©The Author(s) 2018.
World J Gastrointest Pathophysiol. Sep 29, 2018; 9(2): 47-58
Published online Sep 29, 2018. doi: 10.4291/wjgp.v9.i2.47
Published online Sep 29, 2018. doi: 10.4291/wjgp.v9.i2.47
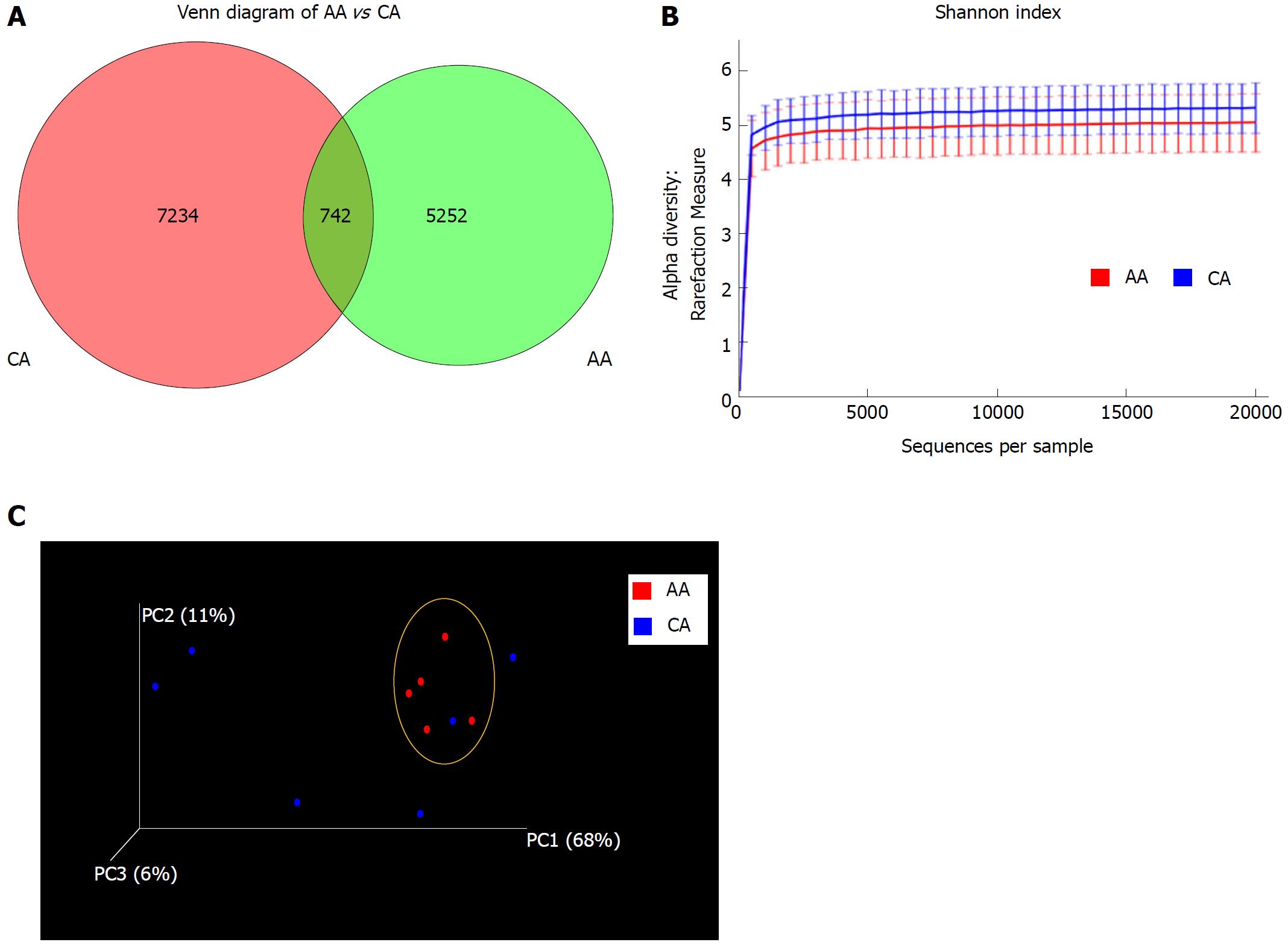
Figure 1 Microbial diversity in colonic effluent from African Americans and Caucasian Americans.
