchard JF, Rawsthorne P, Bernstein CN. Generational differences in the age at diagnosis with Ibd: genetic anticipation, bias, or temporal effects. Am J Gastroenterol. 2002;97:636-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 13. | McInnis MG, McMahon FJ, Chase GA, Simpson SG, Ross CA, DePaulo JR. Anticipation in bipolar affective disorder. Am J Hum Genet. 1993;53:385-390. [PubMed] |
| 14. | Bassett AS, Honer WG. Evidence for anticipation in schizophrenia. Am J Hum Genet. 1994;54:864-870. [PubMed] |
| 15. | McDermott E, Khan MA, Deighton C. Further evidence for genetic anticipation in familial rheumatoid arthritis. Ann Rheum Dis. 1996;55:475-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 16. | Fresko I, Soy M, Hamuryudan V, Yurdakul S, Yavuz S, Tümer Z, Yazici H. Genetic anticipation in Behçet's syndrome. Ann Rheum Dis. 1998;57:45-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 61] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 17. | Wiernik PH, Ashwin M, Hu XP, Paietta E, Brown K. Anticipation in familial chronic lymphocytic leukaemia. Br J Haematol. 2001;113:407-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Armanios M. Telomeres and age-related disease: how telomere biology informs clinical paradigms. J Clin Invest. 2013;123:996-1002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 265] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 19. | Armanios M. Telomerase and idiopathic pulmonary fibrosis. Mutat Res. 2012;730:52-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 179] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 20. | Armanios M, Blackburn EH. The telomere syndromes. Nat Rev Genet. 2012;13:693-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 673] [Cited by in RCA: 738] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 21. | Wang J, Peng X, Chen C, Ning X, Peng S, Li T, Liu S, Hong B, Zhou J, Ma K, Cai L, Gong K. Intra-Familial Phenotypic Heterogeneity and Telomere Abnormality in von Hippel- Lindau Disease: Implications for Personalized Surveillance Plan and Pathogenesis of VHL-Associated Tumors. Front Genet. 2019;10:358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 22. | Holohan B, Wright WE, Shay JW. Cell biology of disease: Telomeropathies: an emerging spectrum disorder. J Cell Biol. 2014;205:289-299. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 23. | Martinez-Delgado B, Yanowsky K, Inglada-Perez L, Domingo S, Urioste M, Osorio A, Benitez J. Genetic anticipation is associated with telomere shortening in hereditary breast cancer. PLoS Genet. 2011;7:e1002182. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 24. | Yashima K, Vuitch F, Gazdar AF, Fahey TJ. Telomerase activity in benign and malignant thyroid diseases. Surgery. 1997;122:1141-5; discussion 1145-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 25. | Wu X, Amos CI, Zhu Y, Zhao H, Grossman BH, Shay JW, Luo S, Hong WK, Spitz MR. Telomere dysfunction: a potential cancer predisposition factor. J Natl Cancer Inst. 2003;95:1211-1218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 353] [Cited by in RCA: 364] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 26. | Srinivas N, Rachakonda S, Kumar R. Telomeres and Telomere Length: A General Overview. Cancers (Basel). 2020;12:558. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 200] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 27. | Risques RA, Lai LA, Brentnall TA, Li L, Feng Z, Gallaher J, Mandelson MT, Potter JD, Bronner MP, Rabinovitch PS. Ulcerative colitis is a disease of accelerated colon aging: evidence from telomere attrition and DNA damage. Gastroenterology. 2008;135:410-418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 139] [Cited by in RCA: 143] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 28. | Friis-Ottessen M, Bendix L, Kølvraa S, Norheim-Andersen S, De Angelis PM, Clausen OP. Telomere shortening correlates to dysplasia but not to DNA aneuploidy in longstanding ulcerative colitis. BMC Gastroenterol. 2014;14:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 29. | Salk JJ, Bansal A, Lai LA, Crispin DA, Ussakli CH, Horwitz MS, Bronner MP, Brentnall TA, Loeb LA, Rabinovitch PS, Risques RA. Clonal expansions and short telomeres are associated with neoplasia in early-onset, but not late-onset, ulcerative colitis. Inflamm Bowel Dis. 2013;19:2593-2602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | Baerlocher GM, Vulto I, de Jong G, Lansdorp PM. Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nat Protoc. 2006;1:2365-2376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 309] [Cited by in RCA: 350] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 31. | Alder JK, Hanumanthu VS, Strong MA, DeZern AE, Stanley SE, Takemoto CM, Danilova L, Applegate CD, Bolton SG, Mohr DW, Brodsky RA, Casella JF, Greider CW, Jackson JB, Armanios M. Diagnostic utility of telomere length testing in a hospital-based setting. Proc Natl Acad Sci USA. 2018;115:E2358-E2365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 179] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 32. | Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754-1760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29052] [Cited by in RCA: 36243] [Article Influence: 2131.9] [Reference Citation Analysis (0)] |
| 33. | Church DM, Schneider VA, Graves T, Auger K, Cunningham F, Bouk N, Chen HC, Agarwala R, McLaren WM, Ritchie GR, Albracht D, Kremitzki M, Rock S, Kotkiewicz H, Kremitzki C, Wollam A, Trani L, Fulton L, Fulton R, Matthews L, Whitehead S, Chow W, Torrance J, Dunn M, Harden G, Threadgold G, Wood J, Collins J, Heath P, Griffiths G, Pelan S, Grafham D, Eichler EE, Weinstock G, Mardis ER, Wilson RK, Howe K, Flicek P, Hubbard T. Modernizing reference genome assemblies. PLoS Biol. 2011;9:e1001091. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 334] [Cited by in RCA: 402] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 34. | Puchner T. [Infections of the lower genital tract in asymptomatic pregnant women--a prospective study]. Z Geburtshilfe Perinatol. 1992;196:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3224] [Cited by in RCA: 2912] [Article Influence: 132.4] [Reference Citation Analysis (0)] |
| 35. | DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491-498. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9842] [Cited by in RCA: 8546] [Article Influence: 569.7] [Reference Citation Analysis (0)] |
| 36. | Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat. 2015;36:425-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 141] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 37. | Boonstra PS, Mukherjee B, Taylor JM, Nilbert M, Moreno V, Gruber SB. Bayesian modeling for genetic anticipation in presence of mutational heterogeneity: a case study in Lynch syndrome. Biometrics. 2011;67:1627-1637. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 38. | Ellis CA, Churilov L, Epstein MP, Xie SX, Bellows ST, Ottman R, Berkovic SF; Epi4K Consortium. Epilepsy in families: Age at onset is a familial trait, independent of syndrome. Ann Neurol. 2019;86:91-98. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 39. | Lanni S, Pearson CE. Molecular genetics of congenital myotonic dystrophy. Neurobiol Dis. 2019;132:104533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 40. | Cahn S, Rosen A, Wilmot G. Spinocerebellar Ataxia Patient Perceptions Regarding Reproductive Options. Mov Disord Clin Pract. 2020;7:37-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 41. | Tahara T, Shibata T, Okubo M, Kawamura T, Sumi K, Ishizuka T, Nakamura M, Nagasaka M, Nakagawa Y, Ohmiya N, Arisawa T, Hirata I. Telomere length in non-neoplastic colonic mucosa in ulcerative colitis (UC) and its relationship to the severe clinical phenotypes. Clin Exp Med. 2015;15:327-332. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 42. | Moran CJ, Klein C, Muise AM, Snapper SB. Very early-onset inflammatory bowel disease: gaining insight through focused discovery. Inflamm Bowel Dis. 2015;21:1166-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 43. | Trotta L, Norberg A, Taskinen M, Béziat V, Degerman S, Wartiovaara-Kautto U, Välimaa H, Jahnukainen K, Casanova JL, Seppänen M, Saarela J, Koskenvuo M, Martelius T. Diagnostics of rare disorders: whole-exome sequencing deciphering locus heterogeneity in telomere biology disorders. Orphanet J Rare Dis. 2018;13:139. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 44. | Cuthbert AP, Fisher SA, Mirza MM, King K, Hampe J, Croucher PJ, Mascheretti S, Sanderson J, Forbes A, Mansfield J, Schreiber S, Lewis CM, Mathew CG. The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease. Gastroenterology. 2002;122:867-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 480] [Cited by in RCA: 471] [Article Influence: 19.6] [Reference Citation Analysis (0)] |