A: Venn diagram showing the unique Operational taxonomic units (OTUs) in different subsets of African American (AA) and Caucasian Americans (CA) including overlap community; B: Rarefaction curves showing the species richness from the average number of OTUs for AA and CA (alpha diversity); C: Principal coordinates analysis for beta diversity showing the very dissimilar individual in the CA group while closely similar to the group indicated by the yellow circle. CD-HIT and R software were used for BIOM format OTU clustering and OTU statistics. For measurement of alpha diversity of observed species, QIIME software and Shannon index were used and the emperor tool generated the PCoA plot.
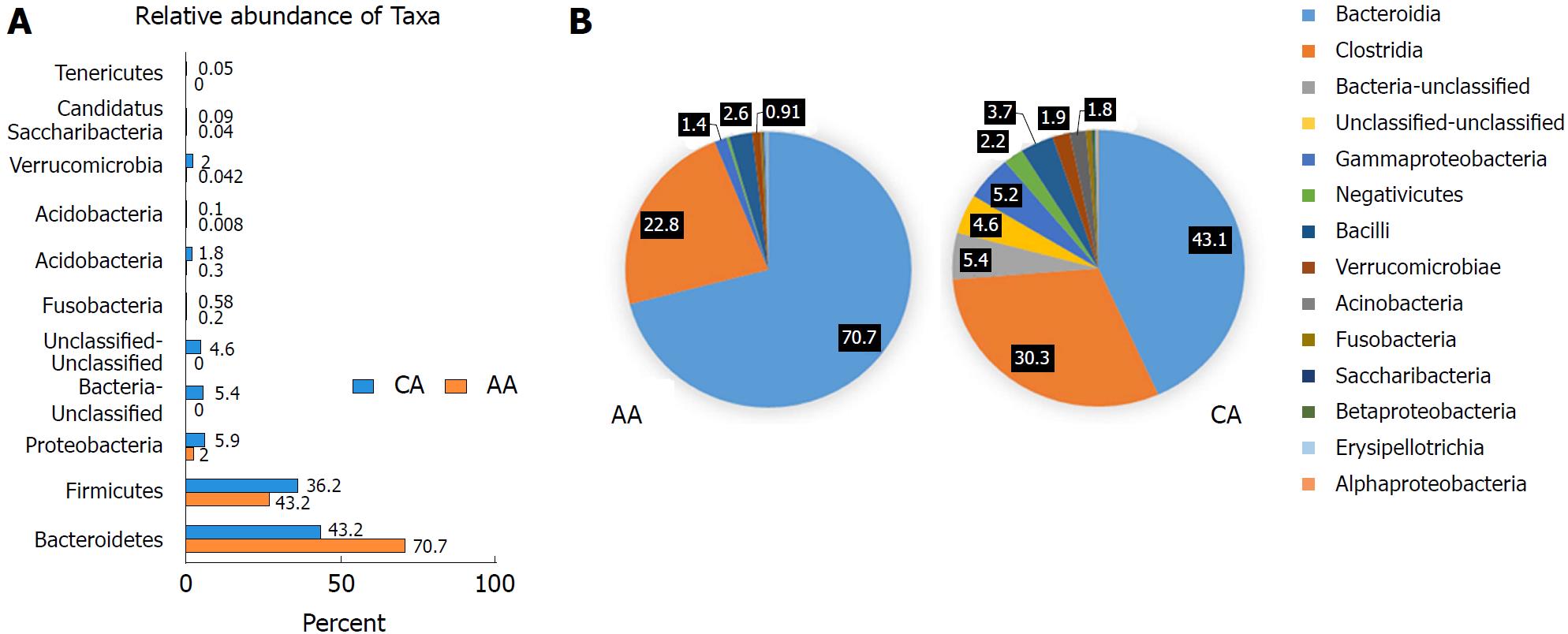
Figure 2 Microbial composition of each group at the phylum and class levels.
A: Bar chart shows the relative abundance of phylum; B: Pie chart show the proportion of predominant different classes of microbial community in African Americans and Caucasian Americans. In order to get more comprehensive and accurate taxonomies, multiple databases, Ribosomal Database Project classifier, Greengenes and NCBI 16S Microbial were used for analysis of plot bars and pie charts.
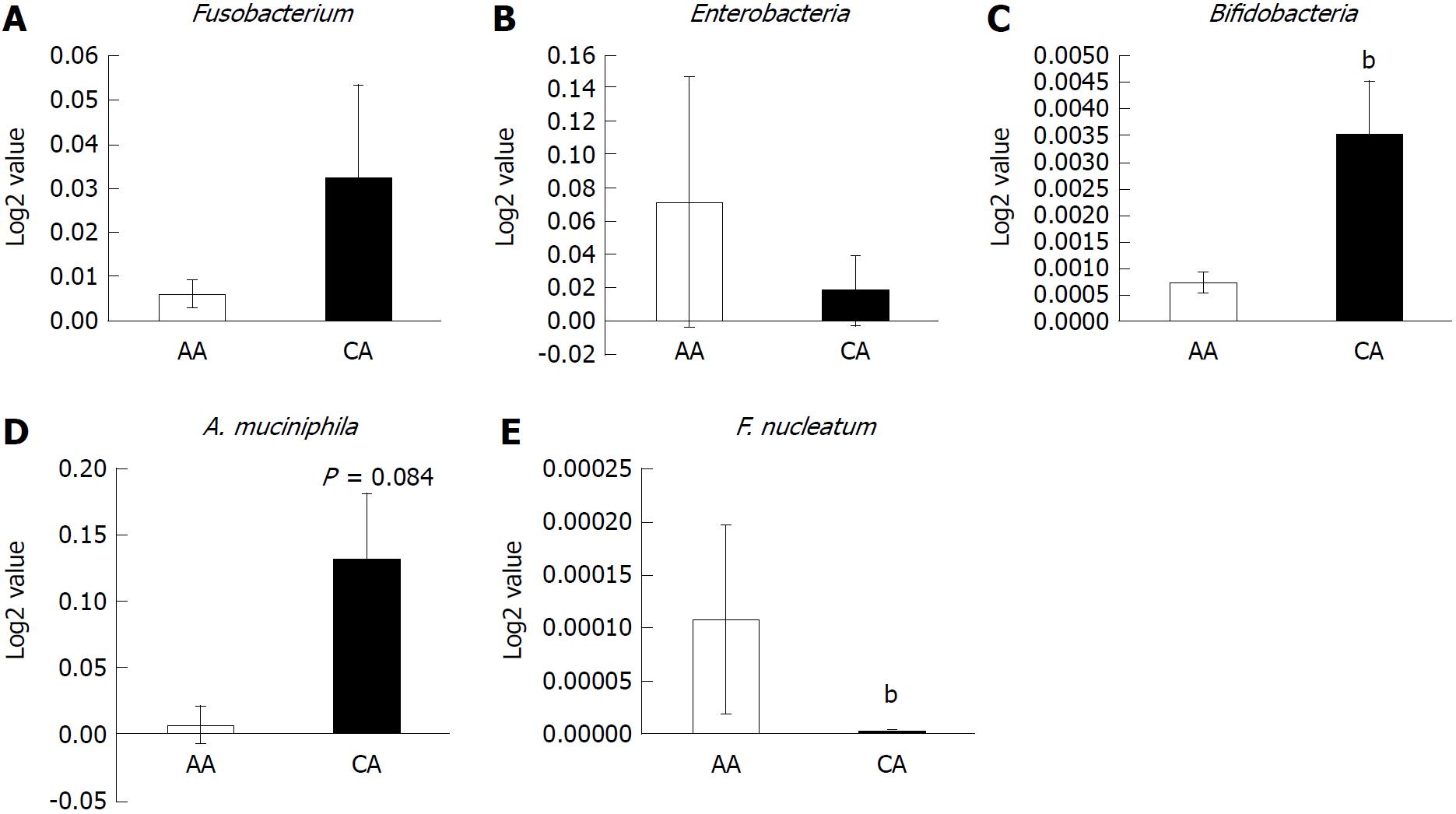
Figure 3 Abundance of inflammatory and probiotic bacteria in colonic effluent from two racial groups using RT-PCR.
A: Genus, Fusobacterium occurrence higher in Caucasian American (CA); B: The relative abundance of pro-inflammatory Enterobacter occurrence is higher in African Americans (AAs); C and D: Probiotic Bifidobacteria and A. muciniphila is higher in CAs; E: Relative abundance of F. nucleatum is higher in AAs than CAs. Data represent mean ± SD of all samples from each group (bP < 0.001).
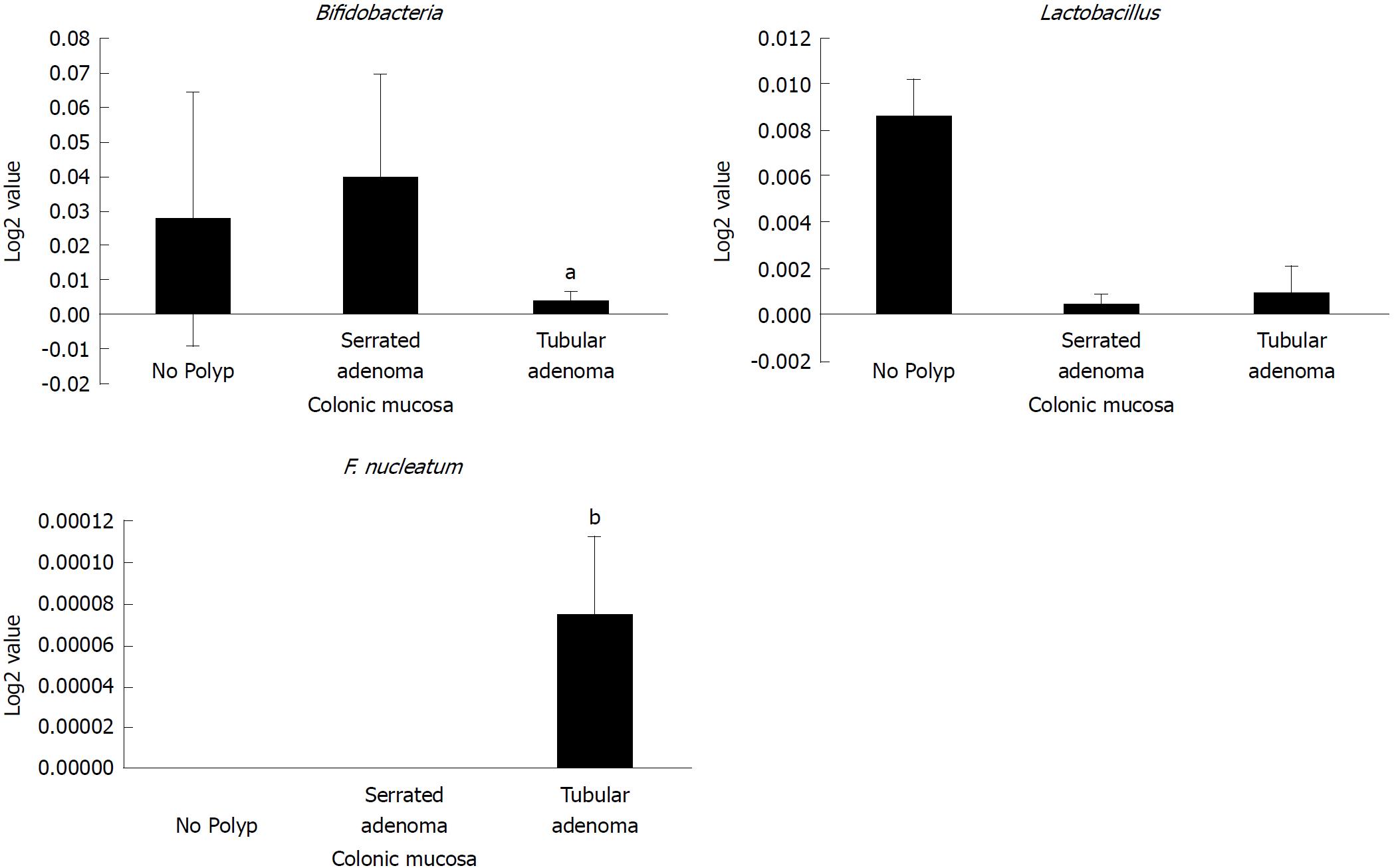
Figure 4 Abundance of probiotic and pro-carcinogenic bacteria in serrated and tubular adenomatous in colonic mucosa.
African American and Caucasian American patients were combined. Data represent mean ± SD of all samples from each group (aP < 0.05, bP < 0.001).
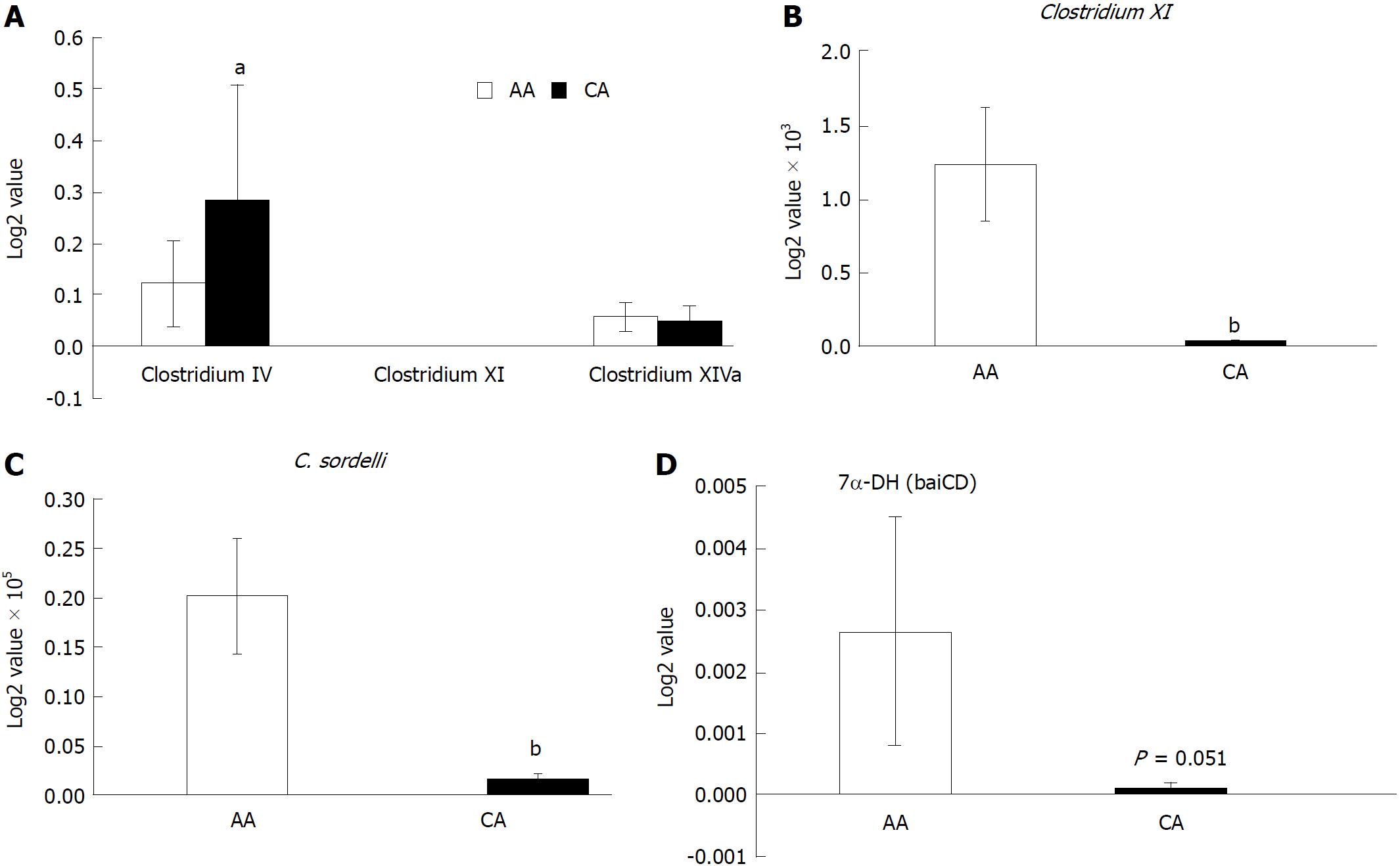
Figure 5 Expression of secondary bile acids transforming enzyme 7-α-DH in the colonic effluent from African Americans and Caucasian Americans using RT-PCR.
A: Distribution of different Clostridium cluster between African Americans (AA) and Caucasian Americans (CA); B: Clostridium XI expression was higher in AAs; C: Secondary bile acids transforming bacteria Clostridium sordelli was higher in AAs than CAs; D: Increased 7α-DH expression in AAs. Data represent mean ± SD of all samples from each group (aP < 0.05, bP < 0.01).
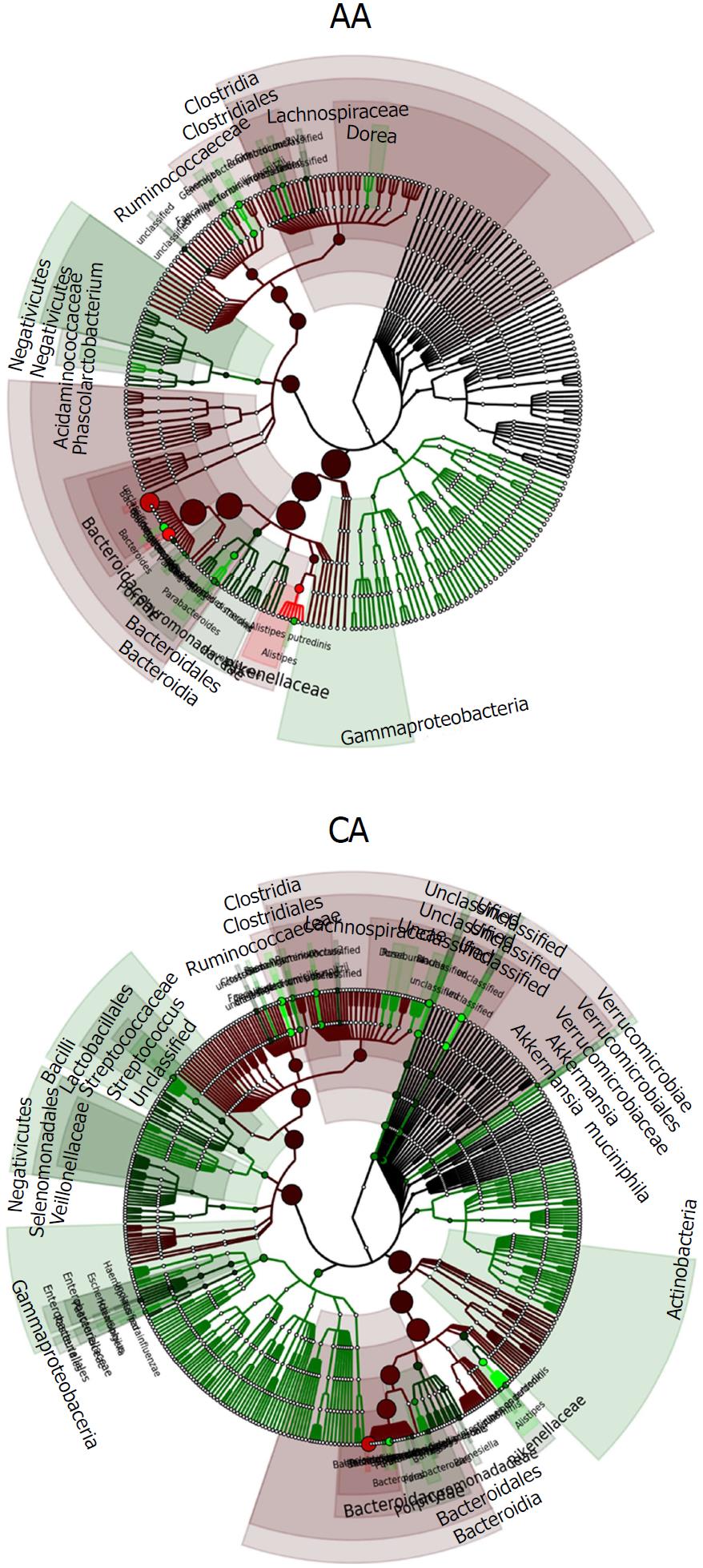
Figure 6 Phylogenetic tree showing the differences and abundance of taxa in African Americans and Caucasian Americans colonic effluents.
The taxon size and color indicate the relative abundance of family. GraPhlAn software was used for taxonomic classification and circular taxonomic phylogenetic trees and Ribosomal Database Project classifier, Quantitative Insights into Microbial Ecology, R software/Tool were used for taxonomic data analysis.
- Citation: Farhana L, Antaki F, Murshed F, Mahmud H, Judd SL, Nangia-Makker P, Levi E, Yu Y, Majumdar AP. Gut microbiome profiling and colorectal cancer in African Americans and Caucasian Americans. World J Gastrointest Pathophysiol 2018; 9(2): 47-58
- URL: https://www.wjgnet.com/2150-5330/full/v9/i2/47.htm
- DOI: https://dx.doi.org/10.4291/wjgp.v9.i2.47